Presynaptic developmental plasticity allows robust sparse wiring of the Drosophila mushroom body
Figures
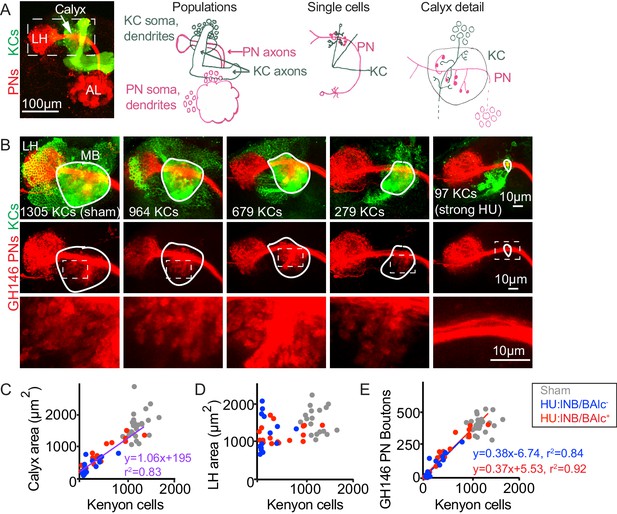
Reduction of Kenyon cells leads to reduced projection neuron innervation of the mushroom body calyx.
(A) Image (left) and models (right) of olfactory PNs and KCs in the adult fly brain. PNs (red) receive input from olfactory sensory neurons in the antennal lobe (AL) and project to the mushroom body calyx and the lateral horn (LH). In the calyx, PNs synapse with KCs (green) in microglomeruli. Dashed box at left indicates region shown in B. (B) Maximum intensity projections of confocal stacks of the brains of adult flies subjected to HU treatment or sham treatment just after larval hatching. PNs labeled red using GH146-Gal4, KCs green using MB247-LexA. White outlines indicate the extent of the MB calyx in each sample; dashed boxes are regions shown magnified below. (C, D) Relationship between number of KCs and calyx (C) or lateral horn (D) maximum cross-sectional area in HU treated (blue, red dots) and sham treated (gray dots) MBs. Treated samples are separated by presence (red) or absence (blue) of lNB/BAlc progeny. Here and throughout, each dot is one hemisphere. Purple trend line in (C) combines all HU-treated samples. (E) Relationship between KC number and GH146+ bouton number in brains of HU- and sham-treated animals.
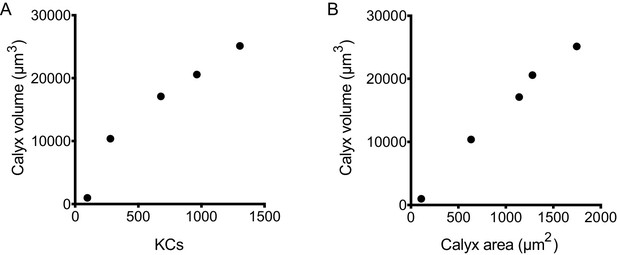
Relationship between KC number, calyx volume, and calyx area.
(A) Correlation between KC number and calyx volume for the samples shown in Figure 1B and Figure 1—video 1. (B) Correlation between calyx maximum cross-sectional area and calyx volume for samples shown in Figure 1B and Figure 1—video 1.
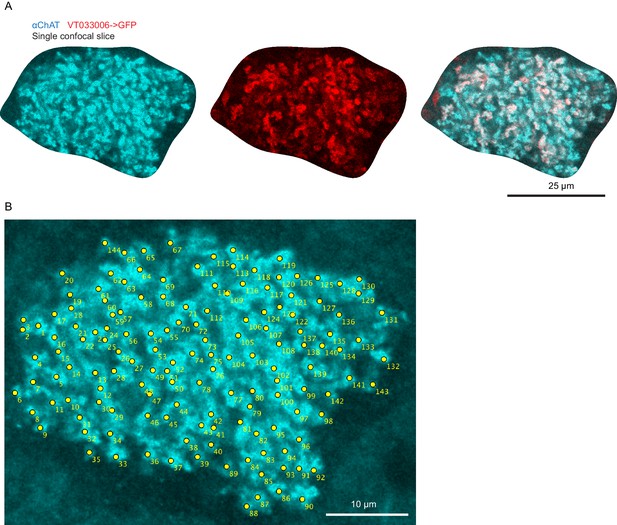
Method of bouton counting.
High-resolution images of calyx boutons (A) and example of bouton counting for a single confocal plane (B). Each yellow dot was scored as a bouton.
Volume renderings of mushroom bodies shown in Figure 1B.
The video cycles through calyces in the order shown in the Figure (1305 KCs, 964 KCs, 679 KCs, 279 KCs, 97 KCs), showing first the full Kenyon cell (GFP) signal (somata, calyx, lobes), and then the isolated calyx of that sample. Red bounding boxes contain the same volume throughout, though the rendering program re-scales certain samples to fill the frame.
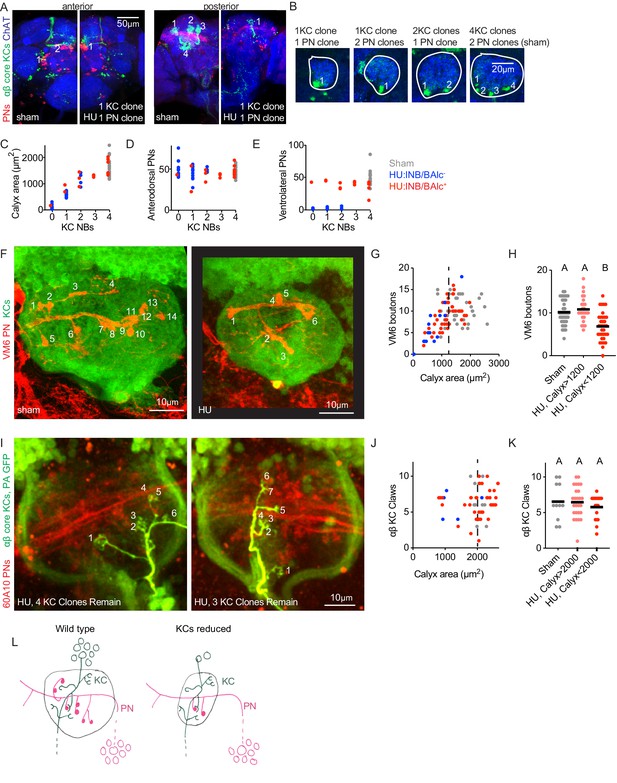
Individual projection neurons scale their bouton repertoire to the Kenyon cell population.
(A) Maximum intensity projections of confocal stacks of the anterior (left) and posterior (right) of HU and sham-treated animals. GH146 labels PNs (red). 58F02 labels late-born αβ ‘core’ KCs (green). Cholinergic neurons labeled with ChAT immunostaining (blue). Numbers indicate PN and KC clones. Anterior and posterior images are of the same brains. (B) Confocal slices of the mushroom body calyx in sham-treated (right) and various HU treated (left) hemispheres. ChAT signal highlights bouton structures, while 58F02 signal allows scoring of the number of KC clones. Numbered neurite bundles innervate the pedunculus when traced through the stack, while green signals not numbered derive from non-KC cell types labeled by 58F02. (C–E) Relationship between KC clone number and maximum calyx cross-sectional area (C), and number of anterodorsal (D) and ventrolateral (E) PNs. (F–H) Example images (F) and quantification (G, H) of MB calyx boutons on the VM6 PN, labeled by 71D09 (red). KCs labeled by MB247 (green). Numbers in (F) indicate counted boutons, and these two calyces are displayed at the same scale. Dashed vertical lines in (G) indicate cutoff between HU-treated calyces similar to controls and those affected by the manipulation. Significance, one-way ANOVA with correction for multiple comparisons. Here and throughout, letters above scatterplots indicate statistically distinguishable versus undistinguishable groups by ANOVA. Black horizontal bars represent medians. (I–K) Example images (I) and quantification (J, K) of GFP photoactivation of individual KCs in HU treated or sham brains. PNs labeled by 60A10 and αβ ‘core’ KCs by 58F02. Examples in (I) show two hemispheres of the same HU treated brain. Note that calyx area is larger in brains imaged ex vivo by two photon (as in I-K) versus fixed, stained, and imaged by confocal (as in F-H). Quantification in (J, K) includes αβ KCs labeled as shown in (I) and those labeled as shown in Figure 2—figure supplement 3; quantification of γ KCs labeled by both methods is shown in Figure 2—figure supplement 3. lNB/BAlc status was scored by examining PN somata near the antennal lobe, as in (A). (L) Model of the effect of KC loss on calyx development. Remaining KCs exhibit wild-type numbers of claws, while PNs reduce their bouton repertoire, reducing the size of the calyx.
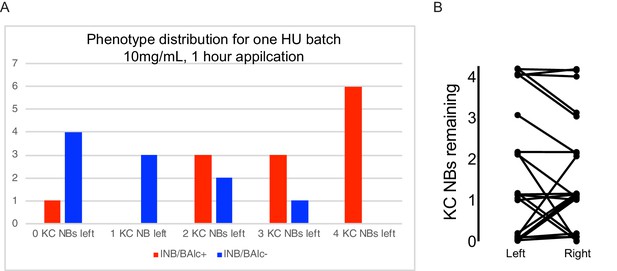
Distribution of HU effects.
(A) Distribution of HU effects for one batch of treated animals. Genotype as in Figure 2A. (B) Relationship of KC neuroblast state across hemispheres. Jitter was added to make each sample apparent.
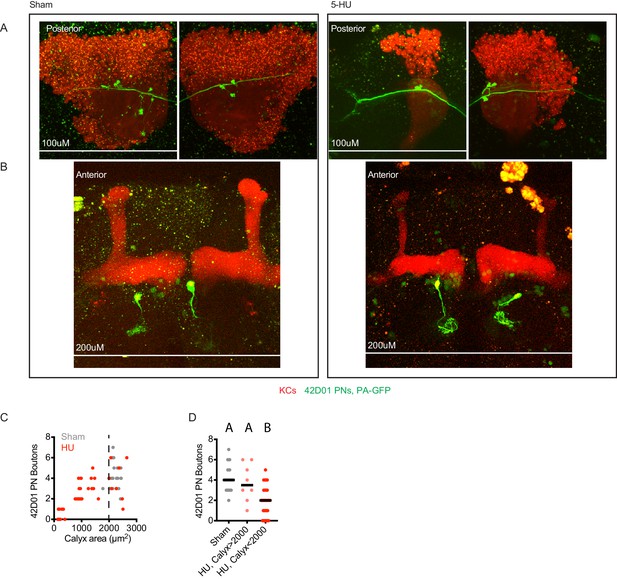
Images and quantification of individual 42D01-labeled PNs highlighted by GFP photoactivation.
Example maximum intensity projections of two photon images (A, B) and quantification (C, D) of PNs labeled by 42D01-Gal4 and highlighted by GFP photoactivation. Each panel shows posterior (A) and anterior (B) images of the same brain, where anterior images show single photoactivated PN somata and glomerular innervation in the antennal lobe, as well as mushroom body lobes, and posterior images show boutons of the same PN in the mushroom body calyx. Speckles are autofluorescence, large yellow blobs are photodamage incurred during imaging. Black horizontal bars in (C,D) represent medians.
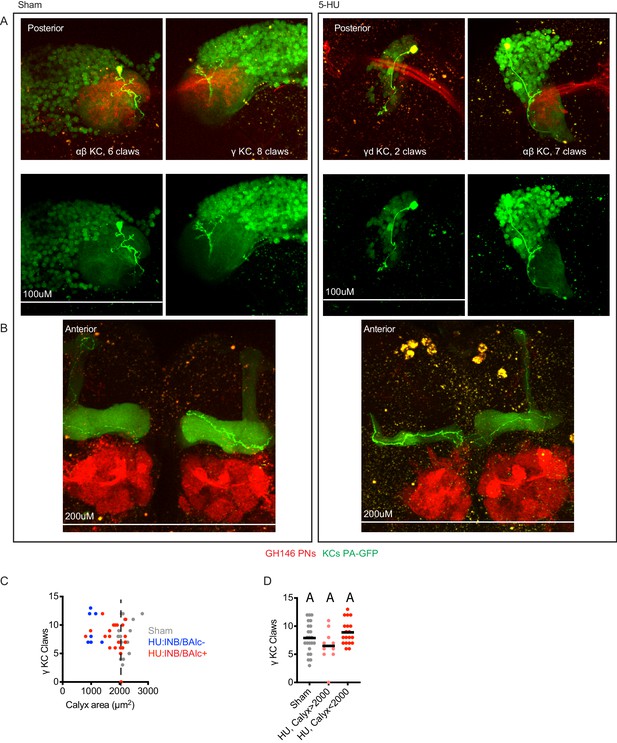
Additional examples and quantification of KCs labeled by GFP photoactivation.
(A, B) Example maximum intensity projections of two photon images of KCs labeled by MB247-LexA and highlighted by GFP photoactivation. Each panel shows posterior (A) and anterior (B) images of a single brain, where anterior images show KC innervation of the mushroom body axonal lobes, as well as the antennal lobe, and posterior images show claws of the same KC in the mushroom body calyx, as well as PN boutons. Axonal lobe innervation allowed us to assign KC type, as noted. Speckles are autofluorescence, large yellow blobs are photodamage incurred during imaging. The left hemisphere of the HU-treated sample is fully ablated and lacks lNB/BAlc progeny, while the right hemisphere is partially ablated and retains lNB/BAlc. (C, D) Quantification of claws of γ KCs labeled using the strategy shown here or through expression of PA-GFP only in γ KCs, driven by 89B01-Gal4. lNB/BAlc status scored by examining GH146-Gal4 or 60A10-LexA, as shown in Figure 2A. Black horizontal bars represent medians.
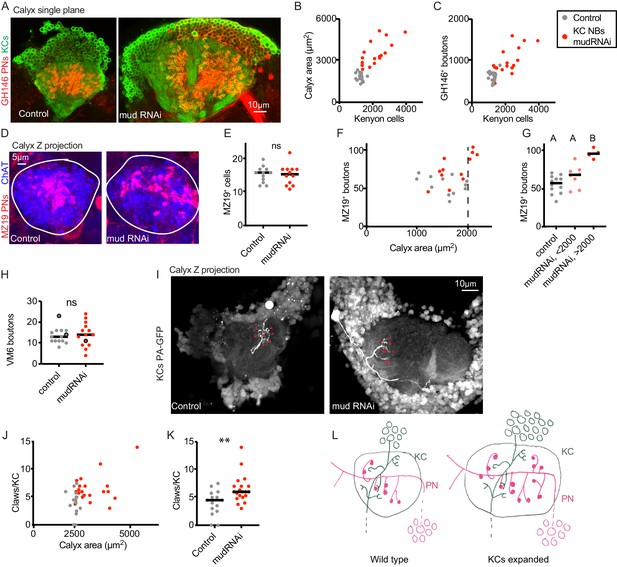
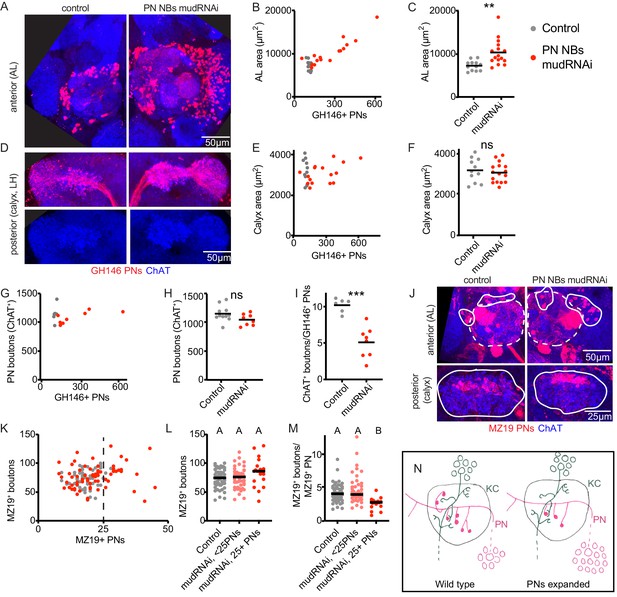
Increase in Kenyon cell number leads to increased projection neuron bouton complement.
(A) Confocal slice of control calyx and calyx subjected to KC expansion by expression of mud-RNAi in KC neuroblasts via OK107-Gal4. GH146 labels PNs (red) and OK107 labels KCs (green). (B, C) Relationship between number of KCs and maximum calyx cross-sectional area (B) and PN bouton number (C) in control (gray) and KC neuroblast>mud RNAi hemispheres (red). (D) Maximum intensity projection of confocal stack of calyx bouton production by ~16 MZ19+ PNs (red) in control and KC neuroblast>mud RNAi. ChAT immunostaining (blue) highlights calyx extent. (E) Quantification of MZ19 cells in the two genotypes. Significance: unpaired t-test. Throughout this figure, black horizontal bars represent medians. (F, G) Relationship between maximum calyx cross-sectional area and MZ19 bouton production. Vertical dashed line indicates calyces larger than 2000 μm2,observed only in the experimental genotype. Significance: one-way ANOVA. (H) Boutons of the VM6 PN, labeled by 71D09-LexA, in control (gray) or brains subjected to KC expansion via OK107>mudRNAi (red). Every hemisphere contained one VM6 neuron in both genotypes except the three circled datapoints, which had two VM6 PNs. Significance: unpaired t-test. (I) Maximum intensity projection of two-photon stack of single KCs highlighted by GFP photoactivation (C3PA driven by OK107) in control or OK107>mudRNAi calyces. Position of out-of-plane somata in control image indicated by white circle. Red arrowheads indicate scored claws. (J, K) Quantification of claws of KCs labeled as in (I). Y axis is average number of claws per KC as multiple KCs were photoactivated in some samples. Significance: unpaired t-test; here and throughout, *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001. (L) Model of the effect of KC expansion on calyx development. KCs retain normal claw numbers while PNs increase their bouton numbers, increasing calyx size.
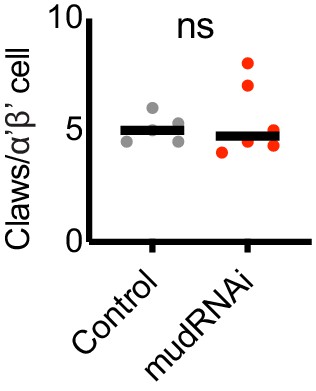
Claw number for α’β’ KCs.
KC claw number in PA-GFP-labeled α’β’ KCs in control or KC-expansion genotype. Significance: unpaired t-test. Black horizonal bars represent medians.
Volume renderings of mushroom bodies shown in Figure 3A.
Using the Kenyon cell (GFP) signal, the video shows the mud-RNAi mushroom body first (3951 KCs), and then the control mushroom body (1227 KCs). For each sample, the full mushroom body is shown first, and then the isolated calyx. Red bounding boxes contain the same volume throughout.
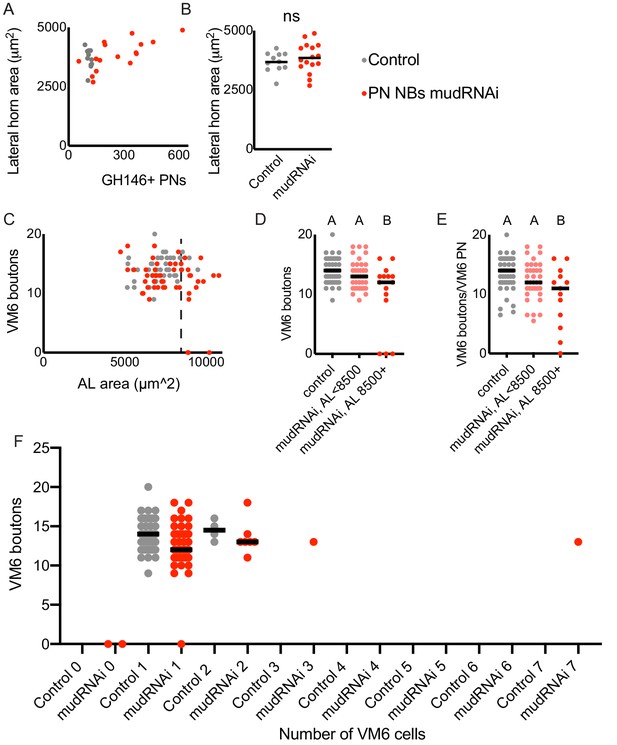
Individual projection neurons scale boutons down as projection neuron population increases.
(A) Maximum intensity projection of confocal stack of anterior of the brain of control or animal subjected to PN expansion by expression of mud-RNAi in PN neuroblasts via 44F03-Gal4. GH146 labels PNs (red) and ChAT staining highlights cholinergic neurons. (B, C) Relationship between PN number (measured by counting GH146+ cells, about 60% of the total PNs) and antennal lobe maximum cross-sectional area in control (gray) or PN neuroblast>mudRNAi hemispheres (red). Significance: unpaired t-test. Throughout this figure, black horizontal bars represent medians. (D) Maximum intensity projection of confocal stacks of the posteriors of the same brains shown in A. Excess PNs can be seen to project to the calyx, due to thickening of the axonal tract passing on top of the calyx (red), and produce increased innervation density in the lateral horn. (E–I) Relationship between PN number and maximum cross-sectional area of the calyx (E, F), PN bouton number (G, H), and ratio of ChAT+ boutons to GH146+ PNs (I) in control and PN neuroblast>mudRNAi brains. Significance: unpaired t-test. (J) Maximum-intensity projections of confocal stacks of antennal lobes (top) and calyces (bottom) of control and PN neuroblast>mudRNAi brains. MZ19+ PNs labeled red. Dashed outlines on anterior show antennal lobe extent; solid outline on anterior, groups of PN somata; solid outlines on posterior, calyx extent. (K–M) Quantification of MZ19+ boutons in control and PN neuroblast>mudRNAi brains. Dashed vertical line indicates 25 MZ19+ PNs, as controls seldom exceed this number. Significance: one-way ANOVA. (N) Model of the effect of increasing PN numbers on calyx development. As PNs increase, PNs reduce bouton production so that the overall number of boutons, and calyx size, stays similar to wild type.
Additional quantification related to Figure 4.
(A, B) Relationship between lateral horn maximum cross-sectional area and PN number in control (gray) and PN neuroblast>mudRNAi (red) brains. Significance: unpaired t-test. Throughout this figure, black horizontal bars represent medians. (C–E) Relationship between antennal lobe maximum cross-sectional area (proxy for PN number) and production of calyx boutons by the VM6 PN, labeled by 71D09. Dashed line indicates 8500 μm2, as antennal lobes larger than this were observed rarely in the control genotype. Significance: one-way ANOVA. In two experimental hemispheres, we observed no VM6 PN. In one hemisphere, we observed one VM6 PN with no boutons in the calyx. (F) VM6 boutons in calyces of brains containing different numbers of VM6 PNs. We observed more than one VM6 PN in 4/50 control and 9/50 PN neuroblast>mudRNAi hemispheres.
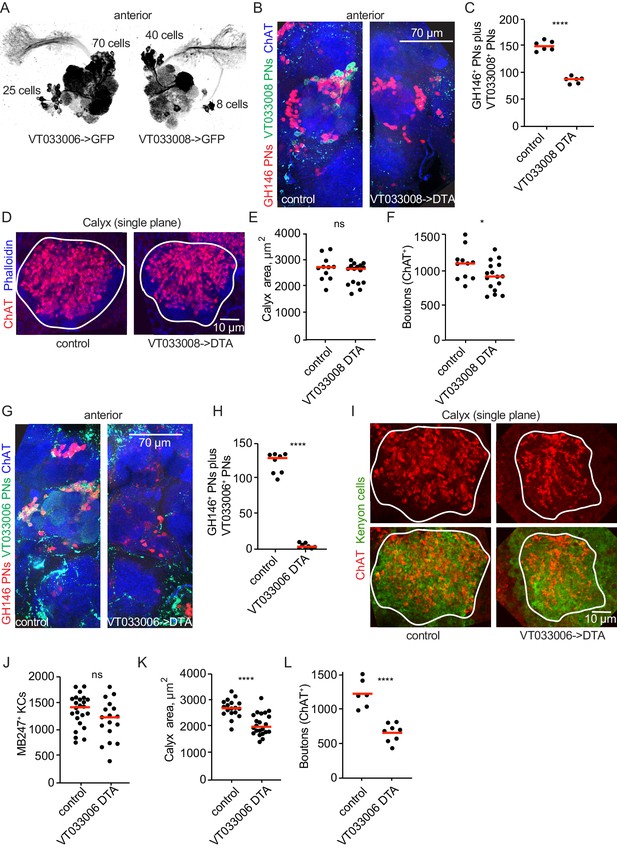
Projection neurons partially compensate for decreases in projection neuron population.
(A) Maximum-intensity projections of two-photon stacks of brains with VT033006 (left) and VT033008 (right) driving GFP. (B) Maximum intensity projection of confocal stack of antennal lobes of control (left) and VT033008>DTA (right) brains. VT033008+ cells shown in green, GH146+ PNs, which partially overlap with VT033008+ PNs, shown in red. (C) Quantitation of PNs positive for GH146, VT033008, or both in control and VT033008>DTA brains. All VT033008+ cells are ablated in this genotype. Significance: unpaired t-test. Throughout this figure, red horizonal bars represent medians. (D) Single confocal slice of control or VT033008>DTA calyx. Phalloidin signal shown in blue and ChAT+ boutons shown in red. (E, F) Quantification of calyx maximum cross-sectional area and bouton number in control and VT033008>DTA animals. Significance: unpaired t-test. (G) Maximum intensity projection of confocal stack of antennal lobes of control (left) and VT033006>DTA (right) brains. VT033006+ cells shown in green, GH146+ PNs, which partially overlap with VT033006+ PNs, shown in red. Green signals outside the antennal lobe in both conditions are autofluorescence. (H) Quantitation of PNs positive for GH146, VT033006, or both in control and VT033006>DTA brains. All VT033006+ cells are ablated in this genotype as well as most GH146+ PNs, even those that are not VT033006+ in the adult. Examination of ChAT+ neurites entering the calyx in VT033006>DTA brains suggests that up to 10 PNs may be labeled by neither VT033006 nor GH146, and that some of these may survive ablation. Significance: unpaired t-test. (I) Single confocal slice of control (left) or VT033006>DTA (right) calyx. KCs labeled using MB247-dsRed (shown green for continuity), ChAT+ boutons shown in red. (J–L) Quantification of the number of MB247+ KCs (J), calyx area (K), and ChAT+ boutons (L) in control or VT033006>DTA brains. Significance: unpaired t-test.
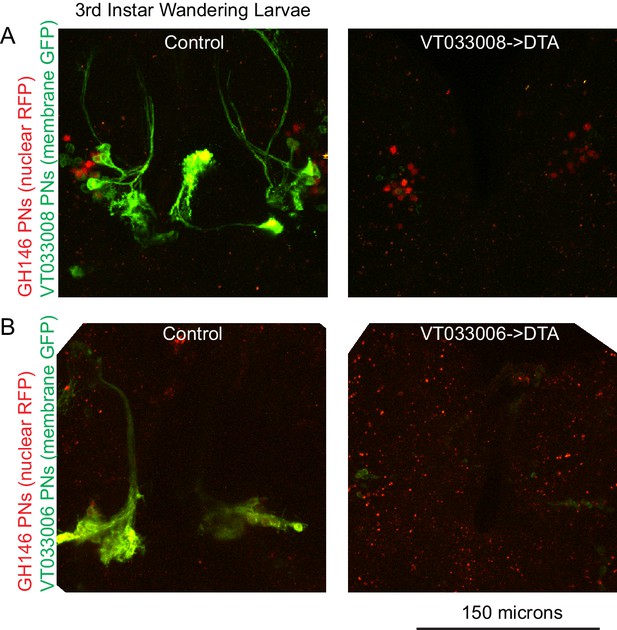
VT033006 and VT033008 action in late larval stage.
Maximum intensity projections of two photon stacks showing expression of VT033008-Gal4 (A) and VT033006-Gal4 (B) in third instar wandering larvae. Labeled cells are lost when these drivers are used to express DTA. The two samples in (A) were imaged and displayed using the same settings as one another, and the two samples in (B) were imaged and displayed using the same settings as one another. Red speckles in (B) are autofluorescence; GH146 signal is weak but detectable in nuclei of VT033006+ PNs in control but is lost in DTA samples. n = 3–5 per genotype.
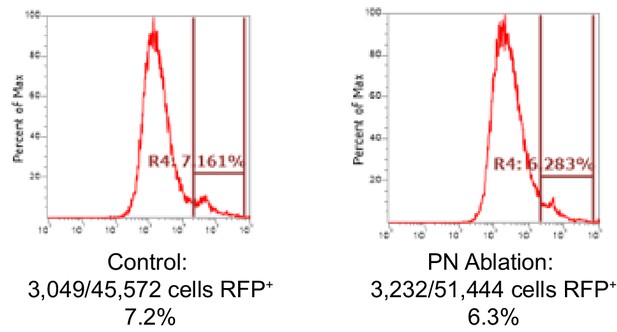
Quantification of Kenyon cells by flow cytometry.
Number of KCs (MB247-dsRed, RFP+) detected in three pooled control or three pooled VT033006>DTA brains dissociated and subjected to flow cytometry. Plots depict cells already gated as singlets positive for a DNA dye, dyeCycle Violet. Representative of two experiments.
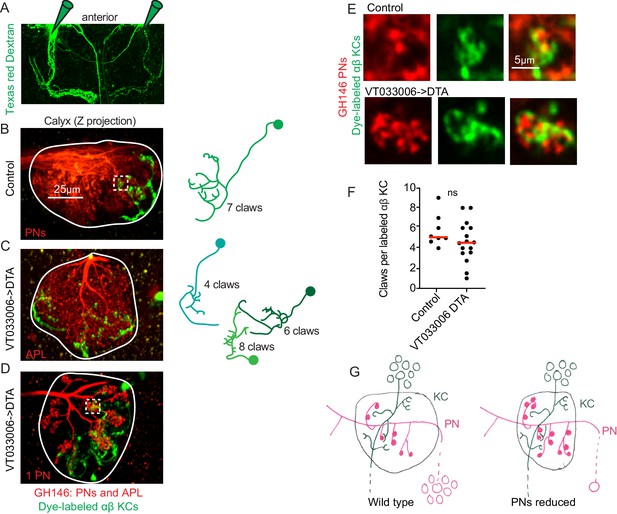
Kenyon cell claw number is unaffected by projection neuron bouton reduction.
(A) Texas red dextran (colored green for continuity with green KCs in other figures) was electroporated into the tips of the mushroom body α lobes to allow specific targeting of αβ KCs. (B–D) Maximum intensity projections of two photon stacks of control (B) and VT033006>DTA (C, D) calyces. GH146, shown in red, labels PNs and APL. In control (B), APL is obscured by dense PN signal. In some VT033006>DTA hemispheres, as in (C), all GH146+ PNs were ablated, making APL signal apparent. In other VT033006>DTA hemispheres, as in (D), 1–2 GH146+ PNs remained. Texas red dextran dye diffusing from KC axons is shown in green. Diagrams show reconstructed anatomies of dye-labeled KCs. In (B), one KC with seven claws is labeled. In C, three KCs with 4, 6, and eight claws are labeled and can be spatially separated from one another. In D, 8 KCs are labeled and cannot be reconstructed. White boxes in (B) and (D) show regions magnified in (E). (E) Texas red dextran dye-labeled KC claws (green) wrapping GH146+ PN boutons (red) in control (top) and VT033006>DTA brains. (F) Quantification of claws per KC in control or VT033006>DTA samples. Each data point is the average number of claws per labeled KC for a spatially separated individual KC or small group of KCs. Significance: unpaired t-test. Red horizontal bars represent medians. (G) Model of the effect on calyx development of ablating large groups of PNs. Remaining PNs increase their production of boutons but cannot fully compensate for lost PNs, reducing the size of the calyx. KC claw numbers are similar to wild type; their source of innervation is unknown.
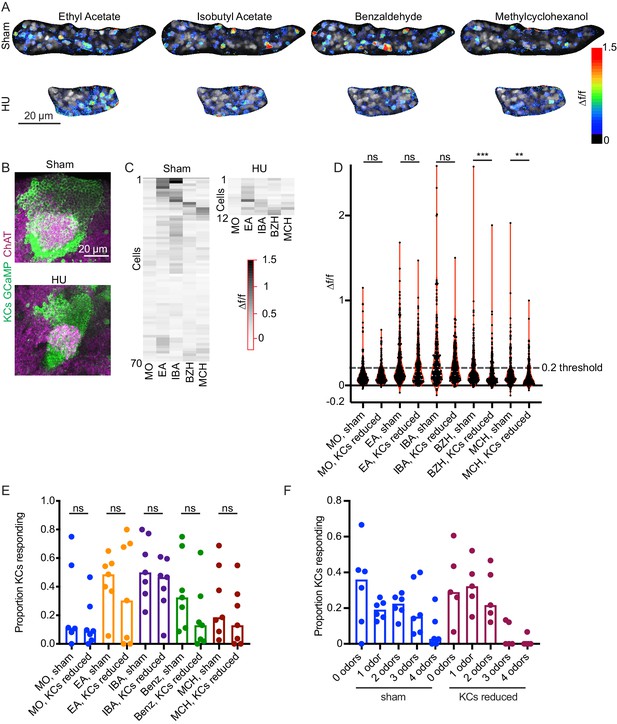
Odor responses in calyces with reduced KCs.
(A) Example KC somatic odor responses in sham-treated hemisphere and HU-treated hemisphere with reduced KCs. (B) Post-imaging immunostaining showing calyces and KC somata of samples shown in (A). (C) Peak post-odor responses of individual KCs from samples shown in (A). Only cells that remained stable in their ROI for the whole stimulus panel are shown. Data was clustered by row such that cells with more similar odor responses are closer to one another. MO: Mineral oil (mechanosensory control), EA: Ethyl Acetate, IBA: Isobutyl Acetate, BZH: Benzaldehyde, MCH: Methylcyclohexanol. (D) Peak odor responses of all cells, aggregated from all analyzed samples. Dashed line indicates 20% Δf/f threshold. Significance: Mann-Whitney test. (E) Fraction of cells in each sample responding to each odor at 20% Δf/f threshold. Significance: unpaired t-test. (F) For samples in which the same cells could be tracked across all odor presentations, fraction of cells responding to 0, 1, or multiple odors is shown. Bar plots in (E, F) show medians.
-
Figure 7—source data 1
Peak response data for all animals in Figure 7.
- https://cdn.elifesciences.org/articles/52278/elife-52278-fig7-data1-v2.xlsx
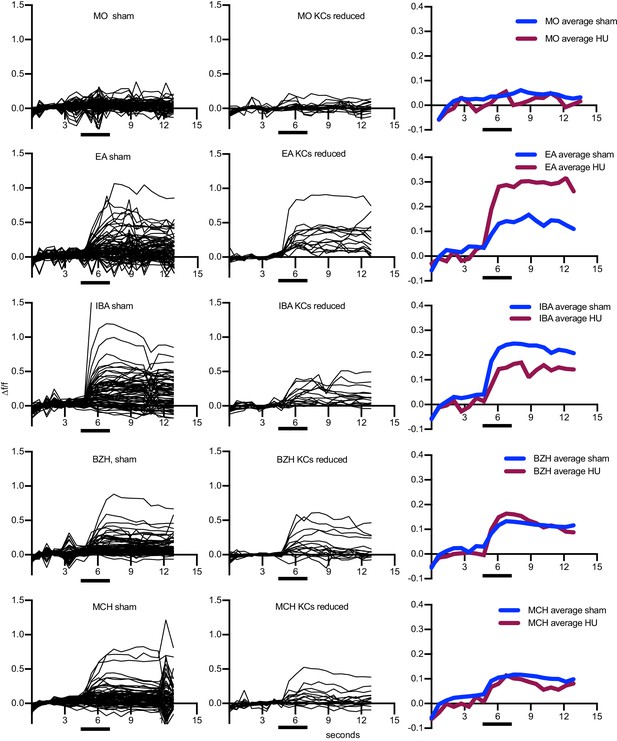
Odor responses over time for cells shown in Figure 7A–C.
x axes show seconds, y axes Δf/f. Black bars indicate odor delivery. Each black line is one cell, with graphs at right showing responses averaged across all cells of the sample. Each cell was normalized to average fluorescence in the 3 s period before stimulus onset. MO: Mineral oil (mechanosensory control), BZH: Benzaldehyde, EA: Ethyl Acetate, IBA: Isobutyl Acetate, MCH: Methylcyclohexanol. An example of a motion artifact can be seen in ‘MCH sham,’ around 12 s.
Full, motion-corrected functional imaging time series for one sham-treated calyx.
Data was collected at 5 Hz, and the series extends over the five minute presentation of four odors and mineral oil control. This is a different sample than shown in Figure 7A–C and Figure 7—figure supplement 1.The intermittently very bright cell in Figure 7—video 2 does not show any correlation with odor presentation and may be damaged.
Full, motion-corrected functional imaging time series for one reduced-KC calyx.
Data was collected at 5 Hz, and the series extends over the five minute presentation of four odors and mineral oil control. This is a different sample than shown in Figure 7A–C and Figure 7—figure supplement 1. The intermittently very bright cell does not show any correlation with odor presentation and may be damaged.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | mud | FLYB: FBgn0002873 | ||
| Genetic reagent (Drosophila melanogaster) | GH146Gal4 (II) | Bloomington Drosophila Stock Center | BDSC 30026 | (Stocker et al., 1997) |
| Genetic reagent (Drosophila melanogaster) | 10XUAS-IVS-myr::tdTomato (attp40) | Bloomington Drosophila Stock Center | BCSC 32222 | (Pfeiffer et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | MB247-LexAVP16 (III) | Scott Waddell, University of Oxford | (Pitman et al., 2011) | |
| Genetic reagent (Drosophila melanogaster) | LexAop-CD2-GFP (III) | Bloomington Drosophila Stock Center | BDSC 66544 | (Lai and Lee, 2006) |
| Genetic reagent (Drosophila melanogaster) | GH146LexA (II) | Tzumin Lee, Janelia Farms Research Campus | (Lai et al., 2008) | |
| Genetic reagent (Drosophila melanogaster) | lexAop-mRFP-nls (II) | Bloomington Drosophila Stock Center | BDSC 29956 | Gunter Merdes |
| Genetic reagent (Drosophila melanogaster) | 58F02-Gal4 (attp2) | Bloomington Drosophila Stock Center | BDSC 39186 | (Jenett et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | UAS-GCaMP6s (attp40) | Bloomington Drosophila Stock Center | BDSC 42746 | (Akerboom et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | UAS-GCaMP6s (VK0005) | Bloomington Drosophila Stock Center | BDSC 42749 | (Akerboom et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | MB247-DsRed (II) | André Fiala | (Riemensperger et al., 2005) | |
| Genetic reagent (Drosophila melanogaster) | 71D09-Gal4 (attp2) | Bloomington Drosophila Stock Center | BDSC 39584 | (Jenett et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | UAS-CD8GFP (III) | Bloomington Drosophila Stock Center | BDSC 5130 | (Lee and Luo, 1999) |
| Genetic reagent (Drosophila melanogaster) | 42D01-Gal4 (attp2) | Bloomington Drosophila Stock Center | BDSC 50152 | (Jenett et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | UAS-C3PA (II) | Vanessa Ruta, Rockefeller University | (Ruta et al., 2010) | |
| Genetic reagent (Drosophila melanogaster) | UAS-C3PA (III) | Vanessa Ruta, Rockefeller University | (Ruta et al., 2010) | |
| Genetic reagent (Drosophila melanogaster) | 60A10-LexA (attp40) | Bloomington Drosophila Stock Center | BDSC 52821 | Pfeiffer, Rubin LexA Collection |
| Genetic reagent (Drosophila melanogaster) | LexAop-tdTomato.Myr (su(Hw)attp5) | Bloomington Drosophila Stock Center | BDSC 56142 | (Chen et al., 2014) |
| Genetic reagent (Drosophila melanogaster) | 89B01-Gal4 (attp2) | Bloomington Drosophila Stock Center | BDSC 40541 | (Jenett et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | 13XLexAop2-IVS-Syn21-mC3PA-GFP-p10 (attp2) | Gerry Rubin, Janelia Farms Research Campus | (Pfeiffer et al., 2012) | |
| Genetic reagent (Drosophila melanogaster) | GH146QF, QUAS-mtdTomato-3xHA (II) | Bloomington Drosophila Stock Center | BDSC 30037 | (Potter et al., 2010) |
| Genetic reagent (Drosophila melanogaster) | UAS-mud-RNAi (attp2) | Bloomington Drosophila Stock Center | BDSC 35044 | (Perkins et al., 2015) |
| Genetic reagent (Drosophila melanogaster) | OK107Gal4 (IV) | Bloomington Drosophila Stock Center | BDSC 854 | (Connolly et al., 1996) |
| Genetic reagent (Drosophila melanogaster) | P(caryP) (attp2) | Bloomington Drosophila Stock Center | BDSC 36303 | (Perkins et al., 2015) |
| Genetic reagent (Drosophila melanogaster) | P(caryP) (attp40) | Bloomington Drosophila Stock Center | BDSC 36304 | (Perkins et al., 2015) |
| Genetic reagent (Drosophila melanogaster) | MZ19QF (II) | Bloomington Drosophila Stock Center | BDSC 41573 | (Hong et al., 2012) |
| Genetic reagent (Drosophila melanogaster) | QUAS-mtdTomato-3xHA (II) | Bloomington Drosophila Stock Center | BDSC 30004 | (Potter et al., 2010) |
| Genetic reagent (Drosophila melanogaster) | 41C07-LexA (attp40) | Bloomington Drosophila Stock Center | BDSC 54791 | Pfeiffer, Rubin LexA Collection |
| Genetic reagent (Drosophila melanogaster) | 89B01-LexA (attp40) | Bloomington Drosophila Stock Center | BDSC 54380 | Pfeiffer, Rubin LexA Collection |
| Genetic reagent (Drosophila melanogaster) | 71D09-LexA (attp40) | Bloomington Drosophila Stock Center | BDSC 54931 | Pfeiffer, Rubin LexA Collection |
| Genetic reagent (Drosophila melanogaster) | LexAop-SPA-T2A-SPA (II) | n/a | (Clowney et al., 2015) | |
| Genetic reagent (Drosophila melanogaster) | 44F03-Gal4 (attp2) | Tzumin Lee, Janelia Farms Research Campus | (Awasaki et al., 2014) | |
| Genetic reagent (Drosophila melanogaster) | VT033006-Gal4 (attp2) | Yoshi Aso, Janelia Farms Research Campus | (Kvon et al., 2014) | |
| Genetic reagent (Drosophila melanogaster) | VT033008-Gal4 (attp2) | Yoshi Aso, Janelia Farms Research Campus | (Kvon et al., 2014) | |
| Genetic reagent (Drosophila melanogaster) | UAS-DTA (UAS-Cbbeta\DT-A.I)[18] (II) | Bloomington Drosophila Stock Center | BDSC 25039 | (Han et al., 2000) |
| Antibody | ChAT (mouse monoclonal) | Developmental Studies Hybridoma Bank | 9E10 | (1:200) |
| Antibody | DsRed (rabbit polyclonal) | Clontech | 632496 | (1:500-1:1000) |
| Antibody | GFP (sheep polyclonal) | Bio-Rad | 4745–1051 | (1:250-1:1000) |
| Antibody | GFP (chicken polyclonal) | Gift from Dawen Cai | n/a | (1:5000) |
| Antibody | Alexa 488 anti-mouse (goat polyclonal) | Fisher | A11029 | (1:500) |
| Antibody | Alexa 647 anti-mouse (goat polyclonal) | Fisher | A21236 | (1:500) |
| Antibody | Alexa 488 anti-sheep (donkey polyclonal) | Fisher | A11015 | (1:500) |
| Antibody | Alexa 488 anti-chicken (goat polyclonal) | Fisher | A11039 | (1:500) |
| Antibody | Alexa 568 anti-rabbit (goat polyclonal) | Fisher | A11036 | (1:500) |
| Chemical compound, drug | Texas Red Dextran | Fisher | D3328 | |
| Chemical compound, drug | Alexa 568 Phalloidin | Fisher | A12380 | |
| Chemical compound, drug | Schneider’s medium | Sigma | S0146 | |
| Chemical compound, drug | Paraformaldehyde | EMS | 15710 | |
| Chemical compound, drug | Hydroxyurea | Sigma | H8627 | |
| Chemical compound, drug | Benzaldehyde | Sigma | 418099 | |
| Chemical compound, drug | Isobutyl acetate | Sigma | 537470 | |
| Chemical compound, drug | Ethyl acetate | Sigma | 270989 | |
| Chemical compound, drug | Methylcyclohexanol | Sigma | 153095 | |
| Peptide, recombinant protein | Collagenase | Sigma | C0130 | |
| Software, algorithm | Suite2p | (Pachitariu et al., 2017) | ||
| Other | Duck EZ Start Packaging tape | Office Depot | 511879 | |
| Other | UV Glue | Loctite | 3106 | |
| Other | JAXMAN 365 nm Flashlight | Amazon | B077GPXBK1 | |
| Other | Electra Waxer | Almore | 66000 | |
| Other | Gulfwax Paraffin | Grocery store | C0130 | |
| Other | hair | self |





