Muscle function and homeostasis require cytokine inhibition of AKT activity in Drosophila
Figures
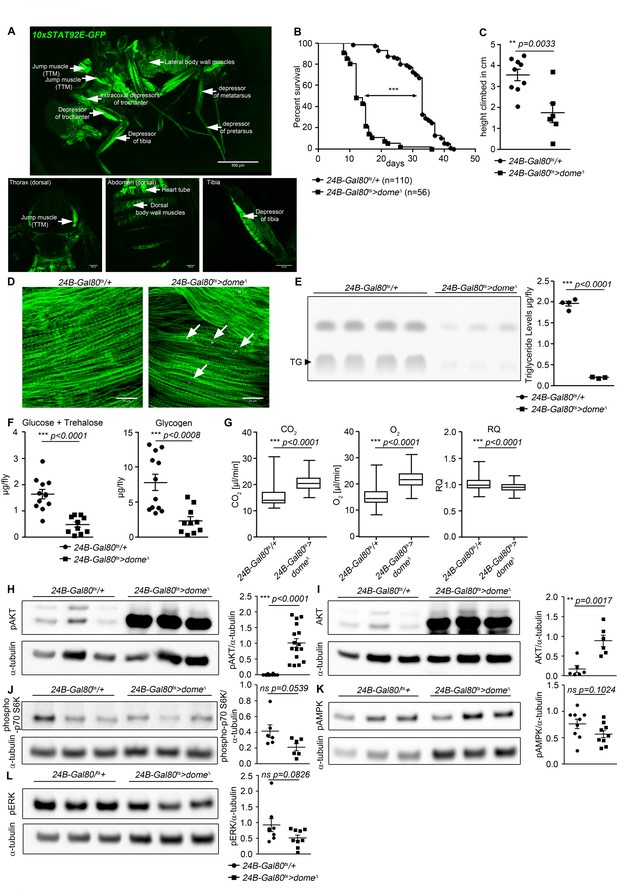
Dome inhibition in adult muscle reduces lifespan, disrupts homeostasis, and causes AKT hyperactivation.
(A) STAT activity in different muscles in 10xSTAT92E-GFP reporter fly. One fly out of 5 shown. Upper panel: lateral view, Scale bar = 500 µm. Lower panels: dorsal thorax (left); dorsal abdomen (middle); tibia (right), Scale bar = 100 µm. (B) Lifespan of 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ at 29°C. Log-Rank test: χ2 = 166, ***p<0.0001; Wilcoxon test: χ2 = 157.7, ***p<0.0001. (C) Negative geotaxis assay of 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. Points represent mean height climbed in individual vials (~20 flies/vial), pooled from three independent experiments. Unpaired T-test: **p=0.0033. (D) Muscle (Phalloidin) and neutral lipid (LipidTox) of thorax samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. One representative fly per genotype is shown of six analysed. Scale bar = 50 µm. (E) Thin layer chromatography (TLC) of triglycerides in 7-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies, n = 3–4 per genotype. One experiment of two is shown. Unpaired T-Test: ***p<0.0001. (F) Glucose and trehalose (left) and glycogen (right) in 7-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies, pooled from two independent experiments. Unpaired T-Test (Glucose +Trehalose): ***p<0.0001 and unpaired T-Test (Glycogen): ***p<0.0001. (G) CO2 produced, O2 consumed, and RQ of 7-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. Box plots show data from one representative experiment of three, with data collected from a 24 hr measurement pooled from 3 to 4 tubes per genotype with 10 flies/tube. P values from Mann-Whitney test. (H–L) Western blots of leg protein from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. (H) Phospho-AKT (S505). One experiment of four is shown. Unpaired T-Test: ***p<0.0001. (I) Total AKT. One experiment of two is shown. Unpaired T-Test: **p=0.0017. (J) Phospho-p70 S6K (T398). One experiment of two is shown. Unpaired T-Test: ns p=0.0539. (K) Phospho-AMPKα (T173). One experiment of three is shown. Unpaired T-Test: ns p=0.1024. (L) Phospho-ERK (T202/Y204). One experiment of three is shown. Unpaired T-Test: ns p=0.0826.
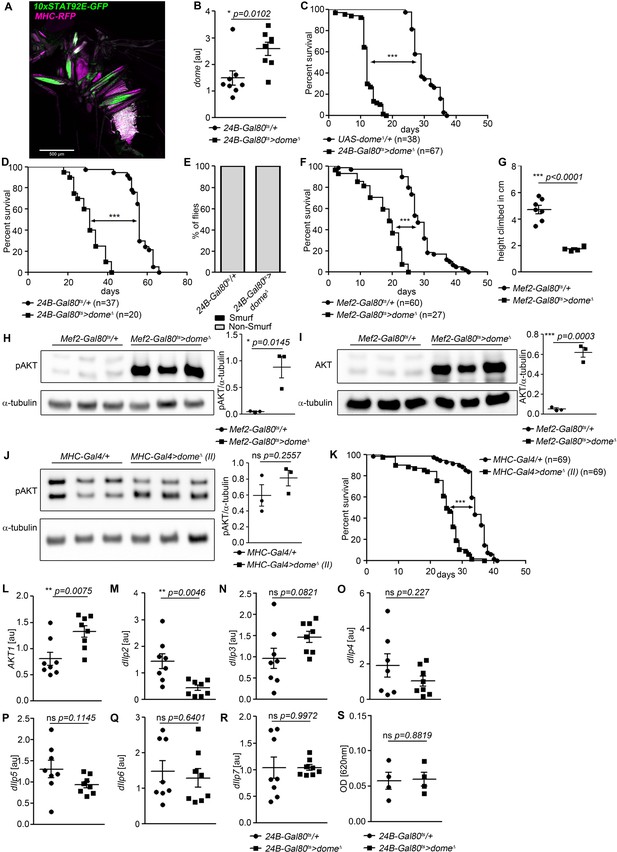
Further characterisation of the requirement for dome in adult muscle.
(A) STAT activity (10xSTAT92E-GFP) and muscle (MHC-RFP) colocalize in adult flies. One fly of 6 shown. Scale bar = 500 µm. (B) dome expression by qRT-PCR in thorax samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. Unpaired T-Test: *p=0.0102. (C) Lifespan of UAS-dome∆/+ and 24B-Gal80ts > dome∆ at 29°C. Log-Rank test: χ2 = 100.8, ***p<0.0001; Wilcoxon test: χ2 = 76.2, ***p<0.0001. (D) Lifespan of 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ at 25°C. Log-Rank test: χ2 = 61.83, ***p<0.0001; Wilcoxon test: χ2 = 55.18, ***p<0.0001. (E) Smurf assay of 14-day-old 24B-Gal80ts/+ (n = 49) and 24B-Gal80ts-dome∆ flies (n = 18). Data pooled from two independent experiments. (F) Lifespan of Mef2-Gal80ts/+ and Mef2-Gal80ts > dome∆ flies at 29°C. Log-Rank test: χ2 = 86.96, ***p<0.0001; Wilcoxon test: χ2 = 78.61, ***p<0.0001. (G) Negative geotaxis assay of 14-day-old Mef2-Gal80ts/+ and Mef2-Gal80ts > dome∆ flies. Points represent mean climbing height of individual vials analysed (~20 flies/vial), pooled from three independent experiments. Unpaired T-Test: ***p<0.0001. (H, I) Western blots of protein from legs of 14-day-old Mef2-Gal80ts/+ and Mef2-Gal80ts > dome∆ flies. One of three independent experiments is shown. (H) Phospho-AKT. Unpaired T-Test: *p=0.0145. (I) Total AKT. Unpaired T-Test: ***p<0.0003. (J) Western blots of Phospho-AKT in leg samples from 14-day-old MHC-Gal4/+ and MHC-Gal4 >dome∆ (II) flies. One of two independent experiments is shown. Unpaired T-Test: ns p=0.2557. (K) Lifespan of MHC-Gal4/+ and MHC-Gal4 >dome∆ (II) flies at 29°C. Log-Rank test: χ2 = 82.9, ***p<0.0001; Wilcoxon test: χ2 = 58.91, ***p<0.0001. (L–R) Expression by qRT-PCR of Akt1 and insulin-like peptides in whole fly samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts-dome∆ flies. All transcript levels are normalized to Rpl1 and shown in arbitrary units [au]. P values in B, G, H-J, L-R from unpaired T-test. (S) Feeding assay of 24-Gal80ts/+ and 24B2-Gal80ts > dome∆ flies at 29°C. n = 4 were analysed per genotype. Absorbance at 620 nm is shown in arbitrary units [au]. Unpaired T-Test: ns p=0.8819.
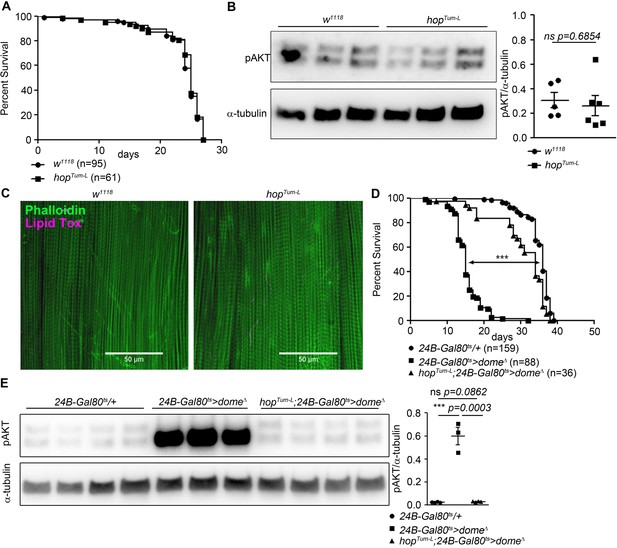
Hop is required, but not sufficient, for Dome to control AKT.
(A) Lifespan of w1118 and hopTum-L flies at 29°C. Log-Rank test: χ2 = 0.3223, ns p=0.5702; Wilcoxon test: χ2 = 0.4756, ns p=0.4906. (B) Phospho-AKT in leg samples from 14-day-old w1118 and hopTum-L flies. One experiment of two is shown. Unpaired T-Test: ns p=0.6854. (C) Actin (Phalloidin) and neutral lipid (LipidTox) in flight muscle from 14-day-old w1118 and hopTum-L flies. One representative fly shown of six analysed per genotype. Scale bar = 50 µm. (D) Lifespan of 24B-Gal80ts/+, 24B-Gal80ts > dome∆, and hopTum-L;24B-Gal80ts > dome∆ flies at 29°C. Log-Rank test (24B-Gal80ts/+ vs. 24B-Gal80ts > dome∆): χ2 = 319.4, ***p<0.0001; Wilcoxon test (24B-Gal80ts/+ vs. 24B-Gal80ts > dome∆): χ2 = 280.2, ***p<0.0001. Log-Rank test (24B-Gal80ts/+ vs. hopTum-L 24B-Gal80ts > dome∆): χ2 = 18.87, ***p<0.0001; Wilcoxon test (24B-Gal80ts/+ vs. hopTum-L 24B-Gal80ts > dome∆): χ2 = 20.83, ***p<0.0001. (E) Phospho-AKT in leg samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > dome∆ and hopTum-L;24B-Gal80ts > dome∆ flies. P values from unpaired T-Test.
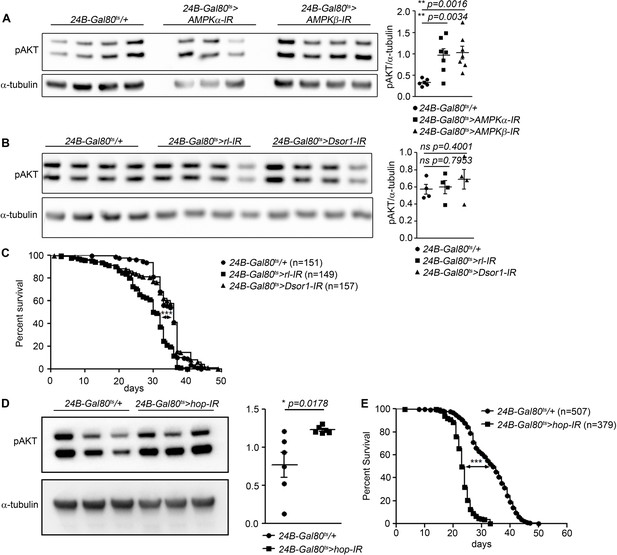
Interactions of dome with AMPK, MAPK, and FOXO signalling in adult muscle.
(A) Phospho-AKT in leg samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > AMPKα-IR, and 24B-Gal80ts > AMPKβ-IR flies. One of three independent experiments is shown. P values from unpaired T-Test. (B) Phospho-AKT in leg samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > rl IR, and 24B-Gal80ts > Dsor1 IR flies. One of three independent experiments is shown. P values from unpaired T-Test. (C) Lifespan of 24B-Gal80ts/+, 24B-Gal80ts > rl IR, and 24B-Gal80ts > Dsor1 IR flies at 29°C. Log-Rank test (24B-Gal80ts/+ vs. 24B-Gal80ts > rl IR): χ2 = 60.29, ***p<0.0001; Wilcoxon test (24B-Gal80ts/+ vs. 24B-Gal80ts > rl IR): χ2 = 58.32, ***p<0.0001; Log-Rank test (24B-Gal80ts/+ vs. 24B-Gal80ts > Dsor1 IR): χ2 = 1.186, ns p=0.2760; Wilcoxon test (24B-Gal80ts/+ vs. 24B-Gal80ts > Dsor1 IR): χ2 = 0.0033, ns p=0.9538. (D) Phospho-AKT in leg samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > hop IR flies. One of two independent experiments is shown. Unpaired T-Test: *p=0.0187. (E) Lifespan of 24B-Gal80ts/+ and 24B-Gal80ts > hop IR flies at 29°C. Log-Rank test (24B-Gal80ts/+ vs. 24B-Gal80ts > hop IR): χ2 = 546.4, ***p<0.0001; Wilcoxon test (24B-Gal80ts/+ vs. 24B-Gal80ts > hop IR): χ2 = 458.1, ***p<0.0001. P values in A, C, E from unpaired T-test.
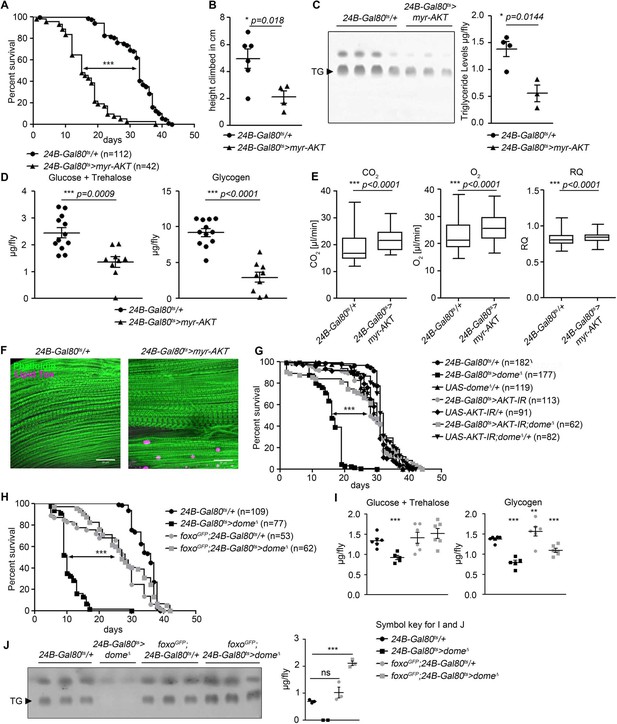
AKT hyperactivation causes pathology in 24B-Gal80ts > dome∆ flies.
(A) Lifespan of 24B-Gal80ts/+ and 24B-Gal80ts > myr-AKT at 29°C. Log-Rank test: χ2 = 115.5, ***p<0.0001; Wilcoxon test: χ2 = 123.6, ***p<0.0001. (B) Negative geotaxis assay of 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > myr-AKT flies. Points represent mean height climbed in individual vials (~20 flies/vial), pooled from two independent experiments. Unpaired T-Test: *p=0.018. (C) TLC of triglycerides in 7-day-old 24B-Gal80ts/+ and 24B-Gal80ts > myr-AKT flies, n = 3–4 per genotype. One experiment of two is shown. Unpaired T-Test: *p=0.0144. (D) Glucose and trehalose (left panel) and glycogen (right panel) in 7-day-old 24B-Gal80ts/+ (n = 12) and 24B-Gal80ts > myr-AKT (n = 9) flies, pooled from two independent experiments. Unpaired T-Test (Glucose +Trehalose): ***p=0.0009 and unpaired T-Test (Glycogen): ***p<0.0001. (E) CO2 produced, O2 consumed, and RQ of 7-day-old 24B-Gal80ts/+ and 24B-Gal80ts > myr-AKT flies. Box plots show data from one representative experiment of three, with data points collected from a 24 hr measurement pooled from 3 to 4 tubes per genotype with 10 flies/tube. P values from Mann-Whitney test. (F) Phalloidin and LipidTox staining of thorax samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > myr-AKT flies. One representative fly per genotype is shown of 3 analysed per group in two independent experiments. Scale bar = 50 µm. (G) Lifespan of 24B-Gal80ts/+, 24B-Gal80ts > dome∆, UAS-domeΔ/+, 24B-Gal80ts > AKT-IR, UAS-AKT-IR/+, 24B-Gal80ts > AKT-IR;dome∆ and UAS-AKT-IR;dome∆/+ flies at 29°C. Log-Rank test (24B-Gal80ts > dome∆ vs. 24B-Gal80ts > AKT-IR;dome∆): χ2 = 101.0, ***p<0.0001; Wilcoxon test (24B-Gal80ts > dome∆ vs. 24B-Gal80ts > AKT-IR;dome∆): χ2 = 59.87, ***p<0.0001. (H) Lifespan of 24B-Gal80ts/+, 24B-Gal80ts > dome∆, foxo-GFP;24B-Gal80ts/+, and foxo-GFP;24B-Gal80ts > dome∆ flies at 29°C. Log-Rank test (24B-Gal80ts > dome∆ vs. foxo-GFP;24B-Gal80ts > dome∆): χ2 = 114.0, ***p<0.0001; Wilcoxon test (24B-Gal80ts > dome∆ vs. foxo-GFP;24B-Gal80ts > dome∆): χ2 = 93.59, ***p<0.0001. (I) Glucose + trehalose and glycogen in 7-day-old 24B-Gal80ts/+, 24B-Gal80ts > domeΔ, foxo-GFP;24B-Gal80/+, and foxo-GFP; 24B-Gal80ts > domeΔ flies. Statistical testing was performed with one-way ANOVA. (J) TLC of triglycerides in 7-day-old 24B-Gal80ts/+, 24B-Gal80ts > domeΔ, foxo-GFP;24B-Gal80ts/+, and foxo-GFP;24B-Gal80ts > domeΔ flies. Statistical testing was performed with one-way ANOVA.
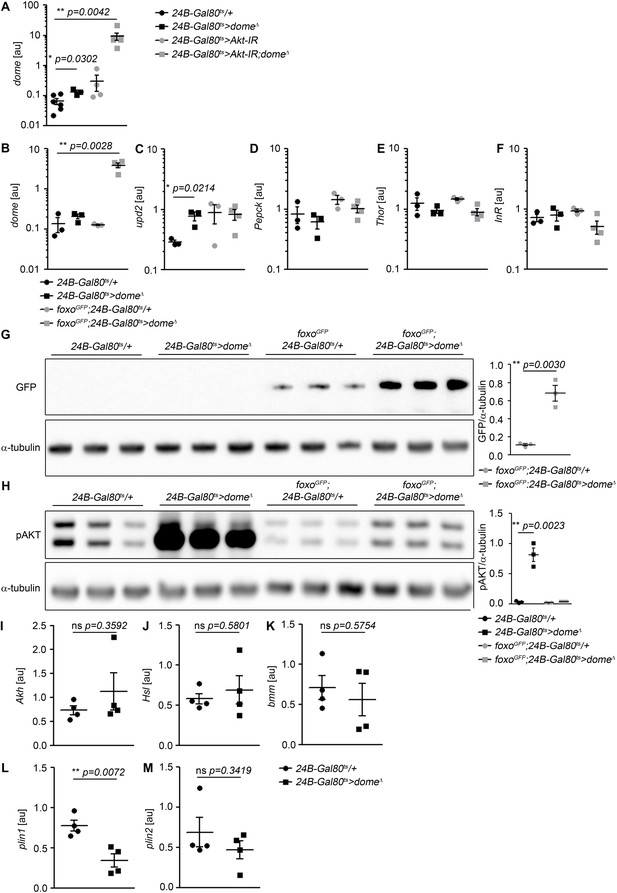
Mutual regulation by AKT, Foxo, and Dome.
(A) dome expression by qRT-PCR in whole fly samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > dome∆, 24B-Gal80ts > Akt-IR, and 24B-Gal80ts > Akt-IR;dome∆ flies. (B–F) Gene xpression analysis of dome, upd2, Pepck, Thor and InR in whole fly samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > dome∆, foxo-GFP;24B-Gal80ts/+, and foxo-GFP;24B-Gal80ts > dome∆ flies. P values from unpaired T-test. (B) dome. (C) upd2. (D) Pepck. (E) Thor. (F) InR. (G) Western blot for GFP to detect the Foxo-GFP fusion protein in leg samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > dome∆, foxo-GFP;24B-Gal80ts/+, and foxo-GFP;24B-Gal80ts > dome∆ flies. (H) Western blot for Phospho-AKT in leg samples from 14-day-old 24B-Gal80ts/+, 24B-Gal80ts > dome∆, foxo-GFP;24B-Gal80ts/+, and foxo-GFP;24B-Gal80ts > dome∆ flies. One of two independent experiments is shown. P values in A-D from unpaired T-test. (I–M) Gene expression analysis of Akh and six confirmed or putative Akh target genes in whole fly samples from 14-day-old 24B-Gal80ts/+ and 24B-Gal80ts > dome∆ flies. Transcript levels are shown in arbitrary units [au]. P values from unpaired T-test. (I) Akh. (J) Hsl. (K) bmm. (L) plin1. (M) plin2.
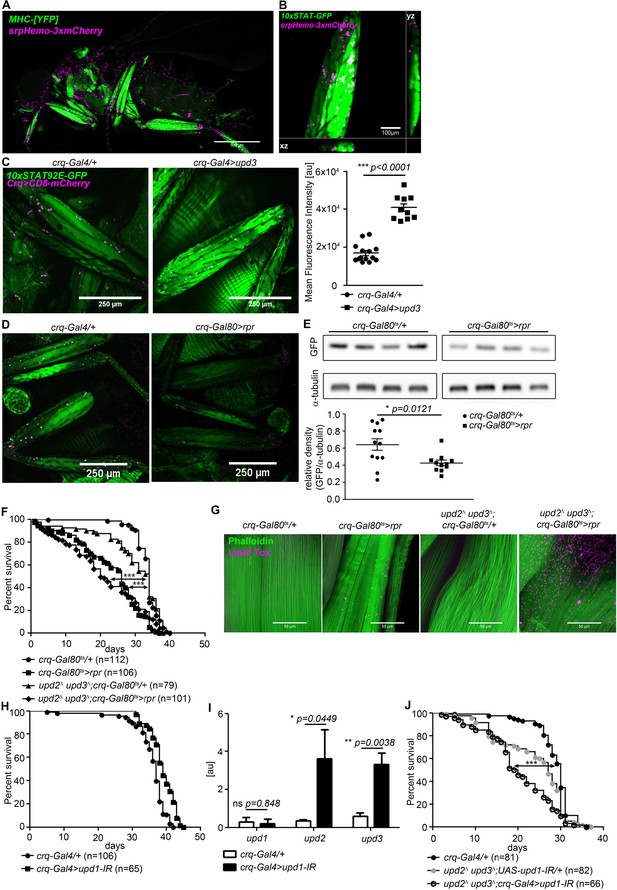
Plasmatocytes promote muscle Dome activity.
(A) Muscle (MHCYFP) and plasmatocytes (srpHemo-3xmCherry) in 7-day-old flies. Plasmatocytes are found in close proximity to adult muscles. One representative fly of 5 is shown. Scale bar = 500 µm. (B) Legs and plasmatocytes in 7-day-old 10xSTAT92E-GFP;srpHemo-3xmCherry flies. Muscle with high JAK-STAT activity (green) is surrounded by plasmatocytes (magenta). One representative fly of 5 is shown. Scale bar = 100 µm. (C) STAT activity and plasmatocytes in legs from control (10xSTAT92E-GFP;crq-Gal4 >CD8-mCherry/+) and upd3-overexpressing (10xSTAT92E-GFP;crq-4>CD8mCherry/UAS-upd3) flies. One representative fly of 10–14 is shown. Scale bar = 100 µm. Graph shows mean fluorescence intensity (MFI). Unpaired T-Test: ***p<0.0001. (D) STAT activity and plasmatocytes in legs from control (10xSTAT92E-GFP;crq-Gal80ts > CD8-mCherry/+) and plasmatocyte-depleted (10xSTAT92E-GFP;crq-Gal80ts > CD8 mCherry/rpr) flies. One representative fly of six is shown. Scale bar = 250 µm. (E) Western blot analysis of STAT-driven GFP in legs from 7-day-old control (10xSTAT92E-GFP;crq-Gal80ts > CD8-mCherry/+) and plasmatocyte-depleted (10xSTAT92E-GFP;crq-Gal80ts > CD8 mCherry/rpr flies). One representative experiment of three is shown. Graph shows STAT-GFP/α-tubulin for control (crq-Gal80ts/+) and plasmatocyte-depleted (crq-Gal80ts > rpr) leg samples. Unpaired T-Test: *p=0.0121. (F) Lifespan of crq-Gal80ts/+, crq-Gal80ts > rpr, upd2Δ upd3Δ;crq-Gal80ts/+, and upd2Δ upd3Δ;crq-Gal80ts > rpr flies at 29°C. Log-Rank test (crq-Gal80ts/+ vs. crq-Gal80ts > rpr): χ2 = 101.7, ***p<0.0001; Wilcoxon test (crq-Gal80ts/+ vs. crq-Gal80ts > rpr): χ2 = 107.8, ***p<0.0001; Log-Rank test (crq-Gal80ts/+ vs. upd2 Δ upd3Δ;crq-Gal80ts > rpr): χ2 = 60.03, ***p<0.0001; Wilcoxon test (crq-Gal80ts/+ vs. upd2 Δ upd3Δ;crq-Gal80ts > rpr): χ2 = 80.97, ***p<0.0001. (G) Actin (Phalloidin) and neutral lipid (LipidTox) in thorax samples from 14-day-old crq-Gal80ts/+, crq-Gal80ts > rpr, upd2 Δ upd3Δ;crq-Gal80ts/+, and upd2 Δ upd3Δ;crq-Gal80ts > rpr flies. One representative fly per genotype shown of 6 analysed per group. Scale bar = 50 µm. (H) Lifespan of crq-Gal4/+ and crq-Gal4 >upd1 IR flies at 29°C. Log-Rank test: χ2 = 31.36, ***p<0.0001; Wilcoxon test: χ2 = 22.17, ***p=0.0001. (I) Expression by qRT-PCR of upd1, upd2 and upd3 in thorax samples of crq-Gal4/+ and crq-Gal4 >upd1 IR flies, data from four independent samples of each genotype.. Unpaired T-Test (upd1): ns p=0.848, unpaired T-Test (upd2): *p=0.0449 and unpaired T-Test (upd3): **p=0.0038. (J) Lifespan of crq-Gal4/+, upd2 Δ upd3Δ;UAS-upd1-IR/+, and upd2Δ upd3Δ;crq-Gal4 >upd1 IR flies at 29°C. Log-Rank test (crq-Gal4/+ vs. upd2 Δ upd3Δ;crq-Gal4 >upd1 IR): χ2 = 41.12, ***p<0.0001; Wilcoxon test (crq-Gal4/+ vs. upd2Δ upd3Δ;crq-Gal4 >upd1 IR): χ2 = 54.47, ***p<0.0001 Log-Rank test (crq-Gal4/+ vs. upd2 Δ upd3Δ;UAS-upd1-IR/+): χ2 = 14.46, ***p<0.0001; Wilcoxon test (crq-Gal4/+ vs. upd2Δ upd3Δ;UAS-upd1-IR/+): χ2 = 19.99, ***p<0.0001. P values in C, E, H from unpaired T-test.
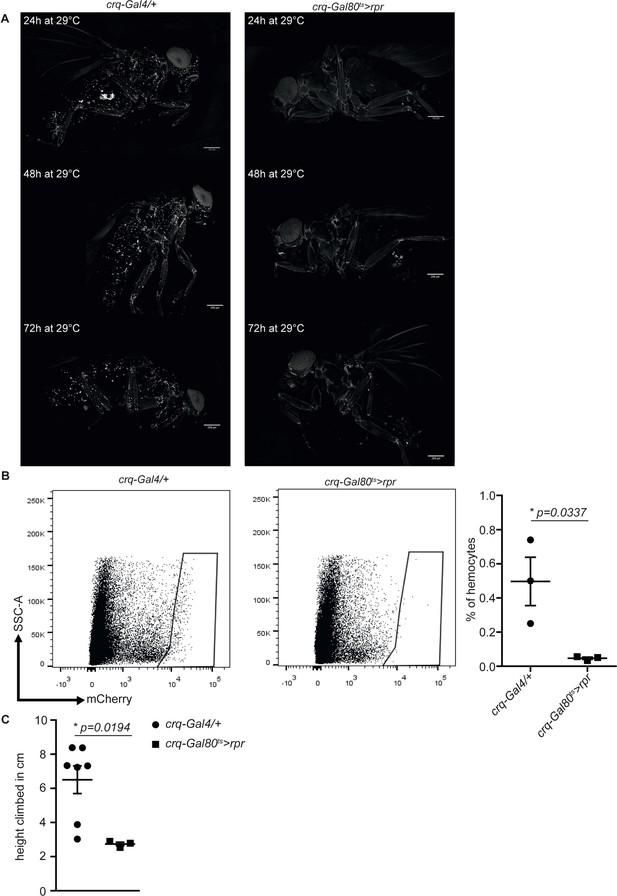
Further characterisation of plasmatocyte-depleted flies.
(A) Plasmatocyte depletion in crq-Gal80ts > rpr flies (right) and controls (left) at 24 hr, 48 hr and 72 hr at 29°C. One out of four flies is shown. Scale bar = 250 µm. (B) Quantification of mCherry-positive plasmatocytes in crq-Gal80ts > rpr flies (right) and controls (left) by flow cytometry after 72 hr at 29°C. n = 3 per group was analysed. Unpaired T-Test: *p=0.0337. (C) Negative geotaxis assay of 14-day-old crq-Gal80ts/+, crq-Gal80ts > rpr, upd2∆upd3∆;UAS-rpr/+ and upd2∆upd3∆;crq-Gal80ts > rpr flies. Points represent mean climbing height of individual vials analysed (~20 flies/vial). Unpaired T-Test: *p=0.0194.
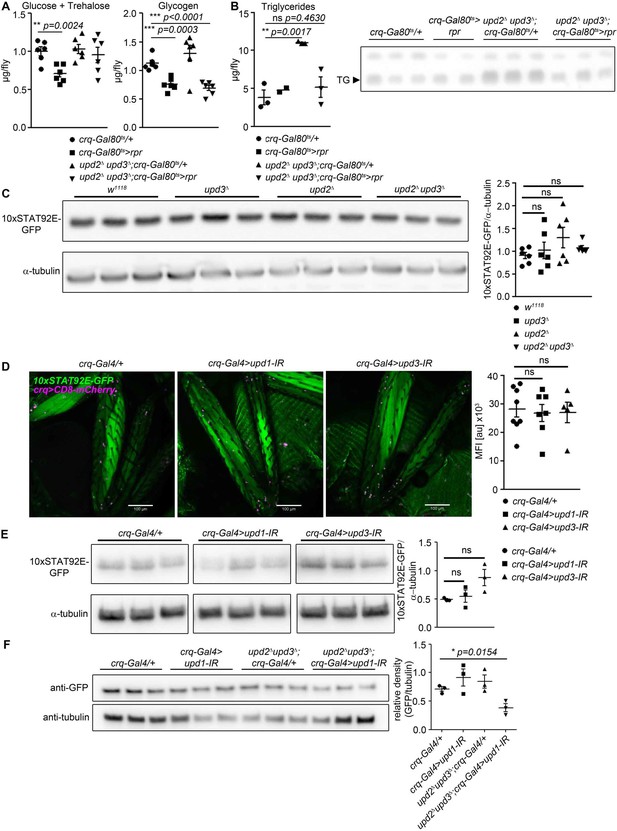
Further characterisation of requirements for specific Upds.
(A) Glucose + trehalose and glycogen in 7-day-old crq-Gal80ts/+, crq-Gal80ts > rpr, upd2Δ upd3Δ;crq-Gal80ts/+, and upd2Δ upd3Δ; crq-Gal80ts > rpr flies. (B) TLC of triglyceride in 7-day-old crq-Gal80ts/+, crq-Gal80ts > rpr, upd2Δ upd3Δ;crq-Gal80ts/+, and upd2Δ upd3Δ;crq-Gal80ts > rpr flies, n = 2–3 samples per genotype. (C) Western blot analysis of STAT-driven GFP in legs from 7-day-old w1118, upd3Δ, upd2Δ, and upd2Δ upd3Δ flies. One representative experiment of two is shown. (D) STAT activity and plasmatocytes in legs from 7-day-old control (crq-Gal4/+), upd1 knockdown (crq-Gal4 >upd1 IR), and upd3 knockdown (crq-Gal4 >upd3 IR) flies. One representative fly is shown of 5–7 imaged for each genotype. Scale bar = 100 µm. Mean fluorescence intensity (MFI) is shown for all flies imaged. (E) Western blot analysis of STAT-driven GFP in legs from 7-day-old control (crq-Gal4/+), upd1 knockdown (crq-Gal4 >upd1 IR), and upd3 knockdown (crq-Gal4 >upd3 IR) flies. One of two independent experiments is shown. (F) Western blot analysis of STAT-driven GFP in thorax from 7-day-old crq-Gal4/+, crq-Gal4 >upd1, upd2Δupd3Δ;crq-Gal4/+ and upd2Δupd3Δ;crq-Gal4 >upd1 IR flies. P values in A-F from unpaired T-test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | w1118; tubulin-Gal80ts/SM6a;24B-Gal4/TM6c, Sb1 | This study | Inserted Elements: P[w[+mC]=tubP-GAL80[ts]]; P[GawB]how24B | |
| Genetic reagent (D. melanogaster) | w1118; tubulin-Gal80ts/SM6a;Mef2-Gal4/TM6c, Sb1 | This study | Inserted Elements: P[w[+mC]=tubP-GAL80[ts]]; P[GAL4-Mef2.R]3 | |
| Genetic reagent (D. melanogaster) | w1118;;UAS-dome∆/TM6c, Sb1 | Brown et al., 2001 | Gift of James Castelli-Gair Hombría | |
| Genetic reagent (D. melanogaster) | w1118;UAS-dome∆/CyO | Brown et al., 2001 | Gift of James Castelli-Gair Hombría | |
| Genetic reagent (D. melanogaster) | w1118;;UAS-myr-AKT/TM6c, Sb1 | Stocker et al., 2002 | Gift of Ernst Hafen | |
| Genetic reagent (D. melanogaster) | w;UAS-AMPKα-IR | Vienna Drosophila Research Center (VDRC) | RRID:FlyBase_FBst0478025; VDRC 106200 | |
| Genetic reagent (D. melanogaster) | w;UAS-AMPKβ-IR | VDRC | RRID:FlyBase_FBst0476347; VDRC 104489 | |
| Genetic reagent (D. melanogaster) | w;UAS-rl-IR | VDRC | RRID:FlyBase_FBst0480887; VDRC 109108 | |
| Genetic reagent (D. melanogaster) | w;UAS-Dsor1-IR | VDRC | RRID:FlyBase_FBst0479098; VDRC 107276 | |
| Genetic reagent (D. melanogaster) | w1118;foxoGFP | BDSC | RRID:BDSC_38644 | Inserted Element: PBac[foxo-GFP.FLAG]VK00037 |
| Genetic reagent (D. melanogaster) | w;UAS-AKT-IR | VDRC | RRID:FlyBase_FBst0475561; VDRC 103703 | |
| Genetic reagent (D. melanogaster) | w1118;10xSTAT92E-GFP | BDSC Bach et al., 2007 | RRID:BDSC_26197 | Inserted Element: P[10XStat92E-GFP]1 |
| Genetic reagent (D. melanogaster) | w1118;MHC-Gal4,MHC-RFP/SM6a | BDSC | RRID:BDSC_38464 | Inserted Element: P[Mhc-GAL4.F3-580]2; P[Mhc-RFP.F3-580]2 |
| Genetic reagent (D. melanogaster) | w upd2 Δ upd3Δ;;; | BDSC | RRID:BDSC_55729 | |
| Genetic reagent (D. melanogaster) | w1118;;crq-Gal4/TM6 c, Sb1 | Gift of Nathalie Franc | ||
| Genetic reagent (D. melanogaster) | w1118;tub-Gal80ts;TM2/TM6 c, Sb1 | BDSC | RRID:BDSC_7108 | |
| Genetic reagent (D. melanogaster) | w1118;;UAS-rpr/TM6 c, Sb1 | BDSC | RRID:BDSC_5824 | |
| Genetic reagent (D. melanogaster) | w1118;UAS-CD8-mCherry | BDSC | RRID:BDSC_27391 | |
| Genetic reagent (D. melanogaster) | w1118;;srpHemo-3xmCherry/TM6c, Sb1 | Gyoergy et al., 2018 | ||
| Genetic reagent (D. melanogaster) | w;UAS-hop-IR | VDRC | RRID:FlyBase_FBst0463355; VDRC 40037 | |
| Genetic reagent (D. melanogaster) | w;UAS-upd1-IR/SM6a | VDRC | RRID:FlyBase_FBst0459787; VDRC 3282 | |
| Genetic reagent (D. melanogaster) | w;UAS-upd3-IR | VDRC | RRID:FlyBase_FBst0456774; VDRC 27134 | |
| Genetic reagent (D. melanogaster) | w1118;;UAS-upd3/TM6 c, Sb1 | Gift of Bruce Edgar | ||
| Genetic reagent (D. melanogaster) | w1118;UAS-2xeGFP/SM6a | BDSC | RRID:BDSC_6874 | |
| Genetic reagent (D. melanogaster) | w1118 hopTum-L/FM7h | BDSC | RRID:BDSC_8492 | backcrossed onto w1118 background |
| Sequence-based reagent | Akt1_forward | This study | PCR primers | 5’-ctttgcgagtattaactggacaga-3’ |
| Sequence-based reagent | Akt1_reverse | This study | PCR primers | 5’-ggatgtcacctgaggcttg-3’ |
| Sequence-based reagent | Ilp2_forward | This study | PCR primers | 5’-atcccgtgattccaccacaag-3’ |
| Sequence-based reagent | Ilp2_reverse | This study | PCR primers | 5’-gcggttccgatatcgagtta-3’ |
| Sequence-based reagent | Ilp3_forward | This study | PCR primers | 5’-caacgcaatgaccaagagaa-3’ |
| Sequence-based reagent | Ilp3_reverse | This study | PCR primers | 5’-tgagcatctgaaccgaact-3’ |
| Sequence-based reagent | Ilp4_forward | This paper | PCR primers | 5’-gagcctgattagactgggactg-3’ |
| Sequence-based reagent | Ilp4_reverse | This paper | PCR primers | 5’-tggaccggctgcagtaac-3’ |
| Sequence-based reagent | Ilp5_forward | This paper | PCR primers | 5’-gccttgatggacatgctga-3’ |
| Sequence-based reagent | Ilp5_reverse | This paper | PCR primers | 5’-agctatccaaatccgcca-3’ |
| Sequence-based reagent | Ilp6_forward | This paper | PCR primers | 5’-cccttggcgatgtatttcc-3’ |
| Sequence-based reagent | Ilp6_reverse | This paper | PCR primers | 5’-cacaaatcggttacgttctgc-3’ |
| Sequence-based reagent | Ilp7_forward | This paper | PCR primers | 5’-cacaccgaggagggtctc-3’ |
| Sequence-based reagent | Ilp7_reverse | This paper | PCR primers | 5’-caatatagctggcggacca-3’ |
| Sequence-based reagent | dome_forward | This paper | PCR primers | 5’-cggactttcggtactccatc-3’ |
| Sequence-based reagent | dome_reverse | This paper | PCR primers | 5’-accttgatgaggccaggat-3’ |
| Sequence-based reagent | upd1_forward | This paper | PCR primers | 5’-gcacactgatttcgatacgg-3’ |
| Sequence-based reagent | upd1_reverse | This paper | PCR primers | 5’- ctgccgtggtgctgtttt −3’ |
| Sequence-based reagent | upd2_forward | This paper | PCR primers | 5’-cggaacatcacgatgagcgaat-3’ |
| Sequence-based reagent | upd2_reverse | This paper | PCR primers | 5’-tcggcaggaacttgtactcg-3’ |
| Sequence-based reagent | upd3_forward | This paper | PCR primers | 5’-actgggagaacacctgcaat-3’ |
| Sequence-based reagent | upd3_reverse | This paper | PCR primers | 5’-gcccgtttggttctgtagat-3’ |
| Sequence-based reagent | Pepck_forward | This paper | PCR primers | 5’-ggataaggtggacgtgaag-3’ |
| Sequence-based reagent | Pepck_reverse | This paper | PCR primers | 5’-acctcctgcgaccagaact-3’ |
| Sequence-based reagent | Thor_forward | This paper | PCR primers | 5’-caggaaggttgtcatctcgga-3’ |
| Sequence-based reagent | Thor_reverse | This paper | PCR primers | 5’-ggagtggtggagtagagggtt-3’ |
| Sequence-based reagent | InR_forward | This paper | PCR primers | 5'-gcaccattataaccggaacc-3' |
| Sequence-based reagent | InR_reverse | This paper | PCR primers | 5'-ttaattcatccatgacgtgagc-3' |
| Sequence-based reagent | Akh_forward | This paper | PCR primers | 5’- agccgtgctcttcatgct-3’ |
| Sequence-based reagent | Akh_reverse | This paper | PCR primers | 5’-aaaggttccaggaccagctc-3’ |
| Sequence-based reagent | Hsl_forward | This paper | PCR primers | 5’-cttggaaatacttgaggggttg-3’ |
| Sequence-based reagent | Hsl_reverse | This paper | PCR primers | 5’-agatttgatgcagttctttgagc-3’ |
| Sequence-based reagent | bmm_forward | This paper | PCR primers | 5’-gtctcctctgcgatttgccat-3’ |
| Sequence-based reagent | bmm_reverse | This paper | PCR primers | 5’-ctgaagggacccagggagta-3’ |
| Sequence-based reagent | plin1_forward | This paper | PCR primers | 5’-gcgttctatggtagccttcag-3’ |
| Sequence-based reagent | plin1_reverse | This paper | PCR primers | 5’-gcgtccggatagaaagctg-3’ |
| Sequence-based reagent | plin2_forward | This paper | PCR primers | 5’-gcagaatggcaagagttctga-3’ |
| Sequence-based reagent | plin2_reverse | This paper | PCR primers | 5’-actgtgtgtaggactggatcctc-3’ |
| Sequence-based reagent | Rpl1_forward | This paper | PCR primers | 5’-tccaccttgaagaagggcta-3’ |
| Sequence-based reagent | Rpl1_reverse | This paper | PCR primers | 5’-ttgcggatctcctcagactt-3’ |
| Peptide, recombinant protein | Trehalase | Sigma Aldrich | T8778 | |
| Peptide, recombinant protein | β-Amyloglucosidase | Sigma Aldrich | 10115 | |
| Antibody | anti-phospho(Ser505)-AKT | Cell Signal Technology (CST) | Cat# 4054; RRID:AB_331414 | WB (1:1000) |
| Antibody | anti-AKT | Cell Signal Technology (CST) | Cat# 4691; RRID:AB_915783 | WB (1:1000) |
| Antibody | anti-phospho(Thr172)-AMPKα | Cell Signal Technology (CST) | Cat# 2535; RRID:AB_331250 | WB (1:1000) |
| Antibody | anti-phospho(Thr389)-p70 S6 kinase | Cell Signal Technology (CST) | Cat# 9206; RRID:AB_2285392 | WB (1:1000) |
| Antibody | anti-GFP | Cell Signal Technology (CST) | Cat# 2956; RRID:AB_1196615 | WB (1:1000) |
| Antibody | anti-phospho-p44/42 MAPK (Erk1/2) | Cell Signal Technology (CST) | Cat# 4370; RRID:AB_2315112 | WB (1:1000) |
| Antibody | anti-α-tubulin | Developmental Studies Hybridoma Bank) | Clone 12G10; RRID:AB_1157911 | WB (1:3000) |
| Antibody | HRP anti-rabbit IgG | Cell Signal Technology (CST) | Cat# 7074; RRID:AB_2099233 | WB (1:5000) |
| Antibody | HRP anti-mouse IgG | Cell Signal Technology (CST) | Cat# 7076; RRID:AB_330924 | WB (1:5000) |
| Commercial assay or kit | First Strand cDNA Synthesis Kit | Thermo Scientific | K1622 | |
| Commercial assay or kit | Sensimix SYBR Green no-ROX | Bioline | QT650-05 | |
| Chemical compound, drug | Bromophenol blue | Sigma Aldrich | SML1656 | |
| Chemical compound, drug | Xylene cyanol | Carl Roth | A513.1 | |
| Chemical compound, drug | Brilliant Blue FCF | Sigma Aldrich | 80717 | |
| Other | HCS Lipid Tox Deep Red | Thermo Fisher | H34477 | IF (1:200) |
| Other | Alexa Fluor 488 Phalloidin | Thermo Fisher | A12379 | IF (1:20) |
| Other | Fluoromount-G | ebioscience | 00-4958-02 | |
| Other | TRIzol | Invitrogen | AM9738 | |
| Other | Fixable Viability Dye 780 | ebioscience | 65-0865-18 | FC (1:1000) |
| Other | Supersignal West Pico Chemiluminescent Substrate | Thermo Scientific | 34077 | |
| Other | Supersignal West Femto Chemiluminescent Substrate | Thermo Scientific | 34094 | |
| Other | Glucose Reagent | Sentinel Diagnostics | 17630H | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | FlowJo | FlowJo | RRID:SCR_008520) |





