Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins
Figures
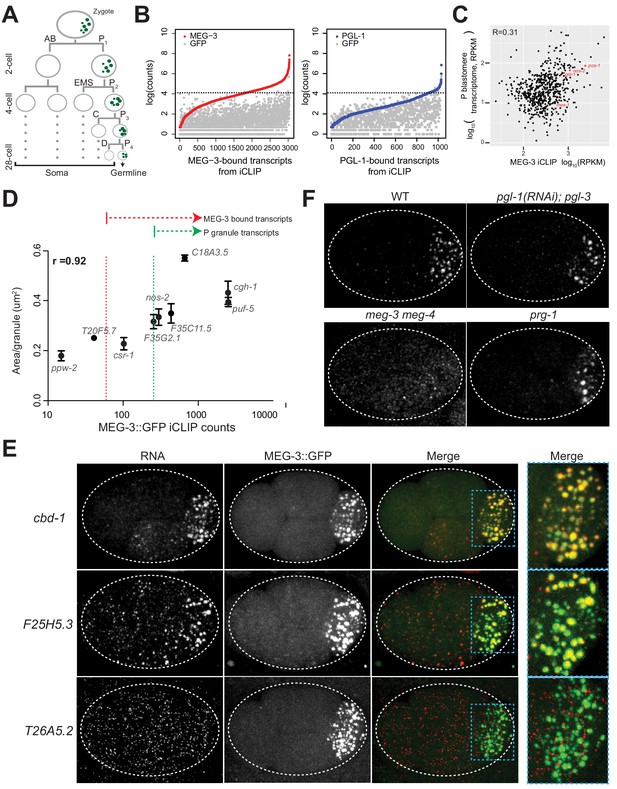
mRNAs are recruited into P granules by the MEG phase.
(A) Abbreviated embryonic lineage showing the somatic (AB, EMS, C, and D) and germline (P) blastomeres. Green dots represent P granules. P4 is the founder cell of the germline. Dotted lines refer to additional divisions not shown. (B) Graphs showing log transformed average read counts (Y axis) from two MEG-3::GFP (red), PGL-1::GFP (blue) and GFP (gray) iCLIP experiments. Genes are arranged along X axis based on the ascending log transformed read counts in the MEG-3::GFP or PGL-1::GFP iCLIP experiments (average of two experiments). Gray dots represent the GFP iCLIP read counts for each rank-ordered gene. The stippled line denotes the GFP background threshold (read counts = 60) above which transcripts were considered true positives (657 transcripts in the MEG-3::GFP iCLIPs and 18 transcripts in the PGL-1::GFP iCLIPs). (C) Graph showing the 657 MEG-3-bound transcripts (black dots) with respect to read counts in the MEG-3::GFP iCLIPs (average FPKM of two replicates, X axis) versus transcript abundance in embryonic P blastomeres (Y-axis, Lee et al., 2017). R is the Pearson correlation coefficient. nos-2, mex-3, gld-1 and pos-1 are highlighted in red. See Figure 1—figure supplement 1 for graphs showing same for all embryonic transcripts. (D) Graph showing average read counts in MEG-3::GFP iCLIPs versus average RNA cluster size as measured from smFISH signal for nine genes. R is the Spearman correlation coefficient. Red stippled line denotes threshold for MEG-3::GFP-bound mRNAs as defined in A. Green stippled line denotes threshold for P granule mRNAs as defined in text. (E) Photomicrographs of embryos expressing MEG-3::GFP hybridized with single molecule fluorescence (smFISH) probes as indicated. cbd-1 and F25H5.3 are examples of transcripts localizing to P granules, as shown by colocalization with MEG-3::GFP in the right-most panels. T26A5.2 is an example of a transcript that does not enrich in P granules. (F) Photomicrographs of 4 cell embryos of the indicated genotypes hybridized with smFISH probes against the nos-2 transcript. All genotypes show localization of nos-2 to P granules in the P2 blastomere, except for meg-3 meg-4. nos-2 is a maternal mRNA that is rapidly degraded in somatic blastomeres (two anterior cells) and therefore present at higher levels in P blastomeres and their newly born sister blastomeres (two posterior cells).
-
Figure 1—source data 1
Data used to generate Figure 1D.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig1-data1-v2.xlsx
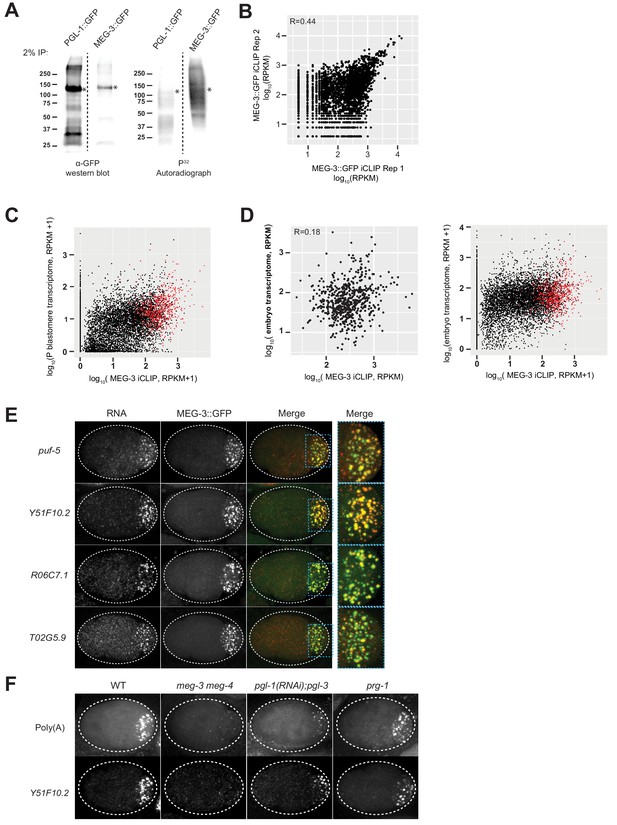
mRNAs are recruited into P granules by the MEG phase.
(A) Anti-GFP western blot and autoradiograph of crosslinked anti-GFP immunoprecipitates separated by SDS-PAGE. The immunoprecipitates were 5’ end radiolabeled to visualize cross-linked protein:RNA complexes. * indicates predicted MEG-3::GFP and PGL-1::GFP bands based on respective sizes. Note that PGL-1::GFP protein is more abundant than MEG-3::GFP, yet does not cross-link with RNA as efficiently. (B) Graph plotting reads for each transcript (black dots) in MEG-3::GFP iCLIP replicate 1 versus replicate 2. R is the Pearson correlation coefficient. (C) Graph plotting all embryonic transcripts (black dots) with respect to average read counts in the MEG-3::GFP iCLIPs (average RPKM of two replicates, X axis) versus transcript abundance in P blastomeres (Y axis). Red dots indicate the 657 MEG-3 bound transcripts. (D) (Left) Graph plotting the 657 MEG-3 bound transcripts with respect to average read counts in the MEG-3::GFP iCLIPs (average RPKM of two replicates, X axis) versus abundance in whole embryo transcriptome. R is the Pearson correlation coefficient. (Right) Graph plotting all embryonic transcripts (black dots) with respect to average read counts in the MEG-3::GFP iCLIPs (average RPKM of two replicates, X axis) versus abundance in whole embryo transcriptome. Red dots indicate the 657 MEG-3 bound transcripts. (E) Photomicrographs of embryos expressing MEG-3::GFP hybridized with single molecule fluorescence (smFISH) probes as indicated. All four are ‘P granule transcripts’ localizing to P granules (MEG-3::GFP), as shown in the higher magnification merged panels. (F) Photomicrographs of 4 cell embryos of the indicated genotypes hybridized with smFISH probes against poly(A) and Y51F10.2. P granule RNA clusters are smaller in pgl-1(RNAi); pgl-3 embryos, consistent with clustering of MEG-3 condensates on the surface of PGL condensates in wild-type embryos (Putnam et al., 2019). The identity of the small polyA clusters in the P2 and EMS blastomeres in meg-3 meg-4 mutant embryos has not been verified, but is likely to correspond to immature processing bodies that form in the P blastomeres and their immediate somatic descendants before maternal RNA degradation is activated (Gallo et al., 2008).
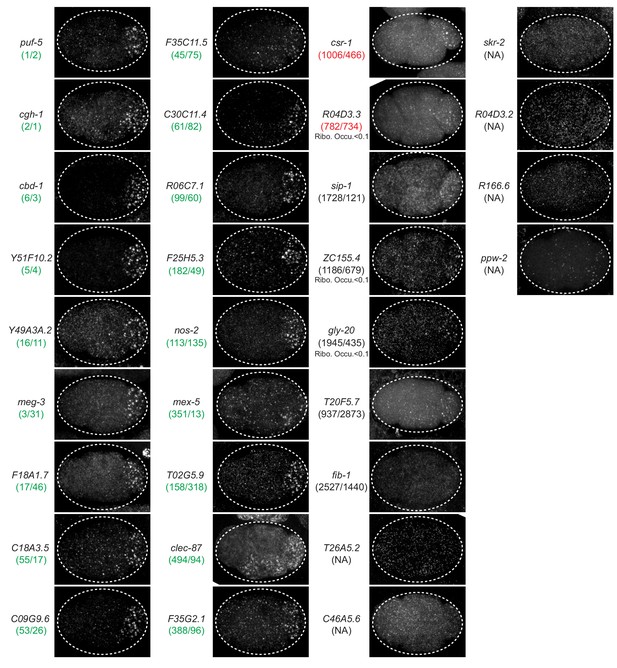
In situ hybridization.
Photomicrographs of 4 cell wild-type embryos hybridized with smFISH probes as indicated. Maximum Z projections of photomicrographs from at least 10 embryos were examined per probe set. Numbers in parenthesis indicate rank order in two MEG-3::GFP iCLIP experiments. Green indicates rankings at or above the nos-2 cluster threshold (‘P granule transcript’ category). Red indicates rankings below the nos-2 cluster threshold (‘MEG-3-bound transcript category’. Black indicates ranking below the GFP background threshold. ‘NA’ indicates no read count in one or both MEG-3::GFP iCLIP experiments.
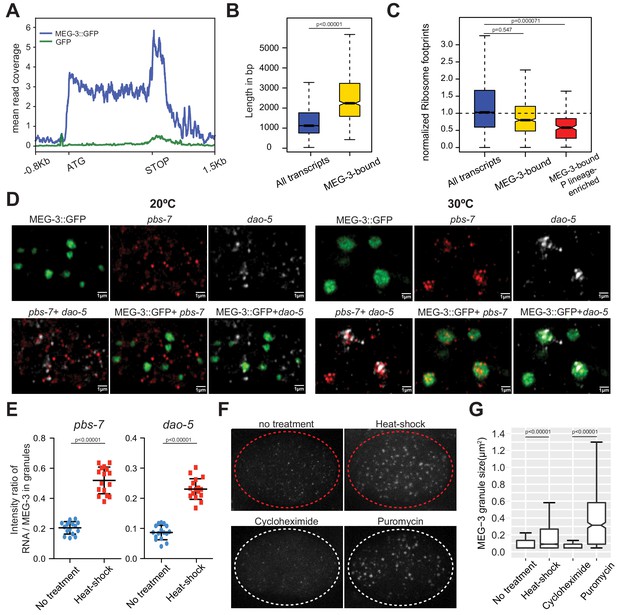
MEG proteins recruit mRNAs into P granules by a sequence non-specific mechanism that favors ribosome-poor mRNAs.
(A) Metagene analysis of the distribution of MEG-3::GFP iCLIP reads on the 657 MEG-3-bound transcripts (blue line) compared to GFP iCLIP (green line) reads. The peak 3’ to the STOP codon correspond to 3’ UTR sequences. (B) Box plot showing the distribution of transcript length for embryonic transcripts (15,345 loci) versus MEG-3-bound transcripts (657 loci). P values were calculated using an unpaired t-test. Each box extends from the 25th to the 75th percentile, with the median indicated by the horizontal line; whiskers extend to the highest and lowest observations. (C) Box plot showing ribosome occupancy in wild-type embryos for three gene categories: embryonic transcripts (15,345 loci), MEG-3-bound transcripts (657 loci) and MEG-3-bound transcripts enriched in P lineage (187 loci). Because ribosome profiling was performed on whole embryos, footprint counts are averages across all cells. Ribosome profiles for the subset of MEG-3::GFP-bound mRNAs that are also enriched in the P lineage (Lee et al., 2017), therefore, are more likely representative of profiles of mRNAs in P granules. See Figure 2—figure supplement 2A for ribosome occupancy of P granule transcripts (MEG-3-bound RNAs above the nos-2 cluster). P values were calculated using an unpaired t-test. Each box extends from the 25th to the 75th percentile, with the median indicated by the horizontal line; whiskers extend to the highest and lowest observations. (D–E) Photomicrographs of P2 blastomeres expressing MEG-3::GFP and hybridized to probes against two non-P granule transcripts (pbs-7 and dao-5) before (20°C) and after 15 min of heat-shock (30°C). Images are single Z sections and are representative of data quantified in (E). See Figure 2—figure supplement 2D for whole embryo images. (E) Graphs showing the intensity ratio of RNA over GFP in MEG-3::GFP granules under no heat-shock (blue dots) or heat-shock (red dots) conditions. Each data point represents the average value for all MEG-3::GFP granules in a single Z section (16 sections were collected from two embryos per condition). P values were calculated using an unpaired t-test. The center horizontal lines indicate the mean and bars represent the SD. (F) Photomicrographs of 4 cell embryos expressing MEG-3::GFP under the indicated treatments. Embryos were derived from mex-5(RNAi) mex-6(RNAi) hermaphrodites, which do not localize P granules (Smith et al., 2016). Embryos treated with cycloheximide or puromycin were derived from hermaphrodites also treated with ptr-2(RNAi) to permeabilize the eggshell. Images are representative of data quantified in (G). (G) Box plot showing the size distribution of MEG-3::GFP granules under different treatments as described in F. P values were calculated using an unpaired t-test. Each box extends from the 25th to the 75th percentile, with the median indicated by the horizontal line; whiskers extend to the highest and lowest observations.
-
Figure 2—source data 1
Data used to generate Figure 2E.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Data used to generate Figure 2G.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig2-data2-v2.xlsx
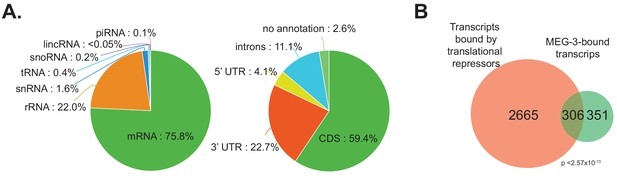
Characterization of MEG-3 bound transcripts.
(A) First pie chart shows distribution of MEG-3::GFP reads relative to different transcript types (total: 521,189 reads). Second pie chart shows distribution of MEG-3::GFP reads mapping to mRNAs relative to gene features (total: 275,083 reads). We also recovered reads mapping to introns in the GFP control IP (20% of reads), the significance of these reads is unclear. (B) Venn diagram showing the overlap between 2971 transcripts bound by translational repressors (GLD-1, CHG-1, OMA-1 and LIN-4 Scheckel et al., 2012; Boag et al., 2008; Tsukamoto et al., 2017) and the 657 MEG-3-bound transcripts defined in this study. Note that the transcripts bound by translation repressors were defined in oocytes, whereas the MEG-3-bound transcripts were defined in embryos, therefore 100% overlap is not expected. The hypergeometric probability p (p<2.57×10−73) is calculated using a statistical software in http://nemates.org/MA/progs/overlap_stats.html.
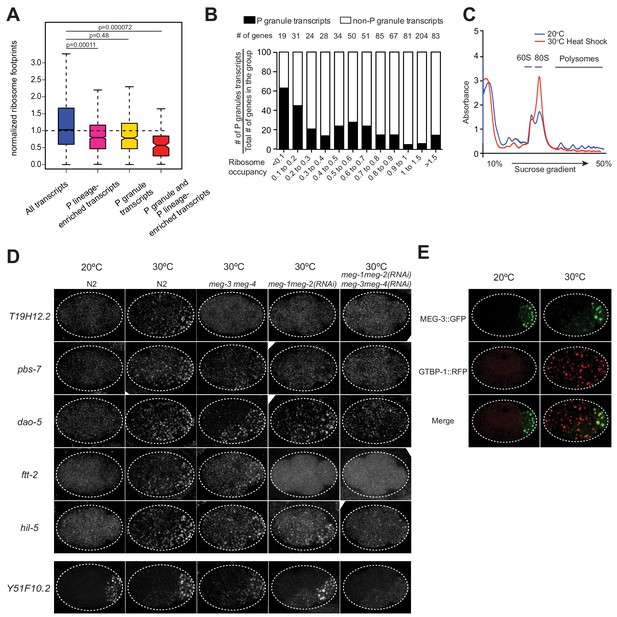
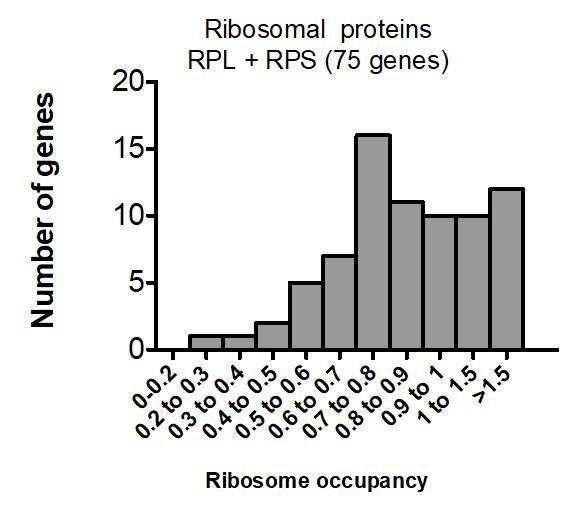
MEG-3 binds ribosome-poor transcripts.
(A) Box plot showing ribosome occupancy in wild-type embryos for all transcripts (15,345 loci), P-lineage enriched transcripts (1347 loci), P granule transcripts (492 loci), and P granule and P-lineage enriched transcripts (148 loci). Because ribosome profiling was performed on whole embryos, footprint counts are averages across all cells. Ribosome profiles for P lineage-enriched transcripts (Lee et al., 2017) are more representative of profiles of mRNAs in P granules. P values were calculated using an unpaired t-test. See Figure 2B for box plot description. (B) Bar graph showing the fraction of P granule transcripts (black) binned by ribosome occupancy. Only P lineage-enriched transcripts (FPM >25) were included in this analysis (Lee et al., 2017). Total number of loci per bin is indicated. (C). Graph showing RNA absorbance of embryonic lysates separated on a sucrose gradient. The lysates were isolated from wild-type embryos raised at 20°C without heat-shock or with heat-shock (30°C for 15 min). (D) Photomicrographs of 4 cell stage embryos of indicated genotypes after no heat-shock (20°C) or after heat-shock (30°C) and hybridized to smFISH probes against the indicated transcripts. Maximum Z projection of photomicrographs from at least 10 embryos were examined per probe set. Note that for dao-5, ftt-2 and hil-5, RNA granules still form preferentially in P2 and EMS in meg-3 meg-4 mutants and is only eliminated upon loss of meg-1 and meg-2, two MEG paralogs that also localize to P granules in embryos (Wang et al., 2014). (E) Single Z plane photomicrographs of a live 4 cell embryo co-expressing MEG-3::GFP (to mark P granules) and GTBP-1::RFP (to mark stress granules) cultured at 20°C and up-shifted to 30°C for 15 min.
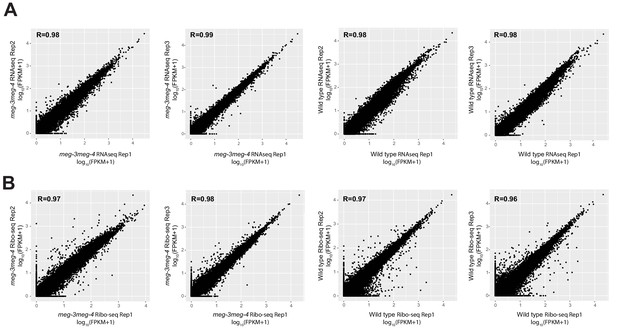
Correlation analyses of sequencing libraries from wild type and meg-3 meg4 embryos.
Sequencing libraries were generated from wild type and meg-3 meg-4 embryonic lysates in triplicates for RNAseq (A) and ribosome profiling (B) experiments. The scatterplots illustrate correlation between replicates. R is Pearson correlation calculated using R.
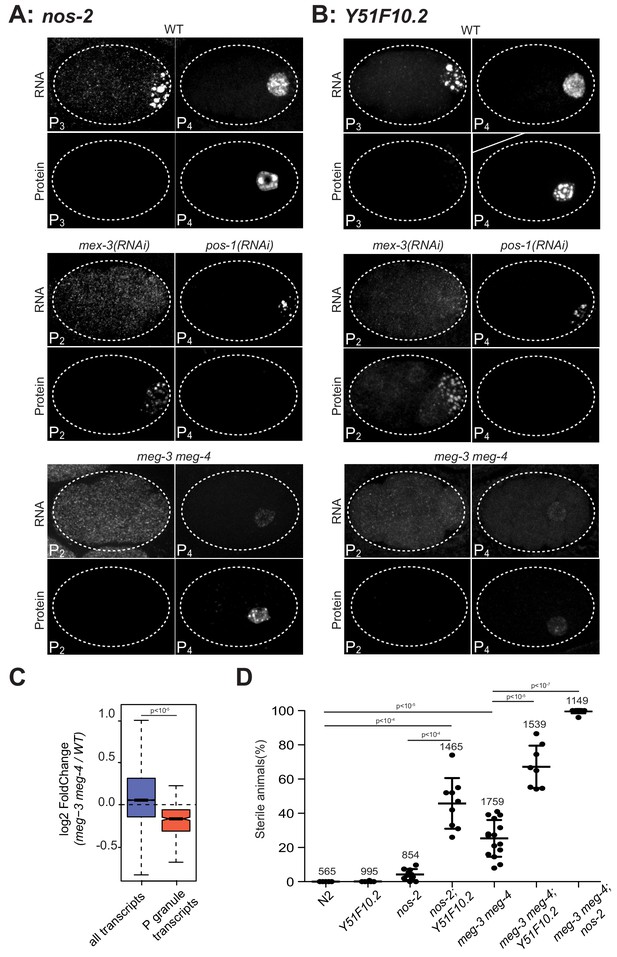
P granules enrich maternal mRNAs required for germ cell development in P blastomeres.
(A and B) Photomicrographs of embryos of indicated stages and genotypes and hybridized to fluorescent probes or antibodies to visualize nos-2 and Y51F10.2 transcripts and proteins. Embryos expressing NOS-2::3xFLAG and Y51F10.2::OLLAS were used for these experiments. Note the correlation between RNA in granules and no protein expression, and RNA in the cytoplasm and protein expression in wild-type, mex-3(RNAi) and pos-1(RNAi) embryos. In mex-3(RNAi) embryos, nos-2 and Y51F10.2 are prematurely translated in P2 where POS-1 is enriched. In pos-1(RNAi) embryos, nos-2 and Y51F10.2 are never translated. In meg-3 meg-4 embryos, nos-2 and Y51F10.2 RNAs are not in P granules and thus are not preferentially segregated to P4, resulting in lower RNA levels in that cell. Protein expression is correspondingly reduced but still activated at the correct stage, demonstrating that P granules are not essential for translational repression or activation. (C) Box plot showing the fold change in abundance between wild-type and meg-3 meg-4 embryos for 15,345 embryonic transcripts and the 492 P granule transcripts. P values were calculated using an unpaired t-test. See Figure 2B for box plot description. The 492 P granule transcripts are present at lower levels overall in meg-3 meg-4 embryos compared to wild-type, consistent with equal segregation to somatic blastomeres which turn over maternal mRNAs. (D) Graphs showing the percentage of sterile animals among progeny of hermaphrodites with the listed genotypes (maternal-effect sterility). Each dot represents the brood from a single hermaphrodite allowed to lay eggs for 24 hr. Total number of progeny scored across all broods is indicated for each genotype. P values were calculated using an unpaired t-test. The center horizontal lines indicate the mean and bars represent the SD.
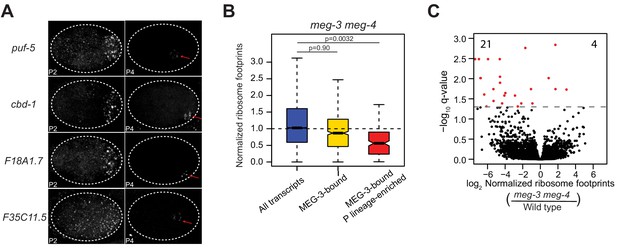
MEG-3 and MEG-4 are not required for translational repression of 623 mRNAs in P granules.
(A) Photomicrographs of P2 and P4 stage embryos hybridized with smFISH probes as indicated. Unlike nos-2 and Y51F10.2 shown in Figure 3, the transcripts shown here remain associated with P granules through the P4 stage (red arrow). (B) Ribosome profiling in meg-3 meg-4 embryos. Compare to Figure 2C. Because ribosome profiling was performed on whole embryos, footprint counts are averages across all cells. Ribosome profiles for MEG-3-bound, P lineage-enriched transcripts (Lee et al., 2017) are more representative of profiles of mRNAs in P granules bound by MEG-3. P values were calculated using an unpaired t-test. See Figure 2B for box plot description. (C) Volcano plot showing the log2 fold-change of normalized ribosome footprints for each gene (dot) in meg-3 meg-4 versus wild-type embryos. Dashed lines mark the significance cutoff of q = 0.05. The number of genes whose normalized ribosome foot-prints were significantly up (4) or downregulated (22) in meg-3 meg-4 embryos compared to wild-type are indicated.
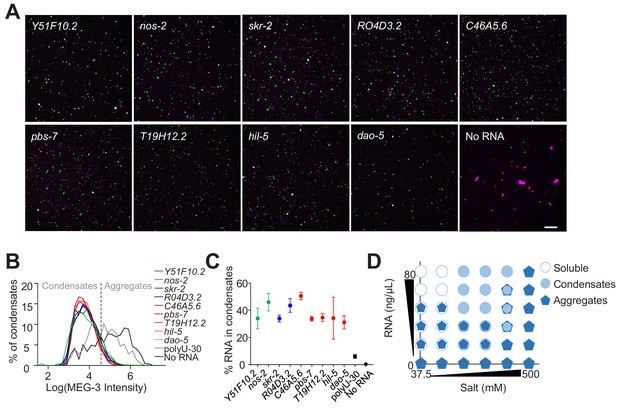
Non-sequence-specific condensation of MEG-3 with RNA.
(A) Representative photomicrographs of condensates of MEG-3 and indicated RNA after incubation in condensation buffer. Reactions contained 500 nM MEG-3 and 20 ng/µL in vitro transcribed RNA. MEG-3 (magenta) was trace labeled with Alexa647 and RNA (green) was trace labeled with Alexa546 (Materials and methods). Scale bar is 20 µm. (B) Histograms of MEG-3 intensity (log10 scale) normalized to the total number of condensates in each reaction assembled as in (A) for each RNA indicated. Each histogram includes condensates from 12 images collected from three experimental replicates. RNAs correspond to transcripts with MEG-3 iCLIP counts above the nos-2 cluster (nos-2, Y51F10.2), below the nos-2 cluster (skr-2, R04D3.2) and not recovered in the MEG-3 iCLIPs (C46A5.6, pbs-7, T19H12.2, hil-5, dao-5). Intersection between No RNA, polyU-30 and mRNA histograms was used to quantify the fraction of MEG-3 in condensates or aggregates as indicated by dashed line. (C) Graph showing the percent of RNA fluorescence in MEG-3 condensates compared to total RNA assembled as in (A). Each data point represents condensates from 12 images collected from three experimental replicates. Circles indicate the mean and bars represent the SD. RNAs corresponding to transcripts with MEG-3 iCLIP counts above the nos-2 cluster (green), below the nos-2 cluster (blue) and not recovered in the MEG-3 iCLIP (red). (D) Phase diagram of MEG-3 condensate composition under varying RNA and salt concentrations. For representative images and quantitation corresponding to positions in the diagram refer to Figure 5—figure supplement 1D–G. MEG-3 was present in three states: i) soluble MEG-3 (no condensates detected, open circles), ii) small uniform condensates (Log(I)≤4.6, filled circles), iii) large irregular aggregates (Log(I)>4.6, pentagons). In conditions with mixed MEG-3 states, the larger object represents the predominant population. See Figure 4—figure supplement 2D for representative images.
-
Figure 4—source data 1
Data used to generate Figure 4B.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Data used to generate Figure 4C.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig4-data2-v2.xlsx
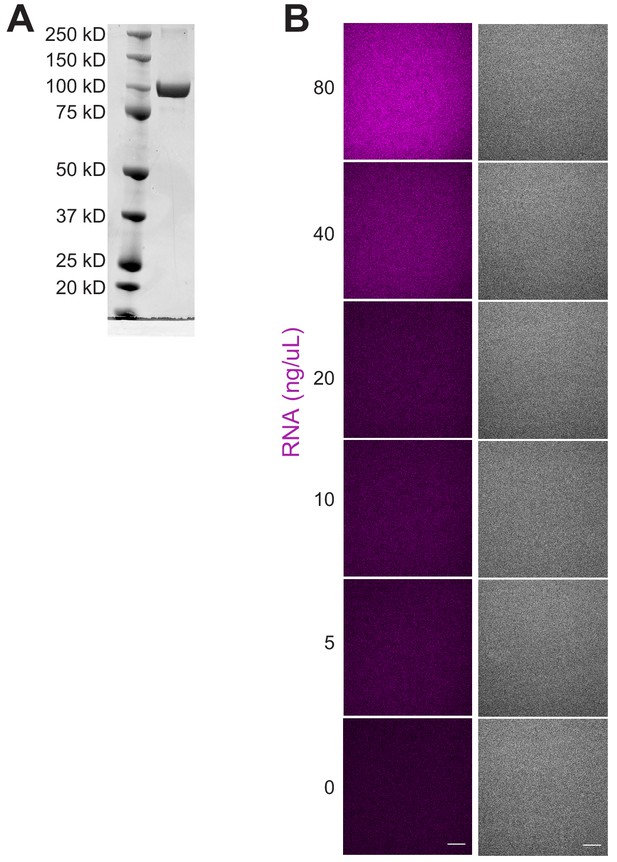
MEG-3 Purification and RNA only condensation.
(A) Gel showing purified recombinant 6Xhis-tagged MEG-3 protein stained with SimplyBlue SafeStain. (B) Representative fluorescence (left panels) or DIC (right panels) photomicrographs of control RNA-only condensation reactions under the same conditions used in Figure 4A (150 mM salt). Reactions contained indicated concentrations of nos-2 RNA trace labeled with Alexa488. No condensates are observed. Scale bar is 20 µm.
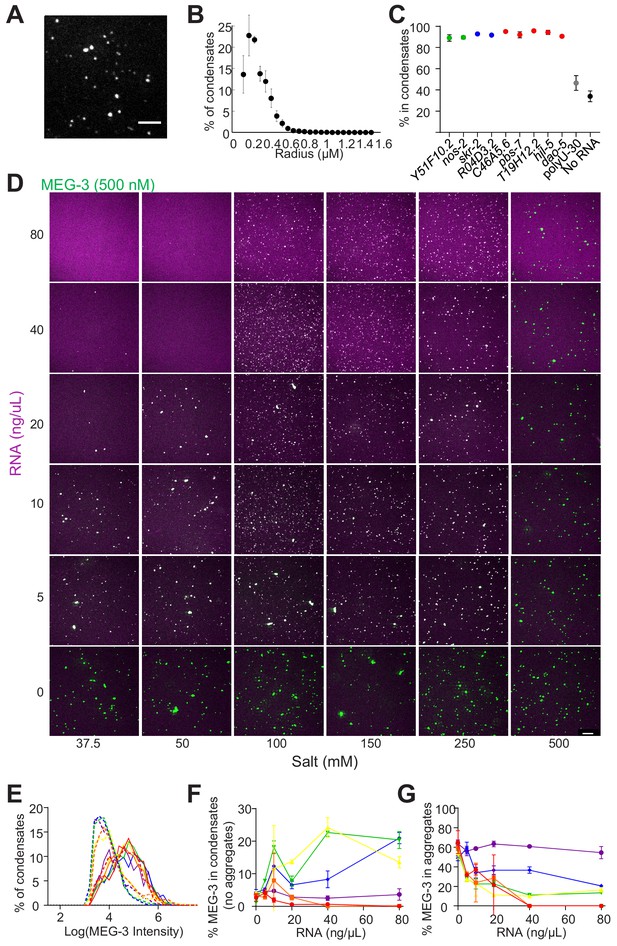
RNA modulates MEG condensation dependent on salt concentration.
(A) Representative photomicrograph of condensates of MEG-3 and nos-2 RNA after incubation in condensation buffer and imaged with a 100X objective. Scale bar is 5 µm. Reactions contained 500 nM MEG-3 and 40 ng/µL nos-2. MEG-3 (white) was trace labeled with Alexa647 and RNA (not shown) was trace labeled with Alexa488. (B) Histograms of MEG-3 condensate radii normalized to the total number of condensates in each reaction assembled as in (A). Each point represents data from 64 images collected from four experimental replicates. Circles indicate the mean and bars represent the SD. (C) Percent of MEG-3 in condensates for each RNA indicated assembled as in Figure 4A. Condensates were defined as objects with a MEG-3 intensity of Log (I) ≤ 4.6 from histograms in Figure 4B (Materials and methods). Each data point represents data from 12 images collected from three experimental replicates. Circles indicate the mean and bars represent the SD. (D) Representative photomicrographs of MEG-3/nos-2 RNA condensation reactions. Reactions contained 500 nM MEG-3 and indicated concentration of nos-2 RNA, and salt. MEG-3 was trace labeled with Alexa647 (green) and RNA was trace labeled with Alexa488 (magenta). Scale bar is 20 µm. Data are summarized in Figure 4D. (E) Histograms of MEG-3 intensity (log10 scale) normalized to the total number of condensates/aggregates in each reaction assembled as (D). Each histogram includes data from eight images collected from two experimental replicates. Lines represent reactions assembled in 150 mM salt (dashed lines) or 500 mM salt (solid lines) for RNA concentrations of 0 ng/µL (red), 5 ng/µL (orange), 10 ng/µL (yellow), 20 ng/µL (green), 40 ng/µL (blue), 80 ng/µL (purple). (F) Percent of MEG-3 in condensates (Log (I) ≤ 4.5 from histograms in (E) plotted vs. RNA concentration in the presence of increasing salt concentrations assembled as in (D). Each data set represents eight images collected from two experimental replicates. Circles indicate the mean and represent reactions assembled in salt concentrations of 37.5 mM (red), 50 mM (orange), 100 mM (yellow), 150 mM (green), 250 mM (blue), and 500 mM (purple). Bars represent the SD. (G) Percent of MEG-3 in aggregates (Log (I) > 4.5 in histograms in (E) plotted vs. RNA concentration in the presence of increasing salt concentration assembled as in (D). Each data set represents 8 images from two experimental replicates. Circles indicate the mean and represent reactions assembled in salt concentrations of 37.5 mM (red), 50 mM (orange), 100 mM (yellow), 150 mM (green), 250 mM (blue), and 500 mM (purple). Bars represent the SD.
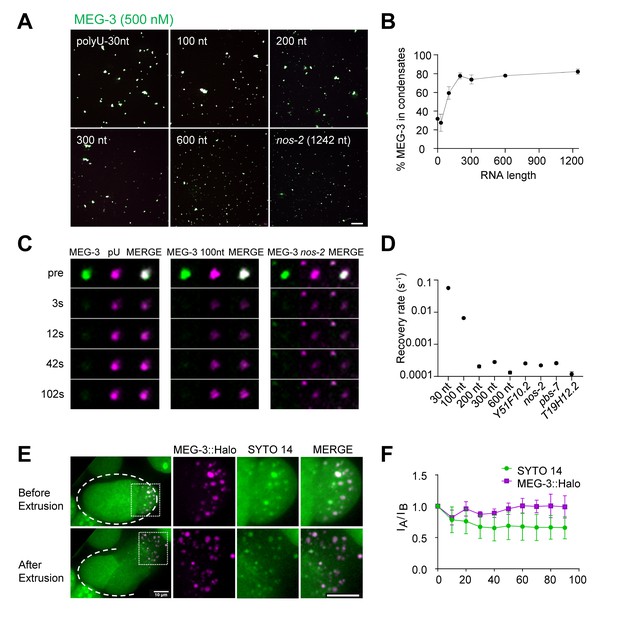
Long RNAs stably associate with the MEG gel phase.
(A) Representative photomicrographs of MEG-3 condensation reactions with indicated RNAs. 100–600 nt RNAs are fragments of nos-2 (Materials and methods). Reactions contained 500 nM MEG-3 and 20 ng/µL RNA, and salt. MEG-3 (green) was trace labeled with Alexa647 and nos-2 RNAs (magenta) were trace labeled with Alexa488 or Alexa546, polyU-30nt was trace labeled with fluorescein (Materials and methods). Scale bar is 20 µm. (B) Percent of MEG-3 in condensates plotted vs. RNA length assembled as in (A). Condensates were defined as objects with a MEG-3 intensity of Log (I) ≤ 4.6 from histograms in Figure 5—figure supplement 1A. Each data set includes condensates from 12 images collected from three experimental replicates. Circles indicate the mean and bars represent the SD. (Materials and methods). (C) Representative images showing fluorescence recovery after partial photobleaching (FRAP) of condensates assembled as in (A) and incubated for 30 min in condensate buffer. (D) Graph showing rates of fluorescence recovery after photobleaching (FRAP) for indicated RNAs in MEG-3 condensates. Values were normalized to initial fluorescence intensity, corrected for photobleaching and plotted. Circles indicate the mean (n > 6) and bars represent the SD. Refer to Figure 5—figure supplement 1B for time traces. (E) Time-lapse photomicrographs of a four-cell embryo expressing MEG-3::Halo and stained with SYTO 14 before and 30 s after laser puncture of the eggshell. MEG-3::Halo and SYTO 14 persist in the granule phase. Scale bar is 10 μm. Quantified in Figure 5F. (F) Graphs showing the fraction of MEG-3::Halo or SYTO 14 retained in the condensate phase after extrusion from embryos normalized to the fraction before extrusion (time 0). Total Halo or SYTO 14 fluorescence in granules was measured before laser puncture (IB) and after laser puncture (IA), corrected for photobleaching and used to calculate a fluorescence ratio (IA/IB). Means are indicated along with error bars representing ± SD calculated from five embryos.
-
Figure 5—source data 1
Data used to generate Figure 5B.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Data used to generate Figure 5D.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Data used to generate Figure 5F.
- https://cdn.elifesciences.org/articles/52896/elife-52896-fig5-data3-v2.xlsx
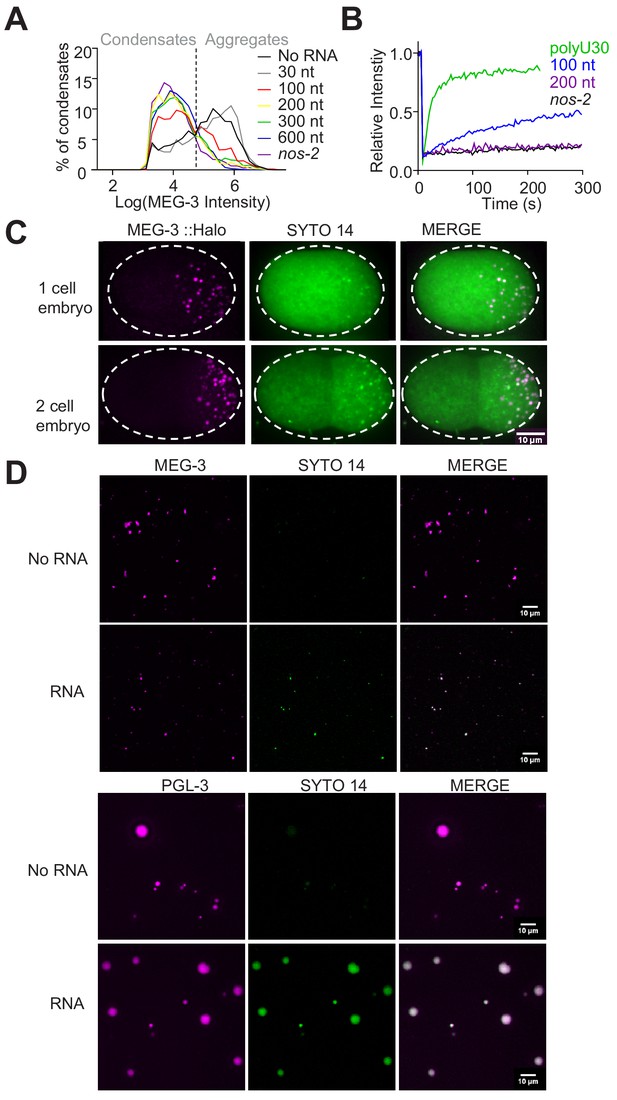
RNA modulates MEG condensation dependent on RNA length.
(A) Histograms of MEG-3 intensity (log10 scale) normalized to the total number of condensates/aggregates in each reaction assembled as in Figure 5A. Each histogram includes data from 12 images collected from three experimental replicates. Lines represent reactions assembled for the indicated RNA. (B) Graph showing fluorescence recovery after photobleaching (FRAP) as described in (Figure 5C, (D). The RNA intensity of condensates (left panels, n > 6) was measured every 3 s for 300 s before and after bleaching. Values were normalized to initial fluorescence intensity, corrected for photobleaching and plotted as an average. Refer to Figure 5D recovery rates. (C) Photomicrographs of one and two-cell embryos expressing MEG-3::Halo and stained with SYTO 14. Scale bar is 10 μm. (D) Representative photomicrographs of condensates of MEG-3 or PGL-3 assembled with and without RNA in the presence of 100 nM SYTO 14 for 30 min. Reactions contained 500 nM MEG-3 or 5 µM PGL-3 and 20 ng/µL nos-2 (Methods). MEG-3 (magenta) and PGL-3 were trace labeled with Alexa647(Materials and methods). Scale bars are 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background C. elegans | JH3503 | Smith et al., 2016 | meg-3(ax3054[meg-3::meGFP]) X. | |
| Strain, strain background C. elegans | JH3269 | Putnam et al., 2019 | pgl-1(ax3122[pgl-1::GFP]) IV. | |
| Strain, strain background C. elegans | JH3193 | Paix et al., 2014 | nos-2(ax2049[3xFLAG::nos-2]) II. | |
| Strain, strain background C. elegans | JH3605 | This study | Y51F10.2(ax4319[Y51F10.2::OLLAS]) I | |
| Strain, strain background C. elegans | EGD364 | Wu et al., 2019 | meg-3(egx4[meg-3::Halo]) X. | |
| Strain, strain background C. elegans | JH3475 | Smith et al., 2016 | meg-3(ax3055) meg-4(ax3052) X | |
| Strain, strain background C. elegans | WM527 | Shen et al., 2018 | prg-1(ne4523 [gfp::tev::flag::prg-1]) I | |
| Strain, strain background C. elegans | JH3357 | Lee et al., 2017 | nos-2(ax3103[nos-2△]) II | |
| Strain, strain background C. elegans | SS608 | Kawasaki et al., 2004 | pgl-3(bn103[pgl-3△]) V | |
| Strain, strain background C. elegans | SX922 | Caenorhabditis Genetics Center | prg-1(n4357[prg-1△])I | |
| Strain, strain background C. elegans | JH3229 | Wang et al., 2014 | meg-1(vr10) meg-3(tm4259)X | |
| Strain, strain background C. elegans | JH3740 | This study | meg-3(ax3055) meg-4(ax3052) X; Y51F10.2(ok1610) I | |
| Strain, strain background C. elegans | JH3743 | This study | nos-2(ax3130) II; Y51F10.2(ok1610) I | |
| Strain, strain background C. elegans | JH3746 | This study | meg-3(ax3055) meg-4(ax3052) X; nos-2(ax3130) II. 100% sterile, no clone was maintained. | |
| Strain, strain background C. elegans | JH1904 | This study | Unc-119(ed3) III; axls1374[Ppie1::GFP] | |
| Strain, strain background C. elegans | JH2878 | Leacock and Reinke, 2008 | meg-1(vr10) X | |
| Strain, strain background C. elegans | JH3562 | This study | meg-3(ax3054[MEG-3::meGFP]) X; K08F4.2 (ax5000[gtbp-1::tagRFP]) IV | |
| Strain, strain background C. elegans | RB1413 | Caenorhabditis Genetics Center | Y51F10.2(ok161) I | |
| Antibody | K76 | DSHB,PMID: 28787592 | RRID:AB_531836 | (1:15) |
| Antibody | Anti-FLAG M2 | Sigma-Aldrich Cat# F3165 | RRID:AB_259529 | (1:200) |
| Antibody | Donky-anti-mouse IgM 647 | Jackson ImmunoResearch Labs | RRID:AB_2340861 | (1:400) |
| Antibody | Goat anti-Rabit IgG (H+L) 568 | Molecular probes cat# A-11011 | RRID:AB_143157 | (1:400) |
| Antibody | Anti-OLLAS-L2 | Novus cat# NBP1-06713 | RRID:AB_1625979 | (1:200) |
| Antibody | Anti-OLLAS | other | gift from Dr. Jeremy Nathans | |
| Antibody | Anti-GFP | Rohe | RRID:AB_390913 | For conjugation |
| Sequence-based reagent | oCYL1089: crRNA to cut Y51F10.2 at 3' end | This study | GTGCTCAAAATAGTAGGCGA | |
| Sequence-based reagent | oCYL1143: repair oligo of Y51F10.2 C-ter Ollas tag (+) | This study | TCCAGCGCCAGCACCACCATTCGAC AACTCCGTCGCCTACTATTTTGGAGGAT CCGGAtccggattcgccaacGAGCTCggac cacgtctcatgggaaagGGAGGATCCGG AGAGCACCAATTTTGA gcttttatatttttttttctc | |
| Sequence-based reagent | oCYL1144: repair oligo of Y51F10.2 C-ter Ollas tag (-) | This study | gagaaaaaaaaatataaaagc TCAAAATTGGTGCTCTCCGGATCCTC CctttcccatgagacgtggtccGAGCTCgtt ggcgaatccggaTCCGG ATCCTCCAAAAT AGTAGGCGACGGAGTTGTCGA ATGGTGGTGCTGGCGCTGGA | |
| Sequence-based reagent | oCYL1096:5' PCR primer 333 bp up of Y51F10.2 TGA stop | This study | GTTTCCAGCCGCTTGACAAG | |
| Sequence-based reagent | GTTTCCAGCCGCTTGACAAG | This study | CTGATCCTCCCCCTTCTTCG | |
| Sequence-based reagent | oCYL1259:5' PCR primer contains T7 promoter for in vitro transcription of T19H12.2 mRNA. | This study | CATGATTACTAATACG ACTCACTATA GGGaccagctcacga aactaacaatg | |
| Sequence-based reagent | oCYL1260:3' PCR primer at the end of T19H12.2 3UTR for T7 in vitro transcription | This study | gaaagcgaaagaaatttt attttacaggagg | |
| Sequence-based reagent | oCYL1261:5' PCR primer contains T7 promoter for in vitro transcription of dao-5 mRNA including utr. | This study | catgattacTAATACGACT CACTATAGGG ggtacccctgatcgctATGAG | |
| Sequence-based reagent | oCYL1262:3' PCR primer at the end of dao-5 3UTR for T7 in vitro transcription | This study | ggaccaaacattttatggat gagacaag | |
| Sequence-based reagent | oCYL1263:5' PCR primer contains T7 promoter forin vitro transcription of hil-5 mRNA. | This study | catgattacTAATACGACT CACTATAGGG actatcacttttcaagtgtttgttcatcg | |
| Sequence-based reagent | oCYL1264:3' PCR primer at the end of hil-5 3UTR for T7 in vitro transcription | This study | agaatctattaatggtttattggaa ggtatatttgttaaaatg | |
| Sequence-based reagent | oCYL1265:5' PCR primer contains T7 promoter forin vitro transcription of pbs-7 mRNA including utr. | This study | catgattacTAATACGACT CACTATAGGG gcatttcattgtcgaaattcacttcctttc | |
| Sequence-based reagent | oCYL1266:3' PCR primer at the end of pbs-7 3UTR for T7in vitro transcription | This study | agaaggattaaatggaag tttatttatcgacttc | |
| Sequence-based reagent | oCYL1267:5' PCR primer contains T7 promoter for in vitro transcription of T07C4.3a mRNA including utr. | This study | catgattacTAATACGACT CACTATAGGG gtttgtgcactcactacgaaatctc | |
| Sequence-based reagent | oCYL1268:3' PCR primer at the end of T07C4.3a 3UTR for T7 in vitro transcription | This study | catcaaaatattctttcatt taacaaaaacagaaacaac | |
| Recombinant DNA reagent | plasmid: 6XHis-MEG-3 | Smith et al., 2016 | ||
| Recombinant DNA reagent | plasmid: MBP-HIS-TEV-PGL-3 | Putnam et al., 2019 | ||
| Chemical compound, drug | SYTO 14 | ThermoFisher Cat#S7572 | In vivo RNA labeling | |
| Chemical compound, drug | Alexa Fluor 647 NHS Ester | ThermoFisher Cat#A37573 | protein labeling | |
| Chemical compound, drug | DyLight 488 NHS Ester | ThermoFisher Cat#46403 | protein labeling | |
| Chemical compound, drug | ChromaTide Alexa Fluor 488–5-UTP | ThermoFisher Cat#C11403 | RNA labeling | |
| Chemical compound, drug | ChromaTide Alexa Fluor 546–14-UTP | ThermoFisher Cat#C11404 | RNA labeling | |
| Recombinant DNA reagent | plasmid: cDNA of pbs-7 | this paper | pbs-7 cDNA, pUC19 vector | |
| Recombinant DNA reagent | plasmid: cDNA of dao-5 | this paper | dao-5 cDNA, pUC19 vector | |
| Recombinant DNA reagent | plasmid: cDNA of T19H12.2 | this paper | T19H12.2 cDNA, pUC19 vector | |
| Recombinant DNA reagent | plasmid: cDNA of hil-5 | this paper | hil-5, pUC19 vector | |
| Recombinant DNA reagent | plasmid: cDNA of Y51F10.2 | this paper | Y51F10.2 cDNA, PCR blunt II topo vector | |
| Recombinant DNA reagent | plasmid: cDNA of nos-2 | this paper | nos-2 cDNA, PCR blunt II topo vector | |
| Recombinant DNA reagent | plasmid: cDNA of skr-2 | this paper | skr-2 cDNA, PCR blunt II topo vector | |
| Recombinant DNA reagent | plasmid: cDNA of R04D3.2 | this paper | R04D3.2 cDNA, PCR blunt II topo vector | |
| Recombinant DNA reagent | plasmid: cDNA of C46A5.6 | this paper | C46A5.6 cDNA, PCR blunt II topo vector | |
| Software, algorithm | DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | RRID:SCR_015687 | |
| Software, algorithm | hisat2 | DOI:10.1038/nprot.2016.095 | RRID:SCR_015530 | |
| Software, algorithm | htseq-count | DOI:10.1093/bioinformatics/btu638 | RRID:SCR_011867 | |
| Software, algorithm | cuffdiff | http://cole-trapnell-lab.github.io/cufflinks/ | RRID:SCR_001647 | |
| Software, algorithm | Slidebook 6 | https://www.intelligent-imaging.com/slidebook | RRID:SCR_014300 | |
| Software, algorithm | Deeptools | https://deeptools.readthedocs.io/en/develop/ | RRID:SCR_016366 | |
| Software, algorithm | icount | https://github.com/tomazc/iCount | RRID:SCR_016712 | |
| Software, algorithm | smatools | http://samtools.sourceforge.net/ | RRID:SCR_002105 | |
| Software, algorithm | BEDTools | https://github.com/arq5x/bedtools2 | RRID:SCR_006646 | |
| Software, algorithm | Galaxy | https://usegalaxy.eu/ | RRID:SCR_006281 | |
| Software, algorithm | Rstusio | http://www.rstudio.com/ | RRID:SCR_000432 | |
| Software, algorithm | STAR | https://github.com/alexdobin/STAR | RRID:SCR_015899 |
Additional files
-
Supplementary file 1
Gene list of MEG-3 bound transcripts, P granule transcripts and PGL-1 bound transcripts.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp1-v2.csv
-
Supplementary file 2
Ribosome footprint for P-blastomere enriched genes.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp2-v2.csv
-
Supplementary file 3
Differential gene expression analysis of wild type and meg-3meg-4 embryos.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp3-v2.xlsx
-
Supplementary file 4
Differential Translation analysis of wild type and meg-3meg-4 embryos.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp4-v2.xlsx
-
Supplementary file 5
Sequencing library information.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp5-v2.xlsx
-
Supplementary file 6
Read counts for iCLIP experiments.
- https://cdn.elifesciences.org/articles/52896/elife-52896-supp6-v2.csv
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52896/elife-52896-transrepform-v2.docx