Tgfβ signaling is critical for maintenance of the tendon cell fate
Figures
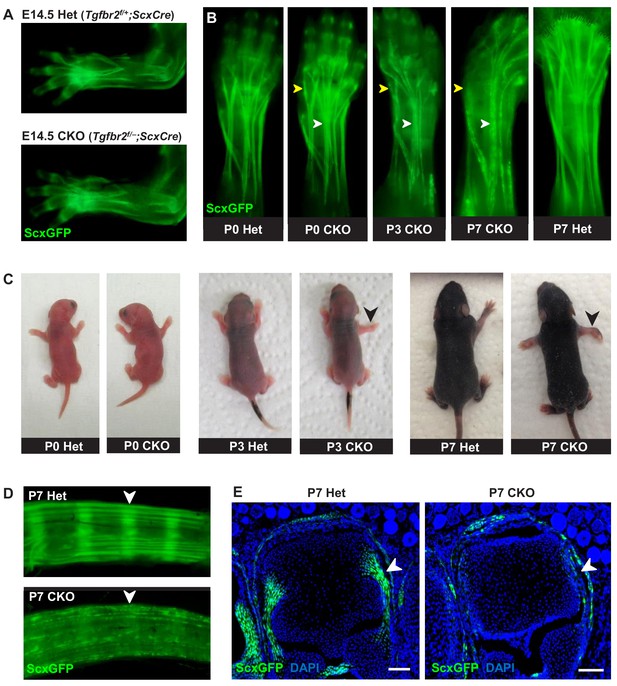
Tendon phenotypes manifested in Tgfbr2;ScxCre mutants.
(A–D) Whole-mount imaging in fluorescent ScxGFP signal or brightfield. (A) Dorsally viewed embryo forelimb shows the formation of a complete network of tendons in both mutant and heterozygous control by E14.5. (B) Tendons of mutant pups appeared intact at birth, but by P3 lateral tendons disintegrated and were eventually eliminated (yellow arrowheads), whereas the majority of other tendons persisted with a substantial loss of the ScxGFP signal (white arrowheads). (C) Mutant pups appeared normal at birth but showed physical abnormalities including abducted paw and splayed limb (black arrowheads) by P3. (D–E) Substantial loss of ScxGFP signal was also detected in all tendons and related tissues. (D) Tail tendons and annulus fibrosus of the intervertebral disc (white arrowheads) in P7 pups. (E) Collateral ligaments of the metacarpophalangeal joint imaged in transverse section through the joints of heterozygous control and mutant pups at P7 (white arrowhead). Scale bar, 100 μm. Mutant: CKO, Heterozygous: Het.
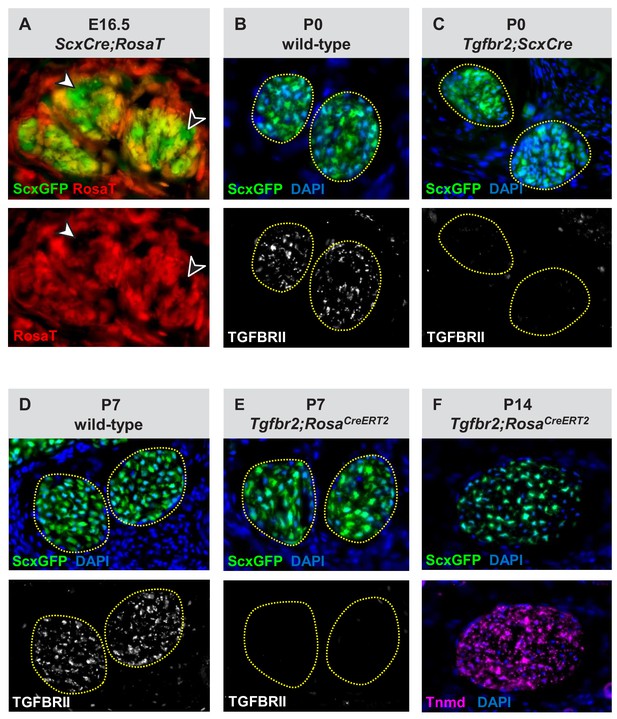
Verification of the Tgfbr2 knockdown efficiency in mutant cells.
(A) Transverse sections through the extensor digitorium communis tendons from an E16.5 ScxCre;RosaT embryo. While some of the tenocytes express the RosaT Cre reporter others do not (arrowheads), reflecting that ScxCre activity is not uniform in embryonic stages. (B–E) Immunostaining for TGFβ type II receptor (TGFBRII) demonstrates a nearly complete loss of receptor expression in (C) Tgfbr2;ScxCre and (E) Tgfbr2;RosaCreERT2 mutant cells, as compared with the robust expression in (B,D) wild-type tendons. (F) Tenocytes in Tgfbr2;RosaCreERT2 mutant retained expression of the tendon markers ScxGFP and tenomodulin (Tnmd), suggesting that a mere loss of TGFβ signaling was not sufficient to cause tenocyte dedifferentiation. All mice also carried the tendon reporter ScxGFP. Dotted lines in (B–E) demarcate tendons. Figures not to scale.
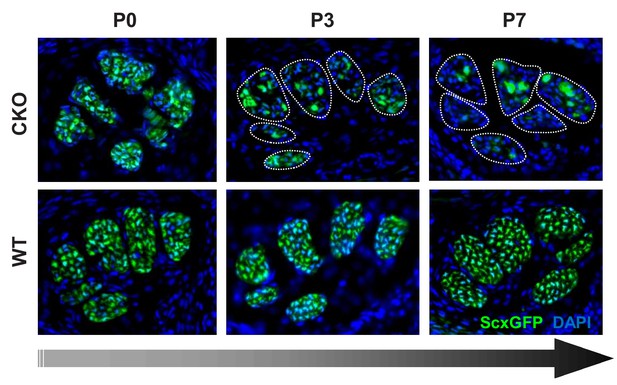
Gradual loss of tendon marker ScxGFP in Tgfbr2;ScxCre mutants at post-natal stages.
Cryosections of the forelimbs of Tgfbr2;ScxCre mutant pups showed positive ScxGFP signal in both wild-type and mutant tendons at P0. A gradual loss of the ScxGFP signal in mutant tendons started around P2-P3, that is before the manifestation of the physical abnormalities in the mutant pups. All mutant tendon cells lost ScxGFP at P7. Most analyses were therefore performed in tendons from this fully-phenotypic stage. Dotted lines demarcate mutant tendons in P3 and P7 pups. Mutant: CKO; Wild-type: WT. Figures not to scale.
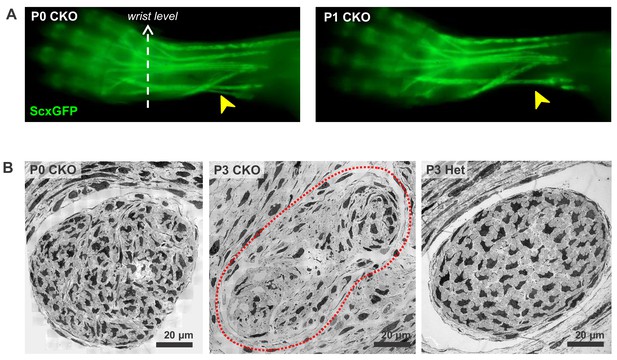
Fragmentation and elimination of lateral tendons in Tgfbr2;ScxCre mutant neonates.
(A) Rapid disruption of lateral extensor tendons in neonatal stages of mutant pups revealed by examination of forelimb tendons using the tendon reporter ScxGFP. The extensor carpi radialis longus tendon (yellow arrowheads) is present in a mutant pup at P0 but lost in a P1 mutant. (B) TEM images of the extensor carpi radialis longus tendon at wrist level. The mutant tendon shows signs of fragmentation already at P0, and by P3 the tendon appears disintegrated accompanied by complete loss of the epitenon and structural definition of the tendon circumference. The red dotted line in (B) demarcates the mutant tendon. Mutant: CKO, Heterozygous: Het.
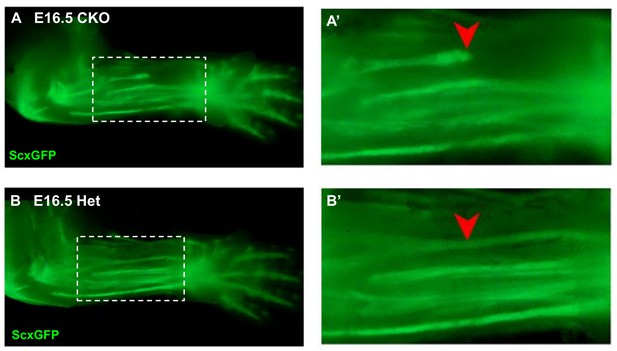
Disruption of the flexor carpi radialis tendon in Tgfbr2;ScxCre mutant embryos.
Examination of flexor tendons in E16.5 (A) mutant and (B) heterozygous littermates using the tendon reporter ScxGFP. Boxed regions in (A) and (B) are shown enlarged in (A’) and (B’). While most tendons appeared normal in mutant embryos, starting at E16.5 the flexor carpi radialis tendon (red arrowheads) was consistently torn in mutant embryo. Mutant: CKO, Heterozygous: Het. Figures not to scale.
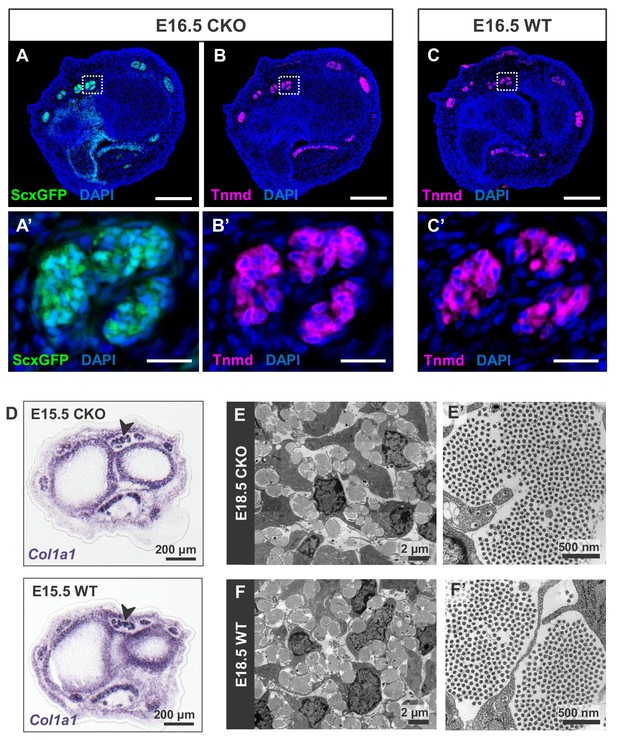
Tendon development in Tgfbr2;ScxCre mutant embryos was not perturbed through embryogenesis.
(A) ScxGFP signal and (B) tenomodulin (Tnmd) immunofluorescence on transverse sections at wrist level of E16.5 mutant embryos demonstrate that the pattern and expression of prototypic tenocyte markers was not disrupted in mutant tendons. (C) Tnmd immunofluorescence in E16.5 wild-type tenocytes. (A’), (B’) and (C’) are higher magnifications of extensor digitorium communis tendons as boxed in (A), (B) and (C). (D) In situ hybridization for Col1a1 on transverse sections of the forelimb from E15.5 mutant and wild-type littermates reveals that expression of the major matrix genes was not altered in mutant embryos (black arrowhead). (E,F) TEM images of tendons from forelimbs of E18.5 mutant and wild-type embryos reveals that organization and accumulation of the tendon extracellular matrix was not disrupted in the mutant. (E’,F’) Higher magnification views of (E) and (F) for direct visualization of the collagen fibers. Scale bars, 200 μm (A–C) and 20 μm (A’–C’). Mutant: CKO, Wild-type: WT.
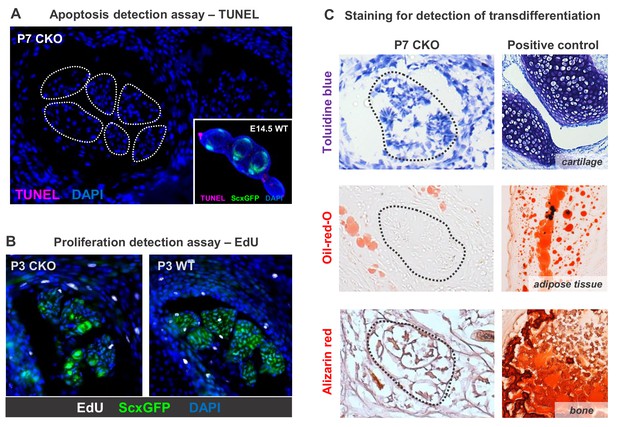
Evaluating cell death, proliferation and transdifferentiation in Tgfbr2;ScxCre mutant tendons.
(A) TUNEL assay did not detect significant cell death in mutant tendons throughout the developmental stages from E14.5 to P7. Shown here is a transverse section of P7 mutant forelimb, in which white line demarcates the extensor digitorium communis tendons. Inset in (A) shows a transverse section of E14.5 forelimb paw that serves as a positive control for TUNEL staining. As expected, cell death is detected only at the distal edge of the autopod, but not in tendons (ScxGFP-positive) at this stage. (B) EdU labeling of proliferating cells in transverse sections of the forelimb from P3 pups. The rate of proliferation was also not altered in mutant tendons compared with the wild-type littermates, an observation that also found in E14.5 to P10 samples. The pups were injected i.p. with 100 μg of EdU in PBS and tissues were harvested 2 hr post-injection. (C) Histological staining for the prototypic markers of chondrocytes (toluidine blue), osteocytes (alizarin red) and adipocytes (oil-red-o) showed that the loss of tendon markers in mutant tenocytes was also not due to transdifferentiation. The positive control tissues for the respective staining are cartilage, adipose tissue and bone from the same section. Dotted lines demarcate tendons. Mutant: CKO, Wild-type: WT. Figures not to scale.
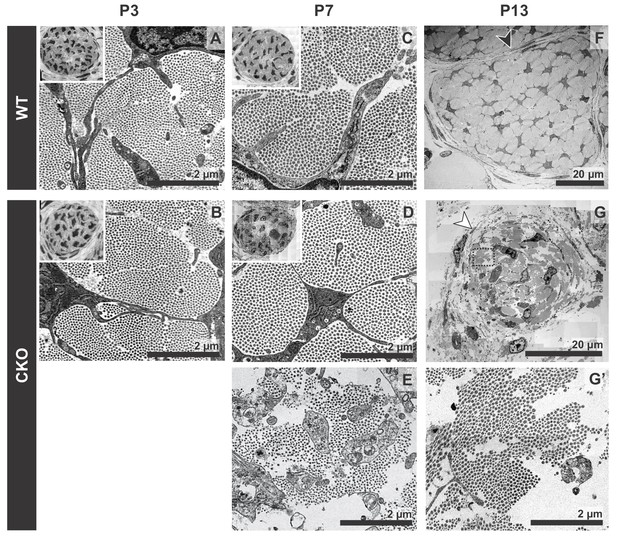
Tendon degeneration observed in Tgfbr2;ScxCre mutants only at later postnatal stages.
TEM images of tendons from forelimbs of mutant and wild-type littermates at P3, P7 and P13. (A,B) Despite detectable functional defects starting around P3 in mutant pups, collagen matrix organization in mutant neonates was indistinguishable from that of their wild-type littermates. (C–E) By P7, the mutant tendon began to show signs of matrix degradation compared to the wild-type tendon. Collagen fibrils remained intact in some areas (D) and showed signs of deterioration in other areas (E). (F,G) Apparent collagen degradation and disrupted epitenon structures (white arrowhead) could be detected in tendons of P13 mutant pups. Black arrowhead indicates epitenon in wild-type pups. Boxed region in (G) is shown enlarged in (G’). Insets show transverse section TEM images of entire tendons at low-magnification (not to scale). Mutant: CKO, Wild-type: WT.
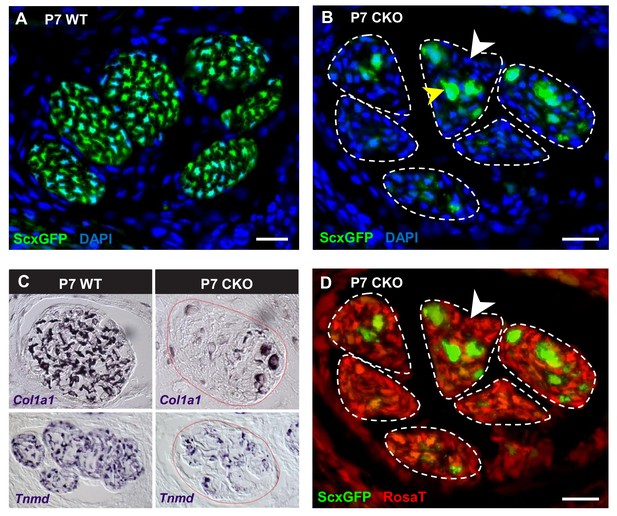
Deletion of Tgfbr2 in Scx-expressing cells (Tgfbr2;ScxCre) results in loss of tenocyte differentiation markers.
(A–D) Transverse sections of extensor digitorium communis tendons of wild-type and mutant pups at wrist level. (A) In P7 wild-type pups, all tenocytes were positive for tendon reporter ScxGFP signal. (B) Conversely, most cells in P7 mutant tendons lost the ScxGFP signal (white arrowhead), whereas the cells positive for ScxGFP signal are newly recruited cells (yellow arrowhead) (Tan et al. in preparation). (C) In situ hybridization shows that the mutant cells also lost gene expression of tendon markers Col1a1 and Tnmd (images not to scale). (D) Lineage tracing using ScxCre shows that all ScxGFP-negative cells in (B) were positive for Ai14 Rosa26-tdTomato (RosaT) Cre reporter (white arrowhead), proving that the ScxGFP-negative cells in mutant tendons were derived from tenocytes. Dashed lines demarcate the mutant tendons. Scale bar, 20 μm. Mutant: CKO, wild-type: WT.
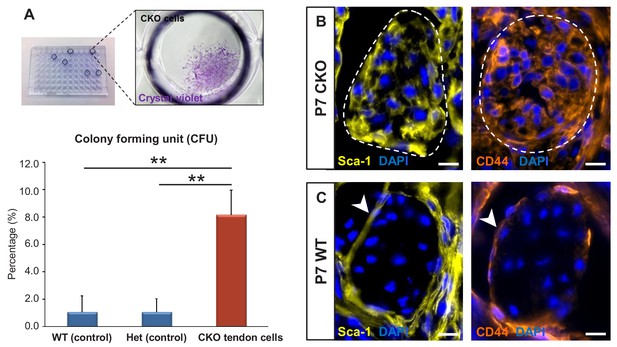
Tgfbr2;ScxCre mutant tenocytes acquired stem/progenitor features.
(A) The colony-forming efficiency of P7 wild-type and heterozygous tenocytes as well as mutant tendon cells were evaluated by seeding one cell per well of the FACS-sorted cells in 96-well plates, and colonies formed were visualized with crystal violet staining. Mutant tenocytes exhibited significantly higher clonogenic capacity compared to wild-type and heterozygous controls. The results shown are mean ± SD (n = 5–6, **p<0.01). (B) Immunofluorescence staining for stem/progenitor markers in transverse sections of mutant tendons shows that mutant tendon cells acquired in postnatal stages expression of stem cell antigen-1 (Sca-1) and CD44. (C) In wild-type littermate controls, expression of both markers was detected in epitenon (white arrowhead), but not in tenocytes. Dashed line demarcates the mutant tendon. Scale bars, 10 μm. Mutant: CKO, Wild-type: WT, Heterozygous: Het.
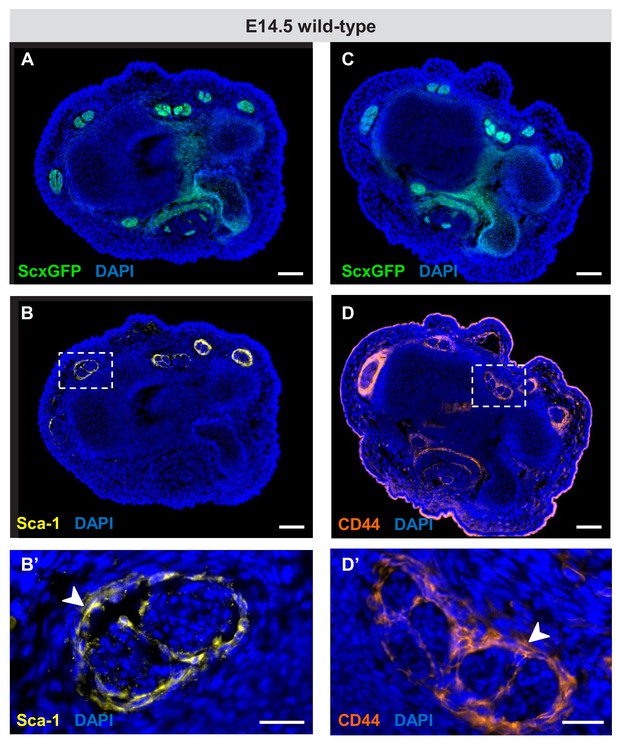
Expression of Sca-1 and CD44 during embryonic tendon development.
(A–D) Immunofluorescence staining for Sca-1 and CD44 on wrist-level transverse sections from E14.5 ScxGFP-carrying wild-type embryos. Robust expression of (B) Sca-1 and (D) CD44 was detected in cells that surround the tendons at E14.5 (boxed areas), likely the precursors of the epitenon/paratenon. (B’,D’) Higher magnification views of the boxed areas in (B) and (D). The epitenon/paratenon layer is indicated by white arrowheads. Note that both markers were not expressed by the tenocytes at E14.5, the onset of tenocyte differentiation or at any other stages during embryonic tendon development (not shown). Scale bars, 100 μm (A–D) and 25 μm (B’,D’).
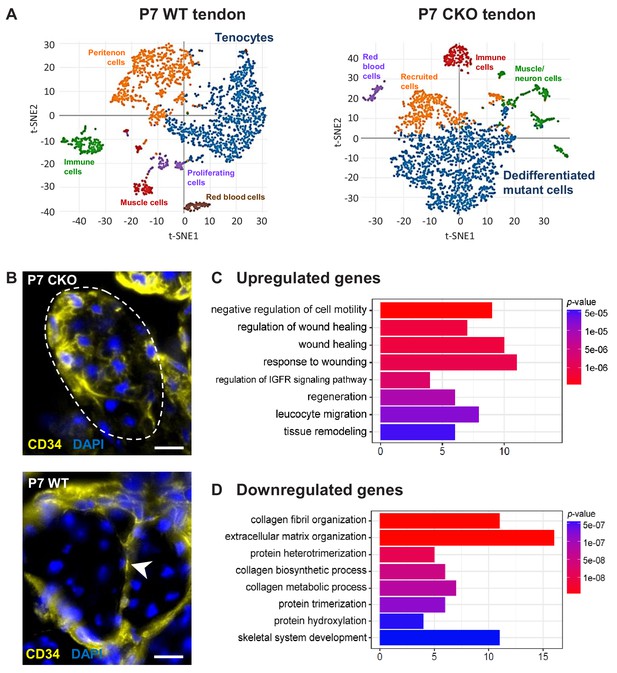
Molecular profile of the dedifferentiated mutant tenocytes.
(A) tSNE plots (K-means clustering) of enzymatically released cells from P7 wild-type and Tgfbr2;ScxCre mutant tendons reveals two major clusters corresponding to tenocytes and dedifferentiated mutant cells in the respective samples. Other cell type assignments are provided in the plots. See Supplementary file 1 for the list of genes highly expressed in these two clusters relative to other clusters. (B) Upregulated expression of Cd34 gene in P7 mutant tenocytes as revealed by scRNASeq analysis (see also Table 2) was determined using immunostaining. Transverse section of forelimb tendons shows that CD34 was indeed expressed by mutant tenocytes, while in wild-type controls CD34 was detected only in epitenon cells (white arrowhead). Dashed line demarcates the mutant tendon. (C,D) Gene ontology (GO) enrichment analysis in terms of biological processes associated with the (C) upregulated and (D) downregulated genes in P7 mutant compared with wild-type tenocytes. Selected GO terms are included in this figure, and genes annotated to the GO terms are available in Supplementary file 3. Scale bar, 10 μm. Mutant: CKO, Wild-type: WT.
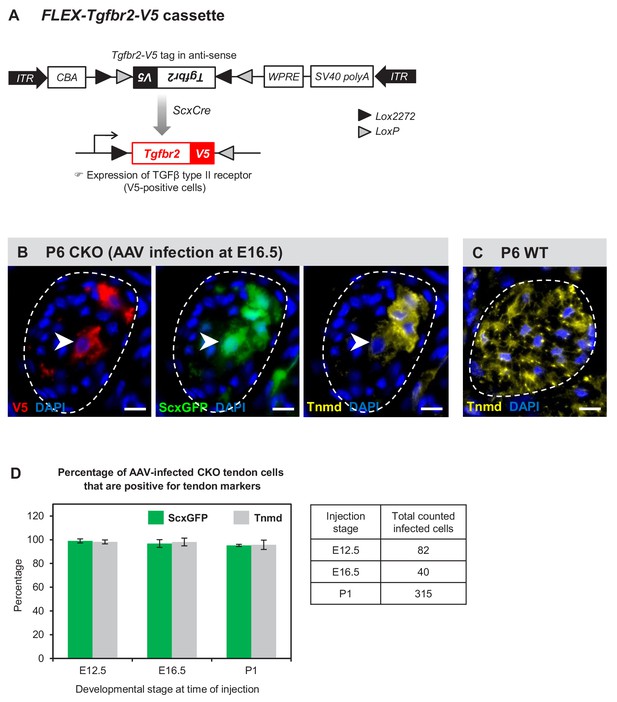
Tenocyte dedifferentiation is dependent on cell autonomous loss of TGFβ signaling.
(A) AAV1-FLEX-Tgfbr2-V5 virus contains the reverse-complement sequence of Tgfbr2 with a C-terminal V5 epitope tag. Cre activity will lead to a permanent inversion of the cassette that will then express the V5-tagged TGFβ type II receptor. (B) Targeted expression of TGFβ type II receptor in E16.5 mutant tendon cells using the AAV1-FLEX-Tgfbr2-V5 prevented the loss of tendon markers in the infected tenocytes. The forelimb of E16.5 mutant embryos was infected with AAV1-FLEX-Tgfbr2-V5 virus and harvested at P6. Transverse forelimb sections were stained with antibodies for V5 (red) to detect AAV-infected cells and tenomodulin (Tnmd; yellow), a prototypic tendon marker expressed by (C) all tenocytes in the wild-type tendon at this stage. Dashed line demarcates the mutant tendon. (D) Quantification shows that about 95–98% of the AAV-infected (V5-positive) mutant tendon cells retained or re-expressed tendon differentiation markers after viral injection at different developmental stages (n = 3 pups for each stage). Note that the embryonic infection was performed with Cre-activated AAV1-FLEX-Tgfbr2-V5 virus and the P1 infection was performed with the constitutive AAV1-Tgfbr2-FLAG virus. Scale bar, 10 μm. Mutant: CKO, Wild-type: WT.
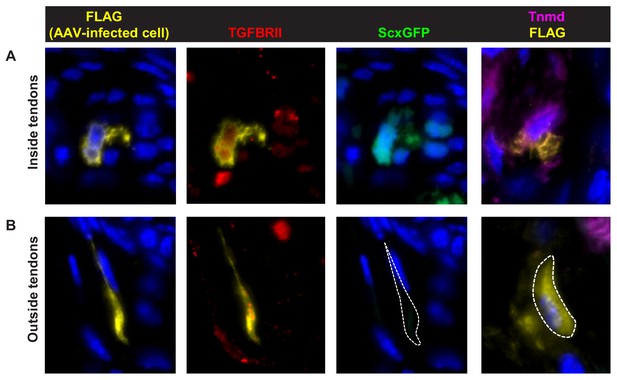
Induction of tendon markers by TGFβ signaling is context dependent.
AAV1-Tgfbr2-FLAG viral infection resulted in constitutive expression Tgfbr2-FLAG expression in cells both within and outside of tendons. The virus was injected locally into P1 mutant limbs and the limbs were harvested at P7. Sections from infected limbs were stained with antibodies to FLAG (yellow) to detect infected cells, and TGFβ type II receptor (TGFBRII) to confirm the re-expression of the receptor. ScxGFP signal and tenomodulin (Tnmd) antibody staining were used to identify induction of tendon markers. (A) Infected mutant tendon cells expressed the tendon markers ScxGFP and Tnmd. (B) In cells located outside of tendons (demarcated lines), the viral infection as detected by positive FLAG and TGFBRII immunofluorescence did not result in induction of the tendon markers ScxGFP and Tnmd. Figures not to scale.
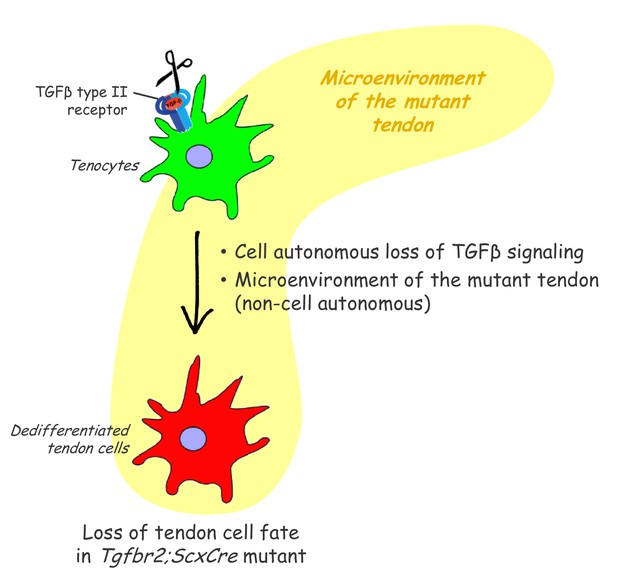
Proposed roles of TGFβ signaling in the maintenance of tendon cell fate.
Targeted disruption of the TGFβ type II receptor (Tgfbr2) by ScxCre resulted in tenocyte dedifferentiation in early postnatal stages. Tenocyte dedifferentiation was reversed by reactivation of TGFβ signaling in individual mutant cells, demonstrating a cell autonomous role for TGFβ signaling for maintenance of the cell fate. Conversely, a mere loss of the receptor in individual tendon cell was not sufficient to cause tenocyte dedifferentiation, suggesting that external factors may also play a critical role in this process. We therefore propose that maintenance of the tendon cell fate is dependent on a combination of a cell autonomous function of TGFβ signaling and an additional, likely non-cell autonomous factor, for example the microenvironment of the tendon in the Tgfbr2;ScxCre mutant (cell-matrix interaction, mechanical loading, cell-cell contacts etc).
Tables
Top 25 downregulated genes in P7 Tgfbr2;ScxCre mutant cells compared with P7 wild-type tenocytes (≥2 fold change, adjusted p<0.05).
See also Supplementary file 2 for a complete list of the downregulated genes.
| Gene symbol | Gene name | Fold change |
|---|---|---|
| Wif1 | Wnt inhibitory factor 1 | 157.4 |
| Col11a2# | Collagen, type XI, alpha 2 | 92.0 |
| Scx# | Scleraxis | 66.2 |
| Col2a1δ | Collagen, type II, alpha 1 | 58.9 |
| Car9 | Carbonic anhydrase 9 | 58.1 |
| Sema3b | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 43.9 |
| Cgref1 | Cell growth regulator with EF hand domain 1 | 33.2 |
| Fmod# | Fibromodulin | 27.9 |
| Cilp2 | Cartilage intermediate layer protein 2 | 24.7 |
| Matn4 | Matrilin 4 | 19.3 |
| P4ha1δ | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha one polypeptide | 13.5 |
| Pcolce2δ | Procollagen C-endopeptidase enhancer 2 | 11.8 |
| Tpm1 | Tropomyosin 1, alpha | 10.0 |
| Wisp1 | WNT1 inducible signaling pathway protein 1 | 9.7 |
| Tnmd# | Tenomodulin | 8.5 |
| Loxl2δ | Lysyl oxidase-like 2 | 8.3 |
| 1500015O10Rik | RIKEN cDNA 1500015O10 gene | 7.1 |
| Col11a1# | Collagen, type XI, alpha 1 | 7.1 |
| Pdgfrlδ | Platelet-derived growth factor receptor-like | 7.0 |
| Mfap4 | Microfibrillar-associated protein 4 | 6.5 |
| Col1a1# | Collagen, type I, alpha 1 | 6.4 |
| Ptgis | Prostaglandin I2 (prostacyclin) synthase | 6.4 |
| Col1a2# | Collagen, type I, alpha 2 | 6.2 |
| Itgbl1 | Integrin, beta-like 1 | 5.7 |
| Tpm2 | Tropomyosin 2, beta | 5.4 |
-
Note:
1) #=Tendon differentiation or specific marker; δ = genes related to tendons.
-
2) Note that the expression level detected for Scx also included that of ScxGFP, and therefore do not reflect the expression level of endogenous Scx.
Top 25 upregulated genes in P7 Tgfbr2;ScxCre mutant cells compared with P7 wild-type tenocytes (≥2 fold change, adjusted p<0.05).
See also Supplementary file 2 for a complete list of the downregulated genes.
| Gene symbol | Gene name | Fold change |
|---|---|---|
| Dlk1 | Delta-like one homolog (Drosophila) | 137.9 |
| Serpine2 | Serine (or cysteine) peptidase inhibitor, clade E, member 2 | 118.2 |
| Dpt | Dermatopontin | 95.7 |
| Ly6a | Lymphocyte antigen six complex, locus A | 54.3 |
| H19 | H19 | 51.1 |
| Cd34 | CD34 antigen | 47.8 |
| Lum | Lumican | 36.6 |
| Lgmn | Legumain | 31.8 |
| Cxcl12 | Chemokine (C-X-C motif) ligand 12 | 26.1 |
| Mfap5 | Microfibrillar associated protein 5 | 22.5 |
| Ly6c1 | Lymphocyte antigen six complex, locus C1 | 21.7 |
| Igf2 | Insulin-like growth factor 2 | 21.4 |
| Serping1 | Serine (or cysteine) peptidase inhibitor, clade G, member 1 | 19.2 |
| Mgst1 | Microsomal glutathione S-transferase 1 | 18.3 |
| Aspn | Asporin | 15.9 |
| Mt1 | Metallothionein 1 | 15.4 |
| Mgst3 | Microsomal glutathione S-transferase 3 | 13.1 |
| Col3a1δ | Collagen, type III, alpha 1 | 13.0 |
| Postn | Periostin, osteoblast specific factor | 13.0 |
| Itm2a | Integral membrane protein 2A | 12.7 |
| Ptn | Pleiotrophin | 10.3 |
| Rps18-ps3 | Ribosomal protein S18, pseudogene 3 | 9.7 |
| Gsn | Gelsolin | 8.3 |
| Ifitm3 | Interferon induced transmembrane protein 3 | 8.2 |
| Col5a1δ | Collagen, type V, alpha 1 | 8.1 |
-
Note: δ = genes related to tendons.
PANTHER protein class differentially expressed in P7 Tgfbr2;ScxCre mutant cells compared with P7 wild-type tenocytes.
A complete list of differentially expressed genes (≥2 fold change, adjusted p<0.05) used for the analysis is available in Supplementary file 2.
| (A) Downregulated protein class | |
|---|---|
| Protein class | Gene list |
| Receptor | Pdgfrl, Col6a3, Kdelr3, Col6a1, Kdelr2, Itgbl1, Ssc5d, Col6a2, Ssr4, Col12a1, Matn4 |
| Signaling molecule | Sdc1, Wisp1, Sparc, Mfap4, Sema3b, Angptl2, Tgfbi |
| Membrane traffic protein | Sec13, Kdelr3, Copz2, Kdelr2, Rabac1, Lman1 |
| Extracellular matrix protein | Sdc1, Crtap, Clec11a, P3h3, Sparc, P3h4 |
| (B) Upregulated protein class | |
| Protein class | Gene list |
| Nuclei acid binding | Ndn, Eif3f, Rpl39, Rpl36a, Rpl3, Rpl9-ps6, Rpl22l1, Rps27, Rps4x, Cirbp, Rps19, Eif3e, Rps18, Rps5, Junb |
| Enzyme modulator | Fstl1, Dbi, Sfrp2, Ctsb, Serpine2, Serping1, Igfbp3, Igfbp4 |
| Cytoskeletal protein | Gsn, Map1lc3b, Tuba1b, Arpc1b, Emp1, Tubb5 |
| Signaling molecule | S100a16, Ptn, Dlk1, Efemp2, Postn, Sfrp2 |
| Transcription factor | Eif3h, Naca, Fos, Id3, Junb |
PANTHER pathway analysis of upregulated genes in P7 Tgfbr2;ScxCre mutant cells compared with P7 wild-type tenocytes.
| PANTHER pathway | PANTHER accession | Gene list |
|---|---|---|
| Integrin signaling pathway | P00034 | Arpc2, Col4a1, Rac1, Col5a2, Rap1b, Cdc42, Arpc5, Col5a1, Rap1a, Rhoc, Fn1, Arpc1b, Col3a1 |
| Inflammation mediated by chemokine and cytokine signaling pathway | P00031 | Arpc2, Rac1, Cdc42, Nfkbia, Arpc5, Rhoc, Arpc1b, Arpc4, Jun, Junb |
| Wnt signaling pathway | P00057 | Fstl1, Sfrp2, Ppp3ca, Csnk1a1 |
| Insulin/IGF pathway | P00032, P00033 | Igf1, Igf2, Fos |
-
Note:
1A complete list of differentially expressed genes (DEGs) used for the analysis is available in Supplementary file 2.
-
2Different values of the filter parameter (mean UMI count and fold change) were applied for enriching DEGs in P7 mutant cells. Only pathways that stood out as relevant for this study are listed.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Tgfbr2f/f | (Chytil et al., 2002) | NA | NA |
| Genetic reagent (M. musculus) | ScxCre | (Blitz et al., 2013) | NA | NA |
| Genetic reagent (M. musculus) | RosaCreERT | (Hameyer et al., 2007) | NA | NA |
| Genetic reagent (M. musculus) | ScxGFP | (Pryce et al., 2007) | NA | NA |
| Genetic reagent (M. musculus) | Ai14 Rosa26-tdTomato (RosaT) | (Madisen et al., 2010) | NA | NA |
| Recombinant DNA reagent | pAAV1-FLEX-Tgfbr2-V5 | GenScript | This paper | NA |
| Recombinant DNA reagent | pAAV1-Tgfbr2-FLAG | GenScript | This paper | NA |
| Antibody | Rat anti-CD34 (Clone RAM34) | BD Biosciences | Cat# 553731 RRID:AB_395015 | IF(1:200), Antigen retrieval |
| Antibody | Rat anti-CD44 (Clone IM7) | BD Biosciences | Cat# 550538 RRID:AB_393732 | IF(1:40), Pre-treated with cold acetone for 10 min at −20°C |
| Antibody | Rabbit anti-FLAG (DYKDDDDK) | Thermo Fisher Scientific | Cat# 740001 RRID:AB_2610628 | IF(1:200), Antigen retrieval |
| Antibody | Rat anti-FLAG (DYKDDDDK) | Novus Biologicals | Cat# NBP1-06712SS RRID:AB_1625982 | IF(1:100), Antigen retrieval |
| Antibody | Goat anti-Sca-1/Ly6 | R and D Systems | Cat# AF1226 RRID:AB_354679 | IF(1:80) |
| Antibody | Rat anti-Sca-1/Ly6 | R and D Systems | Cat# MAB1226 RRID:AB_2243980 | IF(1:50) |
| Antibody | Goat anti-tenomodulin (Clone C-20) | Santa Cruz Biotechnology | Cat# sc-49324 RRID:AB_2205971 | IF(1:50), Antigen retrieval |
| Antibody | Rabbit anti-TGFβ type II receptor | Bioworld Inc | Cat# BS1360 RRID:AB_1663474 | IF(1:250) |
| Antibody | Rabbit anti-V5 | Abcam | Cat# ab206566 RRID:AB_2819156 | IF(1:500), Antigen retrieval |
| Antibody | Rat anti-V5 | Abcam | Cat# ab206570 RRID:AB_2819157 | IF(1:500), Antigen retrieval |
| Antibody | Cy5 donkey anti-goat secondary | Jackson ImmunoResearch | Cat# 705-175-147 RRID:AB_2340415 | IF(1:500) |
| Antibody | AlexaFluor647 donkey anti-rabbit secondary | Jackson ImmunoResearch | Cat# 711-607-003 RRID:AB_2340626 | IF(1:400) |
| Antibody | Cy3 donkey anti-rabbit secondary | Jackson ImmunoResearch | Cat# 711-166-152 RRID:AB_2313568 | IF(1:800) |
| Antibody | AlexaFluor647 donkey anti-rat secondary | Jackson ImmunoResearch | Cat# 712-606-153 RRID:AB_2340696 | IF(1:800) |
| Antibody | Cy3 donkey anti-rat secondary | Jackson ImmunoResearch | Cat# 712-166-150 RRID:AB_2340668 | IG(1:800) |
| Commercial assay or kit | In situ cell death detection kit | Roche | Cat# 12156792910 | Follow the manufacturer’s instruction |
| Commercial assay or kit | Click-iT EdU kit | Life Technologies | Cat# C10340 | Follow the manufacturer’s instruction |
| Other | DAPI stain | Thermo Fisher Scientific | D1306 RRID:AB_2629482 | 1 μg/ml |
-
Note:
* Antigen retrieval: Incubated with warm citrate buffer (10 mM sodium citrate with 0.05% Tween 20, pH 6) at 550W, 50°C for 5 min using a PELCO BioWave.
Additional files
-
Supplementary file 1
Signature genes in tenocytes and dedifferentiated mutant cells in comparison with other clusters.
See also Figure 6A for the tSNE plots of the sample. (A) Top 25 genes highly expressed in the tenocyte cluster relative to other clusters in the P7 wild-type tendon sample (≥1.5 fold change, adjusted p<0.05). (B) Top 25 genes highly expressed in the dedifferentiated mutant cell cluster relative to other clusters in the P7 Tgfbr2;ScxCre mutant tendon sample (≥1.5 fold change, adjusted p<0.05).
- https://cdn.elifesciences.org/articles/52695/elife-52695-supp1-v2.docx
-
Supplementary file 2
Differentially expressed genes in P7 Tgfbr2;ScxCre mutant tendon cells compared with P7 wild-type tenocytes (≥2 fold change, adjusted p<0.05).
Note that the expression level detected for Scx also included that of ScxGFP, and therefore do not reflect the expression level of endogenous Scx.
- https://cdn.elifesciences.org/articles/52695/elife-52695-supp2-v2.xlsx
-
Supplementary file 3
Gene Ontology (GO) term enrichment of differentially expressed genes in P7 Tgfbr2;ScxCre mutant cells compared with P7 wild-type tenocytes.
A complete list of differentially expressed genes (≥2 fold change, p<0.05) used for the analysis is available in Supplementary file 2.
- https://cdn.elifesciences.org/articles/52695/elife-52695-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52695/elife-52695-transrepform-v2.docx





