Ataxin-7 and Non-stop coordinate SCAR protein levels, subcellular localization, and actin cytoskeleton organization
Figures
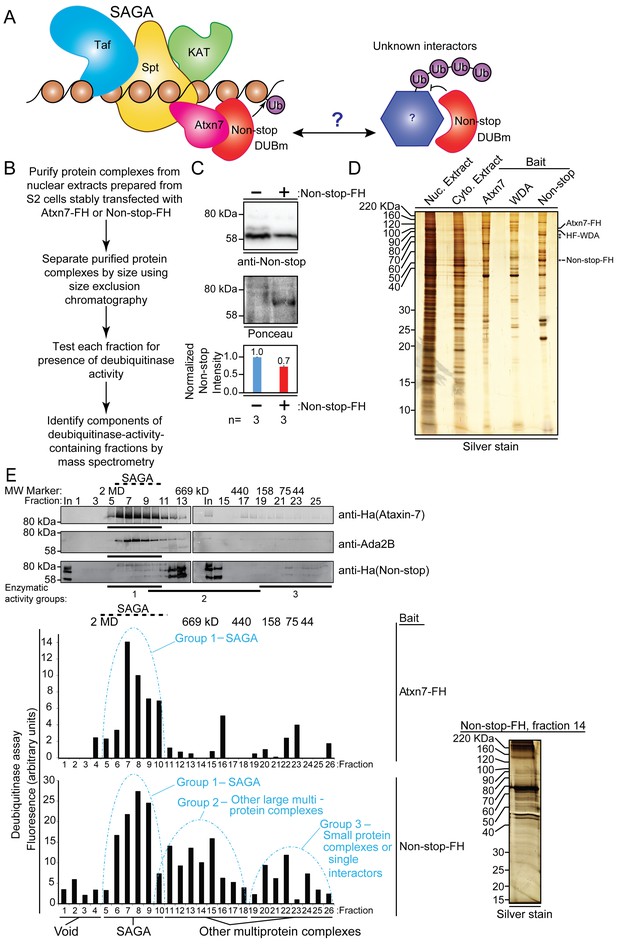
Non-stop (Not) co-purifies with Spt Ada Gcn5 acetyltransferase (SAGA) complex and additional multi-protein complexes.
(A) Working model: The Non-stop-containing SAGA deubiquitinase module (DUBm) functions distally from SAGA. Non-stop is anchored to SAGA by Ataxin-7 (Atxn7) but is released to interact with unknown partners. (B) Scheme to identify Non-stop interactors. (C) Characterization of Non-stop-2xFLAG-2xHA (FH)-expressing S2 cell lines. Stably transfected cells were treated with 10 uM copper sulphate show comparable levels of Non-stop expression in parental and stably transfected cell lines. Parental S2 cells containing no plasmid compared to S2 cells stably transfected with Non-stop plasmid. Quantitation of anti-Non-stop immunoblot signal intensities normalized to Ponceau total protein loading control. Error bars represent standard error. (D) Analysis of purified complexes. Non-stop-containing complexes were analyzed by silver staining. Atxn7 and Will Decrease Acetylation (WDA)-containing complexes are shown for comparison. (E) Protein complexes, purified and fractionated as described in B were analyzed by immunoblotting to observe relative elution by size (top, Atxn7 and Ada2B recreated from Mohan et al., 2014b) followed by measure of deubiquitinase activity contained in each fraction as assayed by ubiquitin-AMC assay in which increased fluorescence correlates directly with deubiquitination activity. The complexity of eluted fractions was analyzed and Group 2 peak fraction, number 14, is shown (right).
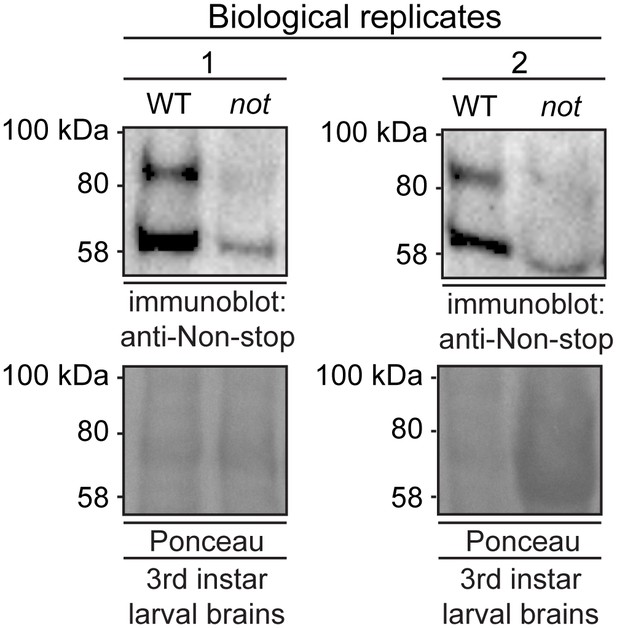
Anti-Non-stop antibody specifically recognizes Non-stop.
Denaturing whole cell protein extracts were prepared from brains isolated from 3rd instar larvae homozygous for P element insertion P{PZ}not02069 in the non-stop gene (not). Immunoblotting shows reductions in immunoreactive bands compared to wild-type Oregon-R brain. Two biological replicates are shown.
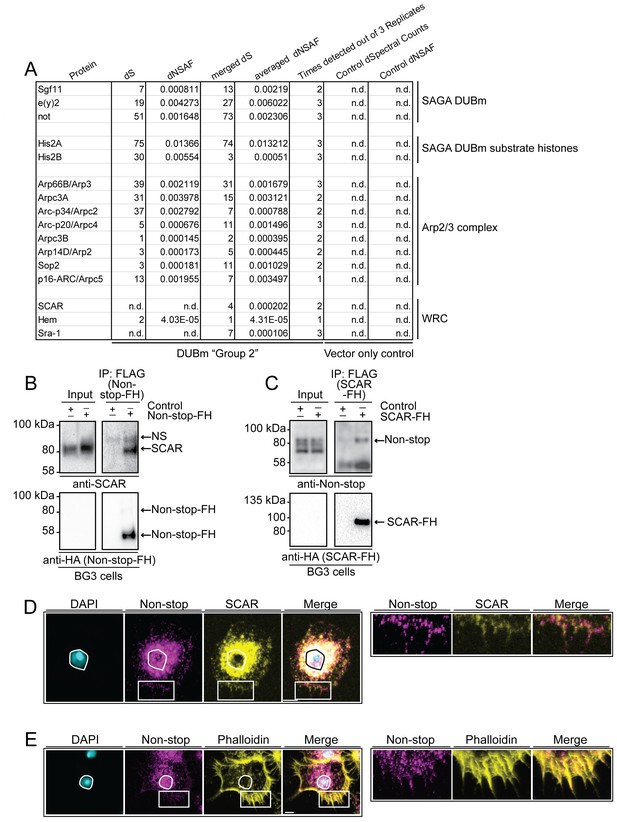
WAVE regulatory complex (WRC) and Arp2/3 interact with Non-stop.
(A) Mass spectrometry analysis of Group 2 fractions revealed Arp2/3 and WRC complexes stably interact with Non-stop. None of these proteins were identified using vector only Control purifications. (B and C) Reciprocal pull-down verifies interaction between Non-stop and WRC subunit suppressor of extracellular cAMP receptor (cAR) (SCAR). (B) BG3 cells were transfected with an expression vector for Non-stop-2xFLAG-2xHA (Non-stop-FH) or no vector (Control). Recombinant Non-stop was immunoprecipitated using anti-FLAG affinity resin and purified interactors analyzed by immunoblotting for the presence of endogenous SCAR. Anti-HA immunoblots verified the presence of Non-stop-FH bait. NS marks a non-specific band. (C) Pull-down was performed as in B, but with SCAR-FLAG-HA (SCAR-FH) expression vector. Immunoprecipitated proteins were probed by immunoblotting to detect endogenous Non-stop. Anti-HA immunoblots verified the presence of SCAR-FH bait. (D) Endogenous Non-stop and SCAR occupy similar subcellular regions. Immunofluorescence of SCAR and Non-stop in BG3 cells. Cells were immunostained with anti-Non-stop (magenta), anti-SCAR (yellow), and DAPI (white). Scale bar is 10 µM. Inset is enlarged to the right. (E) Endogenous Non-stop and F-actin are found in similar subcellular regions. Representative images of BG3 cells immunostained with anti-Non-stop (magenta), phalloidin (F-actin)(yellow), and DAPI (DNA)(white). Scale bar is 10 µM. Inset is enlarged to the right. Images in D and E were adjusted with contrast limited adaptive histogram equalization using the ImageJ CLAHE algorithm (Zuiderveld, 1994).
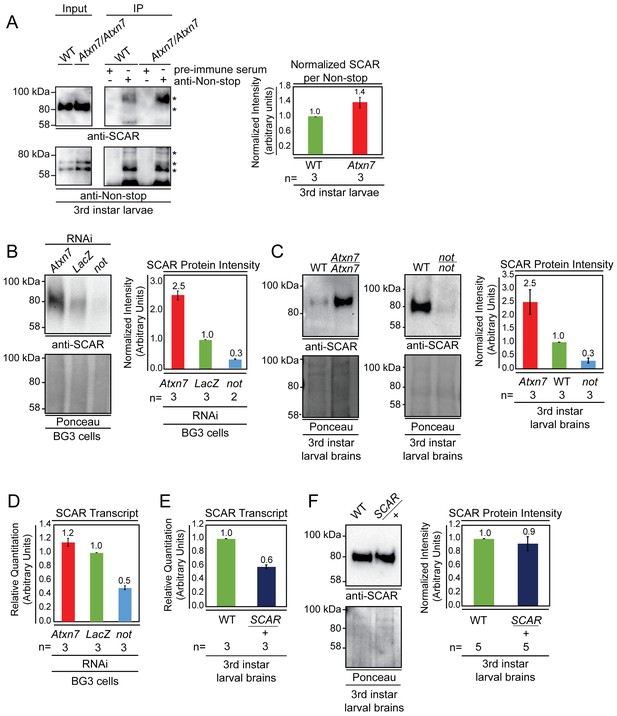
Non-stop and Atxn7 regulate SCAR protein levels.
(A) Endogenous pull-down reveals increased interaction between Non-stop and SCAR in the absence of Atxn7. Whole cell extracts prepared from either OregonR (WT) or homozygous mutant Ataxin7[KG02020] (Atxn7) third instar larvae were subject to immunoprecipitation with pre-immune guinea pig serum or anti-Non-stop antibody as indicated. Immunoprecipitates were analyzed by immunoblotting with anti-Non-stop to verify capture, and with anti-SCAR antibody to assess interaction. To compare the relative proportion of SCAR interacting with Non-stop, immunoblotting signals were quantified by densitometry and the amount of SCAR interacting per given amount of Non-stop was calculated and expressed relative to WT control. Error bars represent standard error. (B and C) Loss of Atxn7 increases, and loss of Non-stop decreases SCAR protein levels. (B) BG3 cells were treated with dsRNA to knock down either Atxn7, LacZ (control), or non-stop (not). Denaturing whole cell extracts were analyzed by immunoblotting to determine SCAR protein levels. Total protein (Ponceau, lower panel) was used as loading control. SCAR intensity was quantified and values were normalized to Ponceau and expressed relative to LacZ control (right panel). Error bars represent standard error. (C) Brain and central nervous systems from WT, non-stop02069 (not), or Ataxin7[KG02020] (Atxn7) third instar larvae were isolated and denaturing whole cell extracts prepared. SCAR protein levels were determined by immunoblotting and quantified as in B. (D, E, F) Atxn7- or Non-stop-mediated gene regulation are not sufficient to change SCAR protein levels. (D) Relative quantification of SCAR transcripts in BG3 cells treated with dsRNA targeting either Atxn7, LacZ (control), or not. Transcripts from not and Atxn7 treated cells are expressed relative to LacZ. (E) Relative quantification of SCAR transcript levels in larval brains dissected from WT or SCAR[Delta37]/cyo-gfp (SCAR/+). SCAR transcript levels are set relative to WT. (F) Immunoblot for SCAR protein in larval brains dissected from OregonR (WT) or SCAR[Delta37]/cyo-gfp (SCAR/+). SCAR protein levels were quantified as in B.
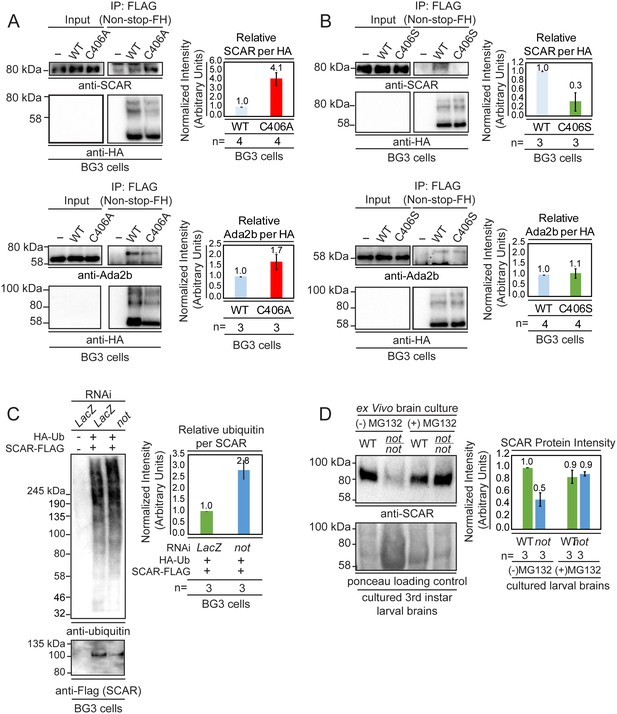
Non-stop binds ubiquitinated SCAR, regulates SCAR ubiquitination, and entry into the proteasome degradation pathway.
(A) Non-stop catalytic mutation which increases substrate binding also increases interaction with SCAR. The Non-stop catalytic cysteine was mutated to alanine creating Non-stop C406A-2xFLAG-2xHA (C406A). The wild type version of Non-stop (WT), the mutant version of Non-stop (C406A), or no plasmid (-) were transfected into BG3 cells and whole cell extracts were prepared. Flag resin was used to immunoprecipitate Non-stop-FH protein. Extracts were immunoblotted for SCAR. Immunoblots for Ada2b control for incorporation into the SAGA complex. Immunoblots for HA verify the presence of the Non-stop-FH constructs. Ada2b and SCAR intensity were quantified and normalized to HA. Values are shown as relative to WT. Error bars represent standard error. (B) Non-stop catalytic mutation which decreases substrate binding also decreases interaction with SCAR. The Non-stop catalytic cysteine was mutated to serine (C406S). BG3 cells were mock transfected (-), transfected with Non-stop-FH, or with Non-stop C406S-FH (C406S) and whole cell extracts were prepared. Flag resin was used to immunoprecipitate the Non-stop-FH constructs. Pull-downs were immunoblotted for SCAR. Incorporation into SAGA was verified by Ada2b immunoblot. HA immunoblots verify the presence of the Non-stop-FH constructs. Ada2b and SCAR intensity was quantified and normalized to HA. Values are shown as relative to wild type. Error bars are standard error. (C) Non-stop counters polyubiquitination of SCAR. BG3 cells were treated with dsRNA targeting either non-stop or LacZ and transfected with HA-ubiquitin (HA-Ub) and SCAR-FLAG (SCAR-F). Control cells were treated with LacZ dsRNA and a mock transfection was performed. After six days, cells were treated with MG132 protease inhibitor for 6 hr and denaturing whole cell extracts were made. Anti-flag resin was used to capture SCAR-FLAG. Immunoblots were performed for ubiquitin (VU-1) and Flag to verify presence of SCAR. Protein intensity for VU-1 was measured and normalized to Flag protein intensity. Values are shown as relative to LacZ. Error bars represent standard error. (D) Non-stop counters proteasomal degradation of SCAR. WT or not brains were dissected from 3rd instar larva and cultured ex vivo. Half of the brains from each genotype were treated for 24 hr with protease inhibitor [50 µm MG132 (MG132+)] while the remaining brains served as untreated control (MG132-). Whole cell extracts were immunoblotted for SCAR. SCAR protein intensity was measured and normalized to Ponceau total protein control. Numbers are expressed as relative to WT untreated control. Error bars represent standard error.
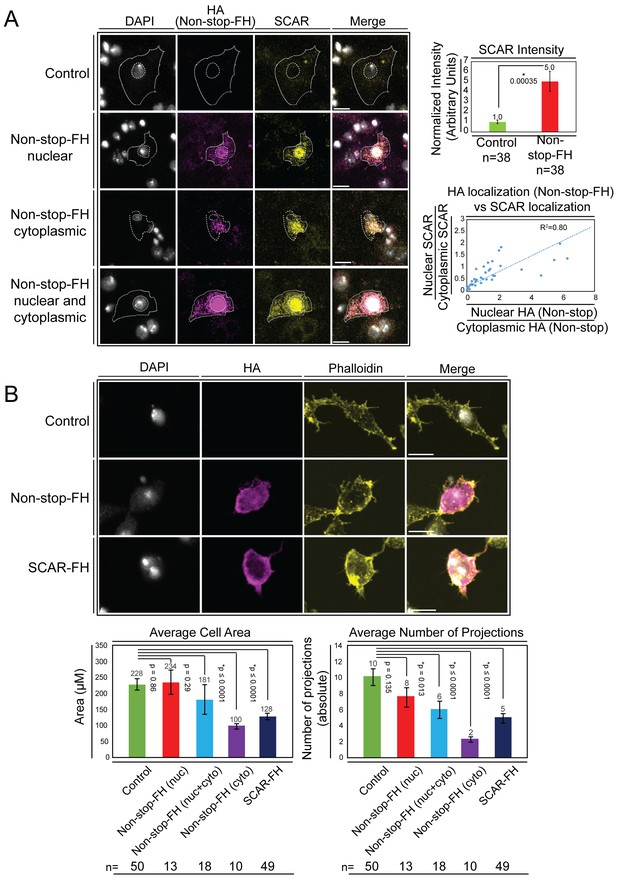
Overexpression of Non-stop increases SCAR levels, directs SCAR localization, and alters cell morphology – reducing cell area and number of cell protrusions.
(A) Augmenting Non-stop increases local levels of SCAR. Drosophila BG3 central nervous system cells were transfected with no plasmid (control) or with Non-stop-FH expression vector as indicated. Exogenous Non-stop and endogenous SCAR were located and quantified through nuclear localization by DAPI (DNA) (white) and indirect immunofluorescence toward: Non-stop-FH (anti-HA) (magenta), and anti-SCAR (yellow). Scale bar is 10 µM. Hashed lines outline DAPI (inner circle) to approximate nuclear location and the outermost SCAR signal to approximate the cell edge (outer circles). Three categories of HA (Non-stop-FH) expression are shown: Non-stop ‘nuclear’, Non-stop ‘cytoplasmic’, and Non-stop ‘nuclear and cytoplasmic.’ Total SCAR signal was measured in HA positive cells (Non-stop-FH) and mock transfected cells (Control). The average increase in endogenous SCAR immunofluorescence intensity was determined relative to control (top, right panel). Error bars are standard error. To examine the relationship between Non-stop localization and increased SCAR protein, SCAR immunofluorescence intensity was measured in the nucleus (as defined by DAPI) and in the cytoplasm. A ratio of nuclear SCAR intensity divided by cytoplasmic SCAR intensity was calculated. HA intensity was similarly measured in the nucleus and cytoplasm. A ratio of nuclear HA intensity to cytoplasmic HA intensity was calculated. The HA ratio was plotted on the X axis and the SCAR ratio was plotted on the Y axis. A trend line was calculated and the R2 was determined to be 0.80. (B) Increasing Non-stop alters F-actin organization similarly to increasing SCAR – reducing cell area and number of cell protrusions. BG3 cells were transfected with no-plasmid (Control), Non-stop-FH, or SCAR-FH. The location of exogenously expressed proteins were determined as above and F-actin was visualized using phalloidin. Cell area and total number of cellular projections were counted by quantifying the phalloidin signal distribution. HA-positive cells (Non-stop-FH or SCAR-FH) were compared to mock transfected (Control) cells (bottom panels). Cells expressing Non-stop-FH were split into three categories of HA (Non-stop-FH) expression as above. T-tests were used to compare samples and p-values are shown. Error bars are standard error.
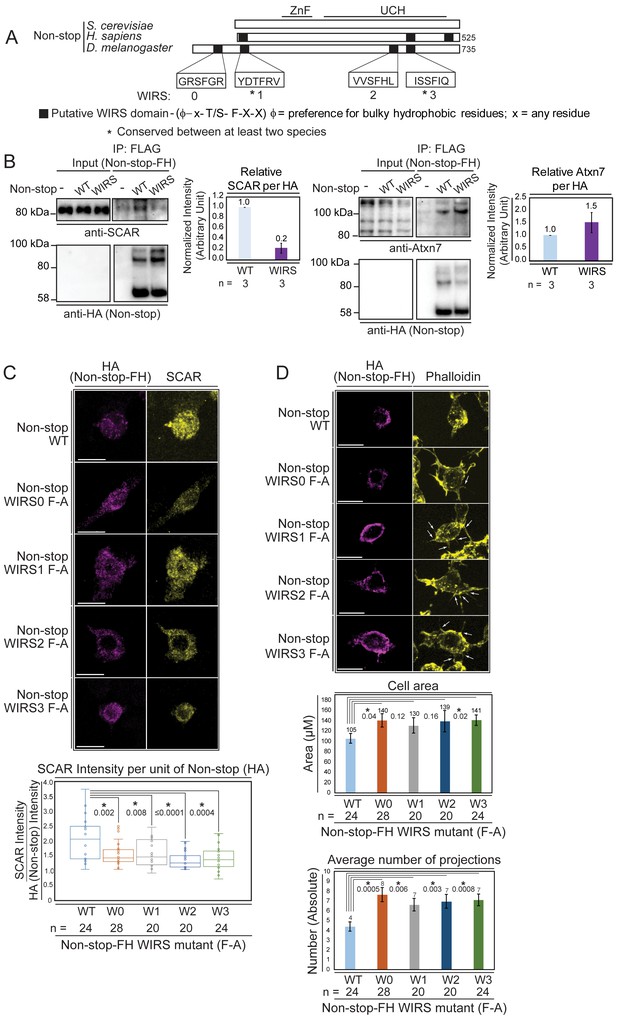
Non-stop bears conserved WIRS motifs.
Mutating these reduces Non-stop’s ability to bind SCAR, increase SCAR levels, reduce cell area, and reduce number of cell protrusions. (A) Alignment of Non-stop protein sequences reveals a conserved distribution of WIRS motifs between higher eukaryotes. (B) A WIRS mutant version of Non-stop-FH was created bearing phenylalanine to alanine point mutations in all four WIRS domains (WIRS). BG3 cells were mock transfected, transfected with Non-stop-FH (WT), or Non-stop WIRS FA-FH (WIRS). Whole cell extracts were prepared and immunoprecipitated using anti-FLAG resin to capture exogenous Non-stop and immunoblotted for SCAR. Immunoblot for Atxn7 verified incorporation into the SAGA complex. Immunoblot for HA verified capture of Non-stop-FH. Protein intensity of Atxn7 and SCAR were normalized to HA intensity. Intensities are shown relative to WT Non-stop. Error bars are standard error. (C) BG3 cells were transfected with either wild type (WT) or Non-stop-FH harboring a point mutation in one of the four putative WIRS motifs (W0–W3) as indicated. Exogenously expressed Non-stop and endogenous SCAR were detected by indirect immunofluorescence. Scale bar is 10 µm. Total fluorescence intensity of SCAR and HA were measured in HA containing cells. Box plots show the SCAR intensity divided by the intensity of HA. T-tests were used to compare samples and p-values are shown. P-values marked with an asterisk are significant. (D) BG3 cells transfected with either wild-type or Non-stop-FH harboring a point mutation in one of the four putative WIRS motifs (W0–W3) were immunostained for HA (magenta) and phalloidin (yellow). Scale bar is 10 µm. Arrows point to localized accumulation of Non-stop WIRS mutant protein, which coincide with aberrant actin protrusions commonly observed upon WRC or Arp2/3 loss of function. Cell area and number of projections protruding from cells was determined for HA positive cells. T-tests were used to compare the samples and p-values are shown. P-values marked with an asterisk are significant. Error bars are standard error.
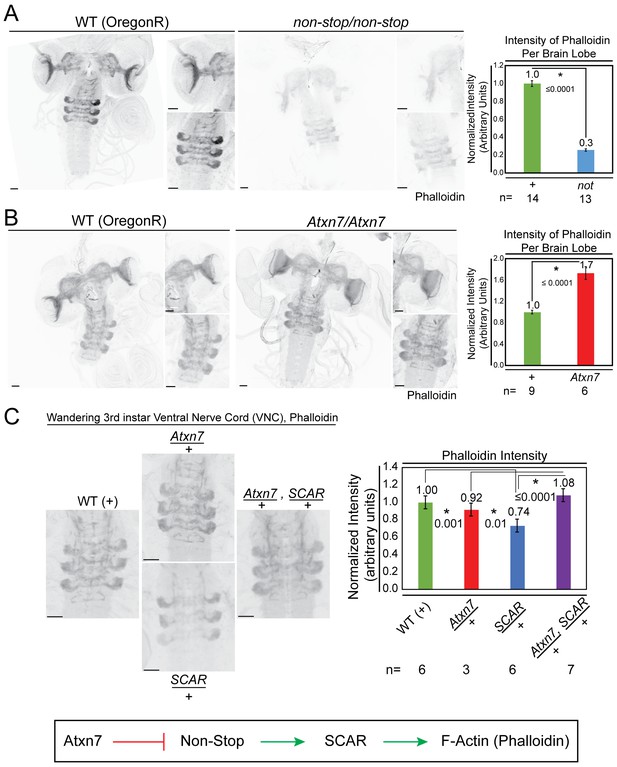
Atxn7 and Non-stop, act through the SCAR pathway in order to regulate the actin cytoskeleton in vivo.
(A) Phalloidin staining in third instar larval brains dissected from not02069 reveals a decrease in neural F-actin. (B) Conversely, phalloidin staining in Atxn7KG02020 shows an increase in F-actin. Microscope acquisition settings were identical to allow comparison. Scale bar is 50 µM. Charts show the averaged phalloidin fluorescence intensity measurements for individual brain lobes. Wild type average intensity was set to one and mutants were normalized to wild -type. A t-test was used to compare the samples and the p-value is shown and asterisks indicate significance. Error bars are standard error. (C) Phalloidin staining in wild type, Atxn7KG02020/+, SCAR[Δ37] /+, and Atxn7KG02020, SCAR[Δ37] /+, + third instar VNC shows that Atxn7 mutation can rescue the defects seen in SCAR heterozygotes. Scale bar is 20 µM. Fluorescence intensity was measured for each VNC. Wild-type average intensity was set to one and mutants were normalized to wild-type. Error bars are standard error. A t-test was used to compare the samples and significant values are listed with an asterisks and p-value. Line diagram outlines an explanation for these observations (bottom).
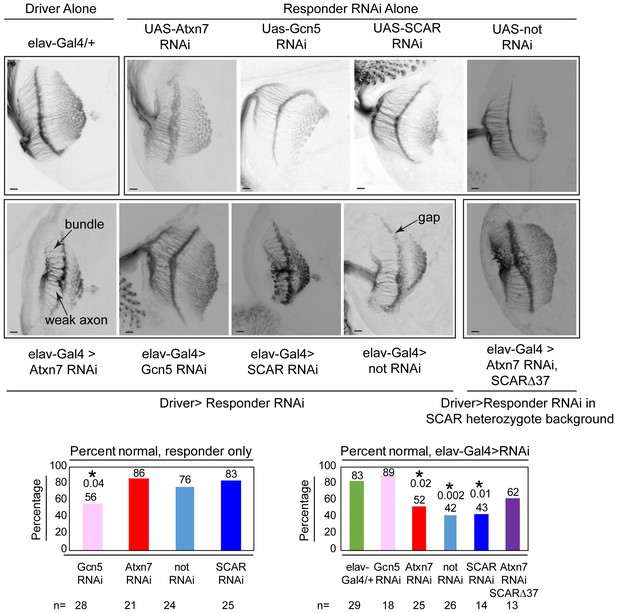
Chaoptin staining in the optic lobe of third instar larval brains reveals similar defects in Atxn7, SCAR, and not knockdowns in vivo.
The elav-Gal4 driver was used to drive UAS-RNAi lines in larvae. Third instar larval brains were dissected and immunostained with a Chaoptin antibody. Images were adjusted with contrast limited adaptive histogram equalization using the ImageJ CLAHE algorithm (Zuiderveld, 1994). Optic lobes were analyzed for the presence of bundles, gaps, or weak axons. Examples of these are indicated in the figure. A brain was determined to be normal if it had three or fewer of these defects. The percentage of normal brains are shown for each genotype. A fisher’s exact test was used to compare samples to the driver alone (elav-gal4/+). Significant p-values are marked with an asterisk. Error bars shows standard error.
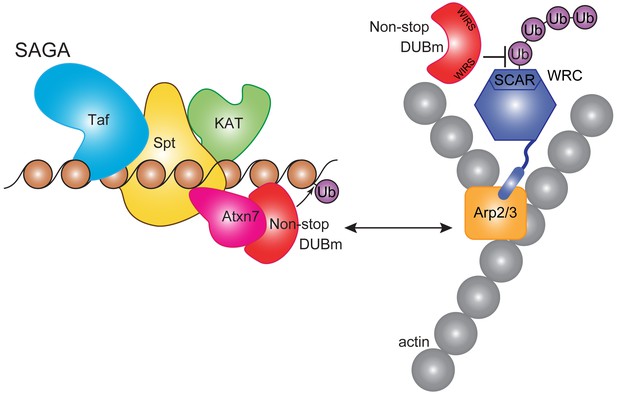
Non-stop regulates SCAR protein levels and location.
Model showing Non-stop interacts with WRC in an Atxn7-dependent manner, where Non-stop then counteracts degradation of WRC subunit SCAR. Loss of Atxn7 leads to increased availability of Non-stop for interaction with WRC.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | not | NA | FLYB: FBgn0013717 | |
| Gene (D. melanogaster) | Atxn7 | NA | FLYB: FBgn0031420 | |
| Gene (D. melanogaster) | SCAR | NA | FLYB: FBgn0041781 | |
| Gene (D. melanogaster) | Gcn5 | NA | FLYB: FBgn0020388 | |
| Gene (D. melanogaster) | Ada2b | NA | FLYB: FBgn0037555 | |
| Strain, strain background (D. melanogaster) | Elav-Gal4 | Bloomington Drosophila Stock Center | BDSC:458 RRID:BDSC_458 | Genotype:P(w[+mW.hs]=GawB)elav[C155] |
| Strain, strain background (D. melanogaster) | Uas-Gcn5 RNAi | Vienna Drosophila Resource Center | VDRC:21786 RRID:FlyBase_FBst0454233 | Construct ID:11218 |
| Strain, strain background (D. melanogaster) | Uas-SCAR RNAi | Bloomington Drosophila Stock Center | BDSC:36121 RRID:BDSC_36121 | Genotype: y[1] sc[*] v[1]; P(y[+t7.7] v[+t1.8]=TRiP.HMS01536)attP40 |
| Strain, strain background (D. melanogaster) | Uas-Not RNAi | Bloomington Drosophila Stock Center | BDSC:28725 rebalanced with Tm6b, Tb RRID:BDSC_28725 | Genotype: y[1] v[1]; P(y[+t7.7] v[+t1.8]=TRiP.JF03152)attP2/TM6B, Tb |
| Strain, strain background (D. melanogaster) | Uas-Atxn7 RNAi | Vienna Drosophila Resource Center | VDRC:102078 RRID:FlyBase_FBst0473949 | Construct ID:110634 |
| Strain, strain background (D. melanogaster) | OreR | DGGR | Catalog number: 109612 RRID:DGGR_109612 | |
| Strain, strain background (D. melanogaster) | Non-stop[02069] | Bloomington Drosophila Stock Center | BDSC:11553 Rebalanced with Tm3 GFP RRID:BDSC_11553 | Genotype: P(ry[+t7.2]=PZ)not[02069] ry[506]/TM3, P(w[+mC]=GAL4 twi.G)2.3, P(UAS-2xEGFP)AH2.3, Sb[1] Ser[1] |
| Strain, strain background (D. melanogaster) | SCAR [Delta37] | Bloomington Drosophila Stock Center | BDSC:8754 Rebalanced with CyoGFP RRID:BDSC_8754 | Genotype: w[*]; SCAR[Delta37] P(ry[+t7.2]=neoFRT)40A/CyO,, P(w[+mC]=GAL4 twi.G)2.2, P(UAS-2xEGFP)AH2.2. Cross BDSC:6662 and BDSC:8754 |
| Strain, strain background (D. melanogaster) | Atxn7[KG02020] | Bloomington Drosophila Stock Center | BDSC: 14255 Rebalanced with CyoGFP RRID:BDSC_14255 | Genotype: y[1] w[67c23]; P(y[+mDint2] w[BR.E.BR]=SUPor P)CG9866[KG02020]/CyO, P(w[+mC]=GAL4 twi.G)2.2, P(UAS-2xEGFP)AH2.2. |
| Strain, strain background (D. melanogaster) | Uas-Atxn7 RNAi, SCARΔ37 | This Paper | Materials and methods Subsection Fly Strains Genotype: w[*]; SCAR[Delta37] P(ry[+t7.2]=neoFRT)40A/CyO, P(w[+mC]=GAL4 twi.G)2.2, P(UAS-2xEGFP)AH2.2.; P{KK110634} VIE-260B/TM6C, cu[1] Sb[1] Tb[1] | |
| Cell line (D. melanogaster) | ML-DmBG3-c2 | Drosophila Genomics Resource Center #68 | FLYB: FBtc0000068; RRID:CVCL_Z728 | FlyBase symbol: ML-DmBG3-c2 |
| Cell line (D. melanogaster) | S2-DRSC | Drosophila Genomics Resource Center #181 | FLYB: FBtc0000181; RRID:CVCL_Z992 | FlyBase symbol: S2-DRSC |
| Antibody | Guinea Pig anti Non-stop | (Mohan et al., 2014a) | Western Blot Dilution (1:1000) IF Dilution (1:150) | |
| Antibody | Mouse anti Chaoptin (Monoclonal) | Developmental Studies Hybridoma Bank | 24B10 RRID:AB_528161 | IF Dilution (1:250) |
| Antibody | Goat anti Rat IGG −568 (Polyclonal) | Invitrogen | A-11077 RRID:AB_141874 | IF Dilution (1:1000) |
| Antibody | Goat anti Mouse IGG-488 (Polyclonal) | Invitrogen | A-11001 RRID:AB_2534069 | IF Dilution (1:1000) |
| Antibody | Goat anti Guinea pig IGG- 488 (Polyclonal) | Invitrogen | A11073 RRID:AB_142018 | IF Dilution (1:1000) |
| Other | Phalloidin-488 | Invitrogen | A12379 | IF Dilution (1:20) |
| Other | Phalloidin-568 | Invitrogen | A12380 RRID:AB_2810839 | IF Dilution (1:20) |
| Other | Vecta Shield | Vector Labs | H-1200 | |
| Antibody | Rat anti HA-HRP (Monoclonal) | Roche | 12013819001 RRID:AB_390917 | Western Blot Dilution (1:500) |
| Antibody | Mouse anti SCAR (Monoclonal) | Developmental Studies Hybridoma Bank | P1C1-SCAR RRID:AB_2618386 | Western Blot Dilution (1:250) IF Dilution (1:100) |
| Antibody | Rabbit anti Atxn7 | (Mohan et al., 2014a) | Western Blot Dilution (1:2000) | |
| Antibody | Guinea Pig anti Ada2b | Gift from Jerry L Workman (Kusch et al., 2003) | Western Blot Dilution (1:1000) | |
| Antibody | Goat anti Guinea Pig HRP (Polyclonal) | Jackson ImmunoResearch INC | 106-035-003, RRID:AB_2337402 | Western Blot Dilution (1:10000) |
| Antibody | Goat anti mouse HRP (Polyclonal) | Jackson ImmunoResearch INC | 115-035-003, RRID:AB_10015289 | Western Blot Dilution (1:5000) |
| Antibody | Goat anti Rabbit HRP (Polyclonal) | Jackson ImmunoResearch INC | 111-035-003 RRID:AB_2313567 | Western Blot Dilution (1:10000) |
| Antibody | Rat anti HA (Monoclonal) | Roche | 11867423001 RRID:AB_390918 | IF Dilution (1:250) |
| Recombinant DNA reagent | pMT-HA-Ub | gift from Jianhang Jia | gift from Jianhang Jia | |
| Recombinant DNA reagent | Scar-FH | Berkley Expression Clone Collection | FMO14142 | |
| Recombinant DNA reagent | Scar-Flag | This paper | Materials and methods Subsection Plasmids Quick change on FMO14142 F Primer:GATGACGACAAGGTCAAACTTGCTGCTTAGACTAGTTCTAGT R Primer: ACTAGAACTAGTCTAAGCAGCAAGTTTGACCTTGTCGTCATC | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA | This Paper | Sequence ID: AAD53181.1 | Materials and methods Subsection Plasmids |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-0FA | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA WIRS0 phenylalanine to alanine F: GCAGTGGCCGAAGCGCCGGCAGGGGAACGGAACGGTGGGC WIRS0 phenylalanine to alanine R: CCGTTCCCCTGCCGGCGCTTCGGCCACTGCTGCTGCTGC | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-1FA | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA WIRS1 phenylalanine to alanine F: CAGCTACGATACAGCCCGGGTCATCGACGCCTACTTCGCTGCTTGCG WIRS1 phenylalanine to alanine R: GGCGTCGATGACCCGGGCTGTATCGTAGCTGTGCTCCTTCACATAGC | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-2FA | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA WIRS2 phenylalanine to alanine F: CCAGCGTGGTGTCGGCCCATTTGAAACGCTTCGAGCACTCAGCTCTG WIRS2 phenylalanine to alanine R: CGAAGCGTTTCAAATGGGCCGACACCACGCTGGGCAGAGTGCGCAG | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-3FA | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA WIRS3 phenylalanine to alanine F: CGCAAGATCTCCTCGGCCATTCAATTCCCCGTGGAGTTCGACATG WIRS3 phenylalanine to alanine R: CCACGGGGAATTGAATGGCCGAGGAGATCTTGCGATCGATCAGAGC | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-WIRSFA | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA all of the above primers sequentially | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-C406A | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA Non-stop C406A F: CTTAATCTGGGCGCCACTGCCTTCATGAACTGCATCGTC Non-stop C406A R: GACGATGCAGTTCATGAAGGCAGTGGCGCCCAGATTAAG | |
| Recombinant DNA reagent | Prmha3-Non-stop-2XFlag-2XHA-C406S | This Paper | Materials and methods Subsection Plasmids Quick change mutagenesis on Prmha3-Non-stop-2XFlag-2XHA Non-stop C406S F: CTTAATCTGGGCGCCACTAGCTTCATGAACTGCATCGTC Non-stop C406S R: GACGATGCAGTTCATGAAGCTAGTGGCGCCCAGATTAAG | |
| Sequence-based reagent | dsRNA Lacz | dsRNA-LacZ-R: GCTAATACGACTCACTATAGGCCAAACATGACCARGATTACGCCAAGCT dsRNA-LacZ-F: GCTAATACGACTCACTATAGGCCAAACGTCCCATTCGCCATTCAGGC | ||
| Sequence-based reagent | dsRNA not | http://www.flyrnai.org | DRSC11378 | Primer F: TAATACGACTCACTATAGGCGCAGGCTGAACTGTTTG Primer R: TAATACGACTCACTATAGGTCTATTCCGGCTCCCGTT |
| Sequence-based reagent | dsRNA not | http://www.flyrnai.org | BKN21994 | Primer F: TAATACGACTCACTATAGGACTTGACCCACGTGTCCTTC Primer R:TAATACGACTCACTATAGGATTGACCAGATCTTCACGGG |
| Sequence-based reagent | dsRNA Atxn7 | http://www.flyrnai.org | DRSC35628 | Primer F:TAATACGACTCACTATAGGCGACATGGAAAAGGTCATCA Primer R:TAATACGACTCACTATAGGGGAAACCTGCCTTCGTGTAA |
| Sequence-based reagent | dsRNA Atxn7 | http://www.flyrnai.org | DRSC23138 | Primer F: TAATACGACTCACTATAGGCTGTTAAGCTGGAGGCCAAGPrimer R: TAATACGACTCACTATAGGGCCCTCTTATTGCACCTCAG |
| Sequence-based reagent | Taqman Assay: not | Thermofisher | TaqManID: Dm01823071_g1 | |
| Sequence-based reagent | Taqman Assay: Atxn7 | Thermofisher | TaqManID: Dm01800874_g1 | |
| Sequence-based reagent | Taqman Assay: SCAR | Thermofisher | TaqManID: Dm01810606_g1 | |
| Sequence-based reagent | Taqman Assay: RPL32 | Thermofisher | TaqMan ID: Dm02151827_g1 | |
| Commercial assay or kit | high capacity cDNA reverse transcription kit | ThermoFisher | 4374966 | |
| Commercial assay or kit | TaqMan Universal PCR Master Mix | ThermoFisher | 4364340 | |
| Chemical compound, drug | MG132 | Sigma-Aldrich | C2211 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij/ |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49677.012





