TorsinB overexpression prevents abnormal twisting in DYT1 dystonia mouse models
Figures
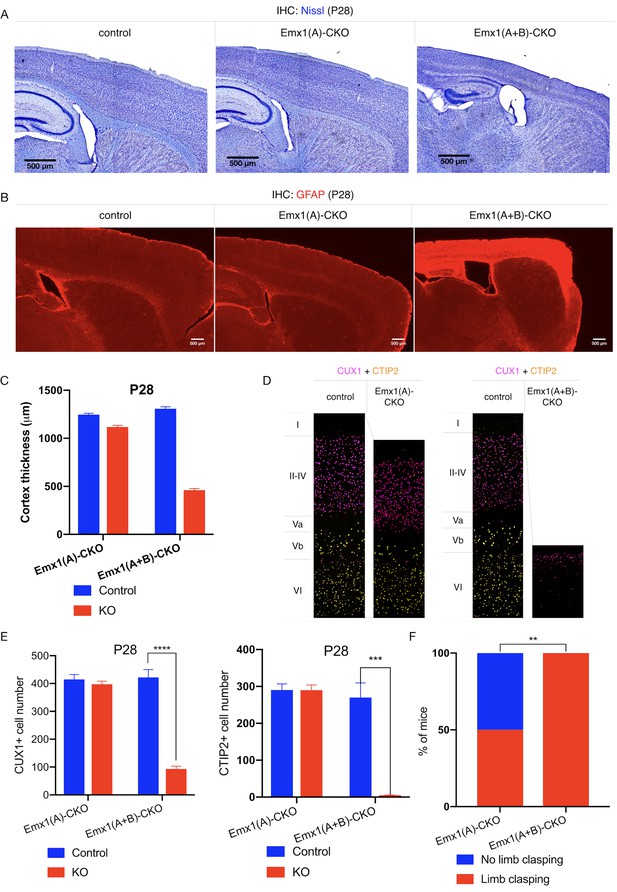
TorsinB deletion worsens torsinA-related motor and neuropathological phenotypes.
(A) Nissl staining of P28 Emx1(A)-CKO and Emx1(A+B)-CKO mice. Emx1(A+B)-CKO mice exhibit significant atrophy of Cre-expressing brain regions including cortex (*) and hippocampus (^). (B) GFAP staining of P28 Emx1(A)-CKO and Emx1(A+B)-CKO brains. Emx1(A+B)-CKO mice exhibit severe reactive gliosis in Cre-expressing regions, including the cerebral cortex and the hippocampus. (C) Cortical thickness of P28 Emx1(A)-CKO and Emx1(A+B)-CKO mice. Cortical thickness is reduced by 10.4% in Emx1(A)-CKO mice (unpaired t-test t16 = 5.834, p<0.0001; control n = 9, Emx1(A)-CKO n = 9). Cortical thickness is reduced by 64.8% in Emx1-dCKO (unpaired t-test t11 = 30.16, p<0.0001; control n = 4, Emx1(A+B)-CKO n = 9). (D) Representative images from CUX1 and CTIP2 stained cerebral cortex of Emx1(A)-CKO and Emx1(A+B)-CKO mice and their respective littermate controls. (E) CUX1 and CTIP2 counts in P28 Emx1(A)-CKO and Emx1(A+B)-CKO mice. CUX1+ neurons are not significantly reduced in Emx1(A)-CKO mice (unpaired t-test t16 = 0.8469, p=0.4095; control n = 9, Emx1(A)-CKO n = 9). CUX1+ cells are significantly reduced in Emx1(A+B)-CKO mice (77.0% reduction; unpaired t-test t8 = 12.86, p<0.0001, control n = 4, Emx1(A+B)-CKO n = 6). CTIP2+ neurons are not significantly reduced in Emx1(A)-CKO sensorimotor cortex (unpaired t-test t16 = 0.02552, p=0.98; control n = 9; Emx1(A)-CKO n = 9). CTIP2+ neurons are significantly reduced in Emx1(A+B)-CKO sensorimotor cortex (98.6%; unpaired t-test t7 = 7.636, p=0.0001, control n = 4, Emx1(A+B)-CKO n = 5). (F) Proportion of Emx1(A)-CKO and Emx1(A+B)-CKO mice exhibiting limb clasping during tail suspension. A significantly greater proportion of Emx1(A+B)-CKO compared to Emx1(A)-CKO mice exhibit limb clasping during tail suspension (Chi square test χ2 = 9.1, p=0.0026; Emx1(A)-CKO n = 12, Emx1(A+B)-CKO n = 14).
-
Figure 1—source data 1
Behavioral and histological data on Emx1(A)-CKO and Emx1(A+B)-CKO mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig1-data1-v2.xlsx
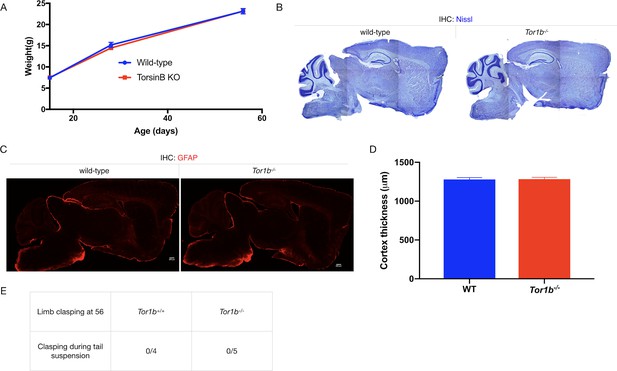
TorsinB null mice exhibit no apparent organismal or neuropathological phenotypes.
(A) Tor1b-/- postnatal growth. Tor1b-/- mutants exhibit no significant changes in body weight from age P15 to P56 compared to controls (two-way repeated measures ANOVA genotype x age, interaction F2, 14 = 0.6325, p=0.5458; wild-type n = 4, torsinB KO n = 5). (B) Nissl analysis of P56 Tor1b-/- brains. Tor1b-/- mutants did not exhibit any differences in brain morphology compared to littermate controls. (C) GFAP staining of P56 Tor1b-/- brains. Tor1b-/- mutants did not exhibit gliosis. (D) Cortical thickness measurements of P56 Tor1b-/- mice. Cortical thickness does not differ significantly between WT and Tor1b-/- mice (unpaired t-test t6 = 0.09577, p=0.9268; wild-type n = 4, torsinB KO n = 4). (E) Proportion of P56 Tor1b-/-- mice and WT littermates that exhibit limb clasping during tail suspension. No mice of either genotype exhibited limb clasping.
-
Figure 1—figure supplement 1—source data 1
Data on histological analysis of torsinB KO mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig1-figsupp1-data1-v2.xlsx
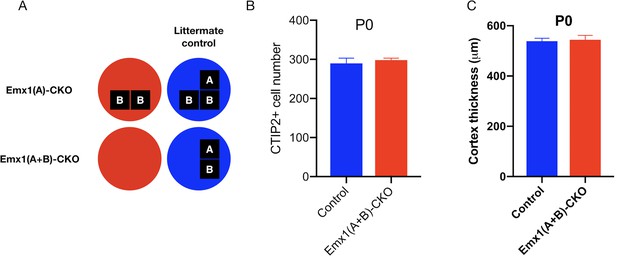
Emx1(A+B)-CKO mice do not exhibit neuropathological abnormalities at birth.
(A) Illustration of examined genotypes. Each row of boxes within the circles denote the presence or absence of Tor1a (top row) or Tor1b (bottom row) alleles following Cre recombination. The presence of a letter (‘A’ or ‘B’) indicates an intact allele, whereas the absence of a letter indicates a deleted allele. (B) CTIP2 neuron counts in P0 Emx1(A+B)-CKO sensorimotor cortex. CTIP2+ neuronal counts in Emx1(A+B)-CKO mice do not differ significantly from littermate controls at P0 (non-parametric Mann-Whitney test U = 38.5, p=0.1911; control n = 13, Emx1(A+B)-CKO n = 9). (C) Cortical thickness measurements at P0 in Emx1(A+B)-CKO mice. Cortical thickness does differ significantly at P0 in Emx1(A+B)-CKO mutants (unpaired t-test t20 = 0.2663, p=0.7927; control n = 13, Emx1(A+B)-CKO n = 9).
-
Figure 1—figure supplement 2—source data 1
Data on P0 characterization of Emx1(A+B)-CKO mouse brains.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig1-figsupp2-data1-v2.xlsx
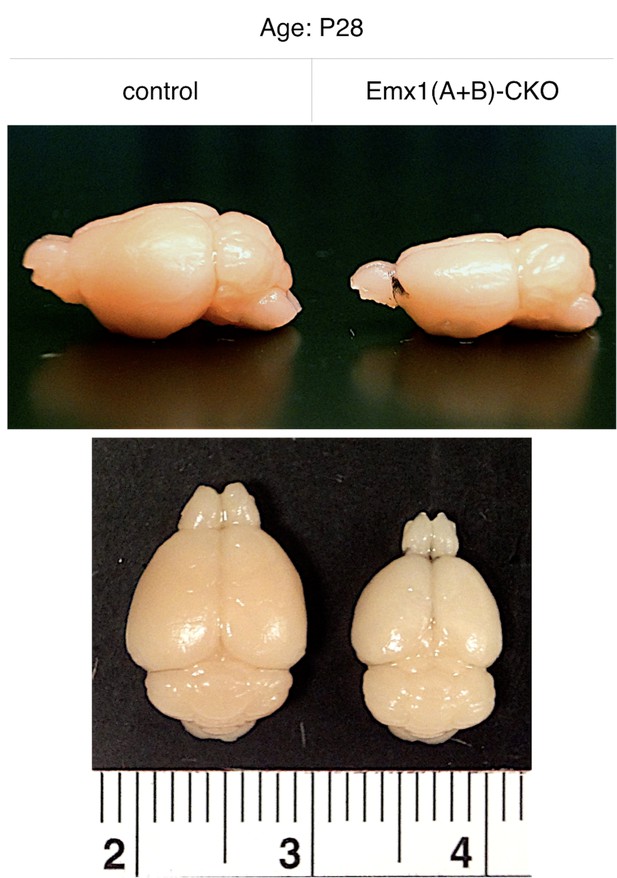
Emx1(A+B)-CKO neuropathological abnormalities that emerge postnatally are restricted to the forebrain.
Gross morphology of Emx1(A+B)-CKO and control brains. Emx1(A+B)-CKO causes grossly apparent atrophy in the forebrain Cre field while hindbrain morphology appears unchanged.
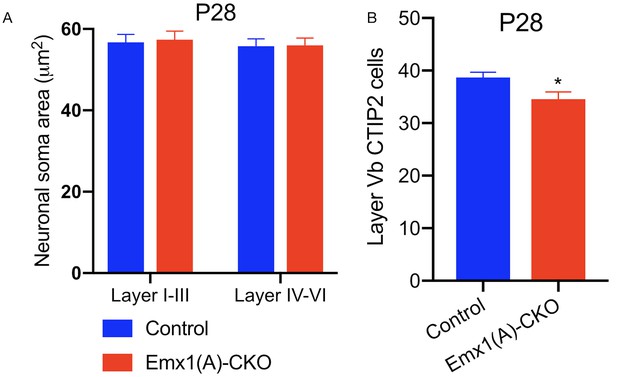
Further assessment of cortical neurons in Emx1(A+B)-CKO mice.
(A) Cortical neuron soma size in Emx1-CKO. Neuronal soma size is unchanged in Emx1(A)-CKO cortical neurons of layer I-III or layer IV-VI (two-way ANOVA main effect of genotype F1, 396 = 0.04729, p=0.8280, main effect of layers F1, 396 = 0.3768 p=0.5397, interaction F1, 396 = 0.01256, p=0.9108, control n = 100, Emx1(A)-CKO n = 100). (B) CTIP2 neuron counts in layer Vb of Emx1-CKO sensorimotor cortex at P28. Emx1-CKO mice undergo a 10.6% reduction of CTIP2 neurons in layer Vb (unpaired t-test t16 = 2.383, p=0.0299, control n = 9, Emx1(A)-CKO n = 9). Source data for Figure 1—figure supplement 4 can be found in Figure 1—figure supplement 4—source data 1. Each tab of the spreadsheet is titled with a letter matching the corresponding figure panel.
-
Figure 1—figure supplement 4—source data 1
Information pertaining to further characterization of cortical neurons in Emx1(A)-CKO mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig1-figsupp4-data1-v2.xlsx
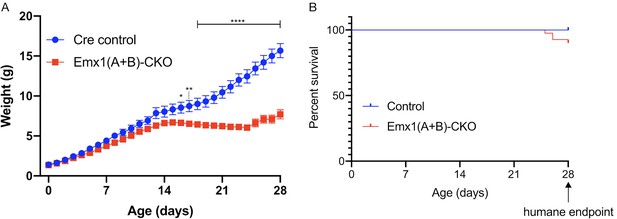
Emx1(A+B)-CKO growth and survival curves.
(A) Growth curves of Cre control and Emx1(A+B)-CKO mice. Emx1(A+B)-CKO mice exhibit reduced postnatal growth (two-way repeated measures ANOVA main effect of genotype F1, 19 = 27.14, p<0.0001, main effect of age F28, 532 = 240.3 p<0.0001, interaction F28, 532 = 46.89, p<0.0001; asterisks denote the following p-values from Sidak’s multiple comparisons tests: *=p < 0.05, **=p < 0.01, **=p < 0.001, ****=p < 0.0001; Cre control n = 9, Emx1(A+B)-CKO n = 12). (B) Survival curves of Cre control and Emx1(A+B)-CKO mice. Emx1(A+B)-CKO mice exhibit lethality starting the 3rd postnatal week, and the humane endpoint for survival and behavioral analysis is P28 (Gehan-Breslow-Wilcoxon method χ2 = 5.256, p=0.0219; Cre control n = 41, Emx1(A+B)-CKO n = 41). Source data for Figure 1—figure supplement 5 can be found in Figure 1—figure supplement 5—source data 1. Each tab of the spreadsheet is titled with a letter matching the corresponding figure panel.
-
Figure 1—figure supplement 5—source data 1
Growth and survival data on Emx1(A+B)-CKO mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig1-figsupp5-data1-v2.xlsx
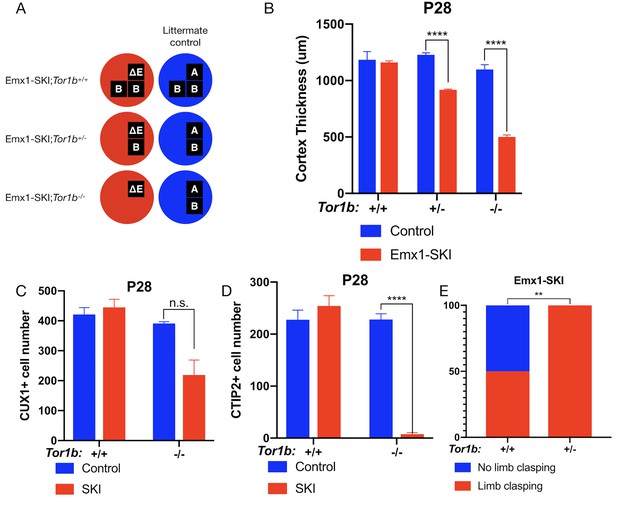
TorsinB dose-dependently worsens a DYT1 model.
(A) Illustration of examined genotypes. Each row of boxes within the circles denote the presence or absence of Tor1a (top row) or Tor1b (bottom row) alleles following Cre recombination. The presence of a letter (‘A’ ‘ΔE’ or ‘B’) indicates an intact allele, whereas the absence of a letter indicates a deleted allele. (B) Cortical thickness of P28 Emx1-SKI;Tor1b+/+, Emx1-SKI;Tor1b+/-, and Emx1-SKI;Tor1b-/- mice. TorsinB loss reduces cortical thickness in Emx1-SKI mice in a dose-dependent manner (2% reduction in Emx1-SKI;Tor1b+/+, 25.2% reduction in Emx1-SKI;Tor1b+/-, and 54.5% decrease in Emx1-SKI;Tor1b-/-; two way ANOVA main effect of background genotype F1,13 = 128.3, p<0.0001, age F2,13 = 62.65, p<0.0001, and interaction F2,13 = 36.98, p<0.0001; Sidak’s multiple comparisons test p=0.9323 for Tor1b+/+, p<0.0001 for Tor1b+/-, p<0.0001 for Tor1b-/-; Tor1b+/+ control n = 3; Emx1-SKI n = 5, Tor1b+/- control n=3, Emx1-SKI n = 3, Tor1b-/- control n=2, Emx1-SKI n = 3). (C) CUX1+ cell counts in sensorimotor cortex of P28 Emx1-SKI;Tor1b+/+ and Emx1-SKI;Tor1b-/- mice. There is no significant reduction in the number of CUX1+ cells in Emx1-SKI;Tor1b+/+ sensorimotor cortex (unpaired t-test t16 = 0.8469, p=0.4095; control n = 3, SKI n = 5). Simultaneous deletion of two torsinB alleles reduces CUX1+ cell counts by 44.0% (unpaired t-test t3 = 2.655, p=0.0766; control n = 2, SKI n = 3) though this reduction does not reach statistical significance. (D) CTIP2+ cell counts in sensorimotor cortex of P28 Emx1-SKI;Tor1b+/+ and Emx1-SKI;Tor1b-/- mice. CTIP2+ neuronal cell counts are not reduced in Emx1-SKI;Tor1b+/+ mice (unpaired t-test t6 = 0.8844, p=0.4105; control n = 3, SKI n = 5). CTIP2+ cell counts in Emx1-SKI;Tor1b-/- are significantly reduced by 96.6% (unpaired t-test t3 = 24.44, p<0.0001; control n = 2, SKI n = 3). (E) Proportion of Emx1-SKI;Tor1b+/+ and Emx1-SKI;Tor1b+/- mice exhibiting limb clasping during tail suspension. A significantly greater proportion of Emx1-SKI;Tor1b+/- mice exhibit limb clasping during tail suspension compared to Emx1-SKI;Tor1b+/+ (Chi square test χ2 = 7.441, p=0.0064; Emx1-SKI;Tor1b+/+n = 12, Emx1-SKI;Tor1b+/- n = 11).
-
Figure 2—source data 1
Raw data on histological and behavioral characterization of Emx1-SKI mice with two, one, and zero intact torsinB alleles.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig2-data1-v2.xlsx
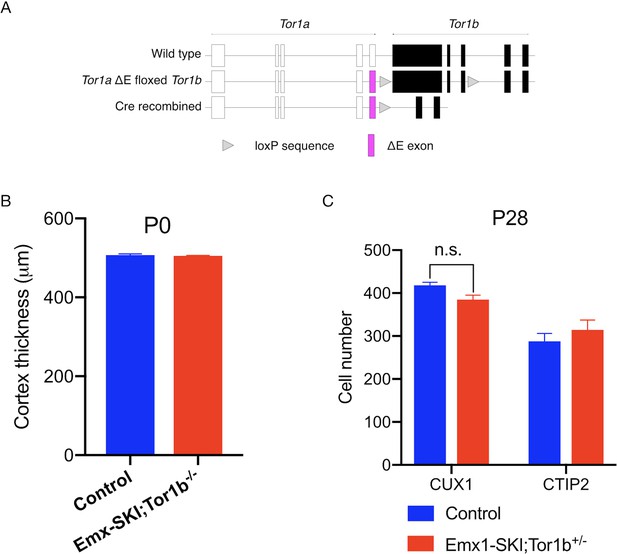
Novel allele design and histological analysis of Emx1-SKI;Tor1b-/- mice and Emx1-SKI;Tor1b+/- mice.
(A) Design of the Tor1aΔE floxed Tor1b allele. (B) Cortical thickness of P0 Emx1-SKI-B0 mice. The thickness of the Emx1-SKI-B0 cortical tissue is unchanged at P0 (unpaired t-test t3 = 0.5425, p=0.6252; control n = 3, Emx1-SKI;Tor1b-/- n = 2). (C) CUX1+ and CTIP2+ counts in the sensorimotor cortex of P28 Emx1-SKI;Tor1b+/- mice. There is a decrease of CUX1+ cells by 8.0% (unpaired t-test t4 = 2.705, p=0.0538; control n = 3, Emx1-SKI;Tor1b+/- n = 5) that does not reach statistical significance in Emx1-SKI;Tor1b+/- mice compared to Cre controls. There is no reduction of total CTIP2+ neurons in the Emx1-SKI;Tor1b+/- mice compared to Cre controls (unpaired t-test t4 = 0.9128, p=0.4130; control n = 3, Emx1-SKI;Tor1b+/- n = 5).
-
Figure 2—figure supplement 1—source data 1
Further histological characterization of Emx1-SKI mice with varying amounts of torsinB.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig2-figsupp1-data1-v2.xlsx
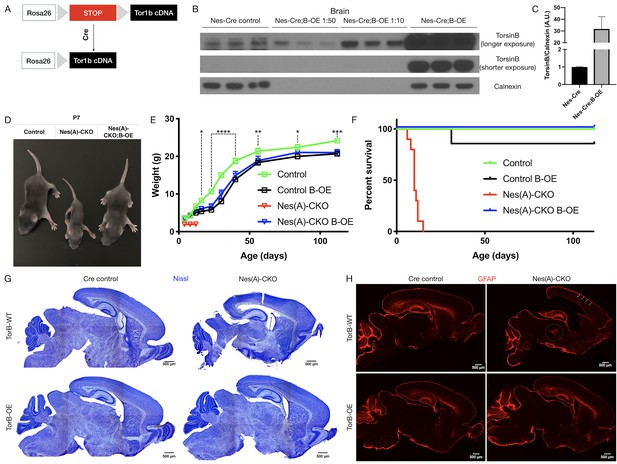
TorsinB augmentation eliminated all phenotypes in CNS conditional torsinA null mice.
(A) Cartoon illustration of ROSA26 locus engineered to express torsinB. Top: In absence of Cre, torsinB expression is prevented by a floxed ‘STOP’ cassette. Bottom: TorsinB is expressed following Cre deletion of the floxed ‘STOP’ cassette. (B) Western blot analysis of whole brain lysates probed with an anti-torsinB antibody. Mice expressing both the Nestin-Cre and B-OE alleles exhibit Cre-dependent overexpression of torsinB. (C) Quantification of western blots of whole brain lysates from Nestin-Cre and Nestin-Cre;B-OE littermates probed with anti-torsinB antibody. The B-OE allele causes significant torsinB overexpression (unpaired t-test t4 = 2.947, p=0.0421; Nestin-Cre n = 3, Nestin-Cre;B-OE n = 3). Quantification was performed by comparing torsinB expression in Cre control samples to that of Nestin-Cre;B-OE samples diluted 1:50. (D) Image of Cre control, Nes(A)-CKO, and Nes(A)-CKO;B-OE littermates at P7 (in order left to right). TorsinB overexpression restores postnatal growth. (E) Growth curves of Cre control, Cre control;B-OE, N(esA)-CKO, and Nes(A)-CKO;B-OE mice. TorsinB overexpression almost entirely restores growth in N-CKO mice (two-way repeated measures ANOVA main effect of genotype F2, 17 = 14.16, p=0.0002, main effect of age F9, 153 = 775.7 p<0.0001, interaction F18, 153 = 6.727, p<0.0001; asterisks denote the following p-values from Tukey’s multiple comparisons tests: *=p < 0.05, **=p < 0.01, **=p < 0.001, ****=p < 0.0001; Cre control n = 7, Cre control;B-OE n = 6, N(A)-CKO;B-OE n = 7). (F) Survival curves of Cre control, Cre control;B-OE, Nes(A)-CKO, and Nes(A)-CKO;B-OE mice. TorsinB overexpression eliminates lethality in N-CKO mice (Gehan-Breslow-Wilcoxon method χ2 = 36.16, p<0.0001; Cre control n = 7, Cre control;B-OE n = 7, Nes(A)-CKO n = 10, Nes(A)-CKO;B-OE n = 7). (G) Nissl staining of P8 brains from Cre control, Cre control;B-OE, Nes(A)-CKO, and Nes(A)-CKO;B-OE mice. TorsinB overexpression eliminates the morphological defects characteristic of Nes(A)-CKO mice. (H) GFAP staining of P8 brains from Cre control, Cre control;B-OE, Nes(A)-CKO, and Nes(A)-CKO;B-OE mice. GFAP immunostaining illustrates the characteristic gliotic changes in Nes(A)-CKO cortex (arrows), thalamus, deep cerebellar nuclei, and hindbrain (circles). TorsinB overexpression eliminates gliotic changes in Nes(A)-CKO mice.
-
Figure 3—source data 1
Biochemical and organismal characterization of Nes(A)-CKO;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig3-data1-v2.xlsx

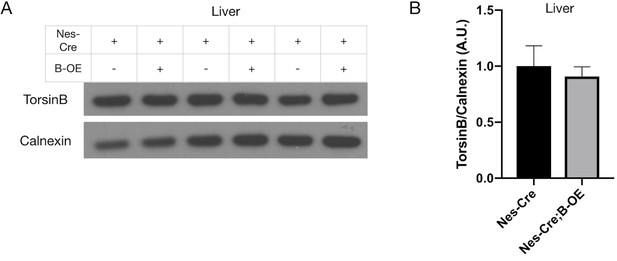
Quantification of torsinB in Nestin-Cre;B-OE liver tissue.
(A) Western blot of whole liver lysates from Nestin-Cre and Nestin-Cre;B-OE littermates probed with anti-torsinB antibody. (B) Quantification of western blots of whole brain lysates from Nestin-Cre and Nestin-Cre;B-OE littermates probed with anti-torsinB antibody. The B-OE allele does not alter torsinB overexpression in liver, which is outside of the Cre field (unpaired t-test t4 = 4560, p=0.6720; Nestin-Cre n = 3, Nestin-Cre;B-OE n = 3).
-
Figure 3—figure supplement 1—source data 1
Information pertaining to torsinA expression in torsinB overexpression brain tissue.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig3-figsupp1-data1-v2.xlsx
Analysis of torsinA expression in Nestin-Cre;B-OE brain tissue.
(A) Western blot of whole brain lysates from Nestin-Cre and Nestin-Cre;B-OE littermates probed with anti-torsinA antibody. (B) Quantification of western blots of whole brain lysates from Nestin-Cre and Nestin-Cre;B-OE littermates probed with anti-torsinA antibody. TorsinA expression is not altered in mice overexpressing torsinB (unpaired t-test t4 = 0.5566, p=0.6075; Nestin-Cre n = 3, Nestin-Cre;B-OE n = 3)).
-
Figure 3—figure supplement 2—source data 1
Information on liver torsinB expression levels.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig3-figsupp2-data1-v2.xlsx
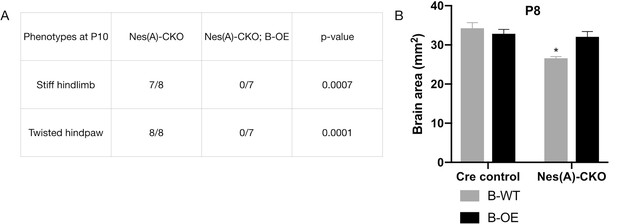
TorsinB overexpression prevents overtly abnormal postures and gross disruption of brain morphology in N-CKO mice.
(A) Table of overtly abnormal postural phenotypes displayed by N(A)-CKO and N(A)-CKO;B-OE mice at P10. TorsinB overexpression eliminates these phenotypes that are prevalent in N(A)-CKO mice (stiff hindlimb: Chi square test χ2 = 11.48, p=0.0007; twisted hindpaw: Chi square test χ2 = 15, p=0.0001). (B) Brain area measurements of P8 Nestin-Cre control, Cre control;B-OE, N(A)-CKO, and N(A)-CKO;B-OE. TorsinB overexpression prevents the reduction in brain size characteristic of N(A)-CKO mice (two-way ANOVA main effect of genotype F1, 8 = 13.54, p=0.0062, main effect of torsinB level F1, 8 = 3.103, p=0.1162, interaction F1,8 = 8.916, p=0.0174; asterisks denote the following p-values from Tukey’s multiple comparisons tests: *=p < 0.05; Cre control n = 3, Cre control;B-OE n = 3; N(A)-CKO n = 3, N(A)-CKO;B-OE n = 3).
-
Figure 3—figure supplement 3—source data 1
Behavioral and brain morphological characterization of Nes(A)-CKO;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig3-figsupp3-data1-v2.xlsx
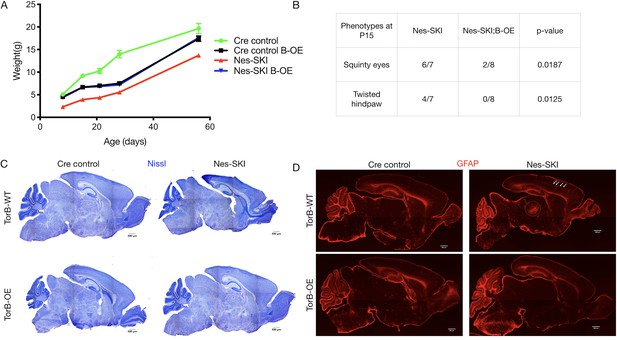
TorsinB overexpression rescues ΔE torsinA phenotypes.
TorsinB overexpression prevents striatal cholinergic interneuron degeneration and dystonic-like movements. (A) Growth curves of Cre control, Cre control;B-OE, Nes-SKI, and Nes-SKI;B-OE mice. Nes-SKI mice exhibited reduced postnatal growth, which was partially rescued by torsinB overexpression (two-way repeated measures ANOVA main effect of genotype F3, 26 = 60.65, p<0.0001, main effect of age F1.828, 47.53 = 794.8 p<0.0001, interaction F12, 104 = 9.831, p<0.0001; Cre control n = 7, Cre control;B-OE n = 8, Nes-SKI n = 7, Nes-SKI;B-OE n = 8). (B) Table of overtly abnormal postural and developmental phenotypes displayed by Nes-SKI and Nes-SKI;B-OE mice at P15. TorsinB overexpression reduces phenotypes that are prevalent in Nes-SKI mice (squinty eyes: Chi square test χ2 = 5.529, p=0.0187; twisted hindpaw: Chi square test χ2 = 6.234, p=0.0125). (C) Nissl staining of P10 brains from Cre control, Cre control;B-OE, Nes-SKI, and Nes-SKI;B-OE mice. TorsinB overexpression eliminates the morphological defects characteristic of Nes-SKI mice. (D) GFAP staining of P10 brains from Cre control, Cre control;B-OE, Nes-SKI, and Nes-SKI;B-OE mice. GFAP immunostaining illustrates the characteristic gliotic changes in Nes-SKI, including in cortex (arrows) and thalamus (circle). TorsinB overexpression eliminates gliotic changes in Nes-SKI mice.
-
Figure 4—source data 1
Information characterizing Nes-SKI;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig4-data1-v2.xlsx
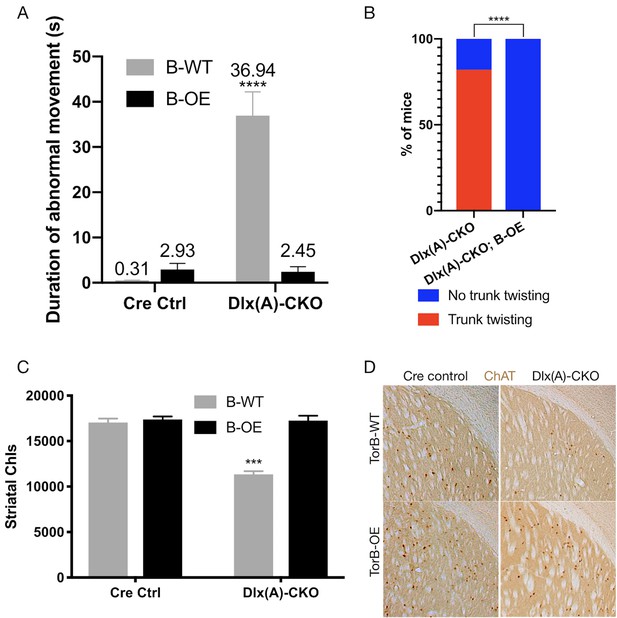
TorsinB overexpression prevents striatal cholinergic interneuron degeneration and dystonic-like movements.
(A) Duration of abnormal movements during one minute of tail suspension in P70 Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE mice. TorsinB overexpression significantly reduces severity of limb clasping (two-way ANOVA main effect of genotype F1,44 = 47.45, p<0.0001, torsinB level F1,44 = 36.86, p<0.0001, and interaction F1,44 = 49.96, p<0.0001; Cre control n = 10, Cre control;B-OE n = 12, Dlx(A)-CKO n = 11, Dlx(A)-CKO;B-OE n = 15). (B) Prevalence of trunk twisting in Dlx(A)-CKO and Dlx(A)-CKO;B-OE mice. TorsinB overexpression significantly reduces prevalence of trunk twisting (Chi square test χ2 = 18.77, p<0.0001; Dlx(A)-CKO n = 11, Dlx(A)-CKO; B-OE n = 15). (C) Stereologic counts of striatal cholinergic interneurons in P70 Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE brains. TorsinB overexpression prevents ChI degeneration characteristic of Dlx(A)-CKO mice (two-way ANOVA main effect of genotype F1,22 = 52.45, p<0.0001, ROSA-Tor1b allele F1,22 = 45.54, p<0.0001, and interaction F1,22 = 41.90, p<0.0001; Cre control n = 6, Cre control;B-OE n = 6, Dlx(A)-CKO n = 7, Dlx(A)-CKO;B-OE n = 7). (D) Representative images of P70 striatum immunostained with antibody to ChAT. ChAT+ cell density is reduced in Dlx(A)-CKO striatum while it appears normal in Dlx(A)-CKO;B-OE striatum.
-
Figure 5—source data 1
Information on behavioral and histological characterization of Dlx(A)-CKO;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig5-data1-v2.xlsx
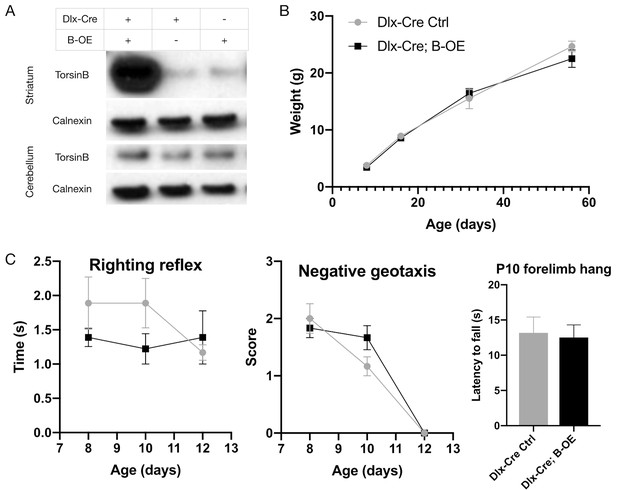
Dlx-Cre;B-OE mice exhibit no apparent motor or organismal phenotype.
(A) Western blots of striatal and cerebellar lysates probed with anti-torsinB antibody. Dlx5/6-Cre selectively activates the B-OE allele in forebrain structures (striatum); torsinB expression in hindbrain structures (e.g., cerebellum) is unaffected. (B) Postnatal weight gain of Dlx5/6-Cre;B-OE mice. Weight gain is normal in Dlx5/6-Cre;B-OE mice compared to littermate controls (two-way repeated measures ANOVA genotype x age interaction F3, 30 = 9439, p=0.4318; Cre control n = 6, Cre; B-OE n = 6). (C) Development of neonatal reflexes in Dlx5/6-Cre; B-OE mice. Neonatal reflexes including righting (two-way repeated measures ANOVA genotype x age interaction F2, 20 = 1.858, p=0.1819; Cre control n = 6, Cre;B-OE n = 6), negative geotaxis (two-way repeated measures ANOVA genotype x age interaction F2, 20 = 2.031, p=0.1574; Cre control n = 6, Cre;B-OE n = 6), and forelimb hang (unpaired t-test t1 = 2317, p=0.8214; Cre control n = 6, Cre;B-OE n = 6) are normal in Dlx5/6-Cre;B-OE mice, indicating normal gross and motor development.
-
Figure 5—figure supplement 1—source data 1
Infomation pertaining to growth and preweaning reflexes of Dlx-Cre;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig5-figsupp1-data1-v2.xlsx
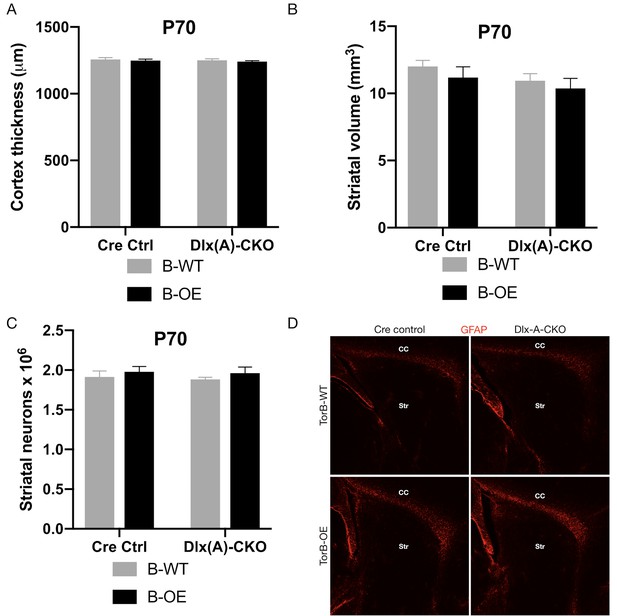
Dlx-Cre;B-OE mice exhibit no apparent neuropathological phenotype.
(A) Cortical thickness of P70 Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE mice. Cortical thickness is unchanged (two-way ANOVA main genotype x torsinB level interaction F1,12 = 0.0002485, p=0.9877; n = 4 for all genotypes). (B) Striatal volume of P70 Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE mice. Striatal volume is unchanged (two-way ANOVA genotype x torsinB level interaction F1,12 = 0.03657, p=0.9515; n = 4 for all genotypes). (C) Striatal medium and small neuron counts in P70 Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE mice. The overall number of striatal neurons is unchanged two-way ANOVA genotype x torsinB level interaction F1,12 = 0.01144, p=0.9166; n = 4 for all genotypes). (D) GFAP staining from Cre control, Cre control;B-OE, Dlx(A)-CKO, and Dlx(A)-CKO;B-OE mice. Labels show striatum (Str) and corpus callosum (CC). GFAP staining shows no reactive gliosis within the striatum of any mice.
-
Figure 5—figure supplement 2—source data 1
Information on neuropathological characterization of Dlx(A)-CKO;B-OE mice.
- https://cdn.elifesciences.org/articles/54285/elife-54285-fig5-figsupp2-data1-v2.xlsx
Tables
Features of mouse models used in this study.
Characteristics of DYT1 dystonia mouse models including extent of Cre field, behavioral phenotype, and neuropathological phenotype.
| Model | Genotype | Cre field | Organismal/behavioral phenotype | Histologic findings | Rationale for use | Impact of torsinB modulation |
|---|---|---|---|---|---|---|
| Emx1(A)-CKO | Emx1-Cre; Tor1aKO/flx | Forebrain excitatory, prominent in cortex and hippocampus | Normal appearing Limb clasping in a subset of mice | Forebrain-selective neurodegeneration | Forebrain motor loop involvement Mild behavioral and neuropathology findings to assess combined torsinA and B LOF | Reduction: Worsened neuropathology and behavior |
| Emx1-SKI | Emx1-Cre; Tor1aΔE/flx | Forebrain excitatory, prominent in cortex and hippocampus | Normal appearing Limb clasping in a subset of mice | Forebrain-selective neurodegeneration milder than that seen in Emx1-CKO | Forebrain motor loop involvement Presence of disease mutant torsinA | Reduction: Dose-dependent worsening of neuropathology and behavior |
| Nes(A)-CKO | Nestin-Cre; Tor1aKO/flx | Entire nervous system | Lack of postnatal weight gain Early lethality by 3rd postnatal week Overtly abnormal postures at rest | Degeneration in multiple sensorimotor regions | Clear and robust phenotypes Widespread involvement of nervous system | Overexpression: Prevention of lethality, restored weight gain Prevention of degeneration and gliosis |
| Nes-SKI | Nestin-Cre; Tor1aΔE/flx | Entire nervous system | Reduced postnatal weight gain Overt postural and developmental phenotypes at rest | Degeneration in multiple sensorimotor regions | Widespread involvement of nervous system Presence of disease mutant torsinA | Overexpression: Restored weight gain Prevention of degeneration and gliosis |
| Dlx(A)-CKO | Dlx5/6-Cre; Tor1aKO/flx | Forebrain GABAergic and cholinergic, including all striatal neurons | Limb clasping and trunk twisting; symptoms respond to drugs used in human patients | Selective degeneration of dorsal striatal cholinergic interneurons | Predictive validity Time course of motor abnormalities mimics that of human patients | Overexpression: Prevention of abnormal limb clasping and twisting movements Prevention of cholinergic interneuron degeneration |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit polyclonal α-GFAP | Dako | Cat#Z0334 RRID:AB_10013382 | IHC(1:2000) |
| Antibody | Rabbit polyclonal α-CUX1 | Santa Cruz Biotechnology | Cat#sc-13024 RRID:AB_2261231 | IHC(1:200) |
| Antibody | Rat monoclonal α-CTIP2 | Abcam | Cat#ab18465 RRID:AB_2064130 | IHC(1:500) |
| Antibody | Goat polyclonal α-ChAT | Millipore | Cat#AB144P RRID:AB_90560 | IHC(1:200) |
| Antibody | Donkey α-rabbit IgG, Alexa Fluor 555 conjugated | Invitrogen | Cat#A-31572 RRID:AB_162543 | IHC(1:800) |
| Antibody | Donkey α-rat IgG, Alexa Fluor 488 conjugated | Invitrogen | Cat# A-21208 RRID:AB_141709 | IHC(1:800) |
| Antibody | Donkey α-goat IgG, biotin-SP conjugated | Jackson Immunoresearch | Cat#705-065-147 RRID:AB_2340397 | IHC(1:800) |
| Antibody | Rabbit polyclonal α-torsinA | Abcam | #ab34540 RRID:AB_2240792 | WB(1:10,000) |
| Antibody | Rabbit polyclonal α-torsinB | Kind gift of Schlieker lab | N/A | WB(1:1,000) |
| Antibody | Goat α-rabbit IgG, HRP-linked | Cell Signaling | Cat#7074 RRID:AB_2099233 | WB(1:20,000) |
| Antibody | Rabbit polyclonal α-Calnexin | Enzo Life Sciences | Cat#ADI-SPA-860-F RRID:AB_11178981 | WB(1:20,000) |
| Sequence-based reagent | Lox-gtF | Integrated DNA Technologies | PCR primers | CTG ACA CAG TGA GTG AAG GTG C |
| Sequenced-based reagent | Lox-gtR | Integrated DNA Technologies | PCR primers | GGT GCT GAG GAA GTG CTG TG |
| Sequenced-based reagent | Frt-gtF | Integrated DNA Technologies | PCR primers | AGG GGC CAT AGA GTG GTT AGG |
| Sequenced-based reagent | Frt-gtR | Integrated DNA Technologies | PCR primers | CTT AGC CGC TTT GTG CTG |
| Sequenced-based reagent | Rosa-gtF | Integrated DNA Technologies | PCR primers | AGT CGC TCT GAG TTG TTA TCA G |
| Sequenced-based reagent | Rosa-gtR | Integrated DNA Technologies | PCR primers | CTG ACA CAG TGA GTG AAG GTG C |
| Commercial assay or kit | ABC Kit | Vectastain | Cat#PK6100 RRID:AB_2336819 | Use kit directions |
| Commercial assay or kit | Prolong Gold Antifade | Thermo Scientific | Cat#P36930 | Use kit directions |
| Software, algorithm | Stereoinvestigator | MBF Bioscience | https://www.mbfbioscience .com/stereo-investigator _ | N/A |
| Software, algorithm | ImageJ | NIH | https://imagej.nih.gov/ij/; RRID:SCR_003070 | N/A |
| Software, algorithm | Prism | Graphpad | http://www.graphpad.com; RRID:SCR_002798 | N/A |
| Strain, strain background (M. musculus, male and female) | Tor1b+/- | University of Connecticut Center for Mouse Genome Modification | N/A | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | Tor1atm1Wtd/J | The Jackson Laboratory | RRID:IMSR_JAX:006251 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | Tor1atm3.1Wtd/J | The Jackson Laboratory | RRID:IMSR_JAX:025832 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | STOCK Tor1atm2Wtd/J | The Jackson Laboratory | RRID:IMSR_JAX:025637 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | Tor1abΔE floxed Tor1b/+ | This paper (University of Connecticut Center for Mouse Genome Modification) | N/A | B6;129 hybrid Mouse line will be submitted and available from The Jackson Laboratory |
| Strain, strain background (M. musculus, male and female) | Emx1tm1(cre)Krj/J | The Jackson Laboratory | RRID:IMSR_JAX:005628 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | Tg(dlx5a-cre)1Mekk/J | The Jackson Laboratory | RRID:IMSR_JAX:008199 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | Tg(Nes-cre)1kln/J | The Jackson Laboratory | RRID:IMSR_JAX:003771 | B6;129 hybrid |
| Strain, strain background (M. musculus, male and female) | B-OE | This paper (from Biocytogen) | N/A | B6;129 hybrid Mouse line will be submitted and available from The Jackson Laboratory |
Genotyping PCR programs and band sizes.
| Gene | Primer name | Cycle | Band sizes |
|---|---|---|---|
| Tor1a ΔE floxed Tor1b | Lox-gtF | 94°C, 3 min; 94°C, 30 s; 61.5°C, 30 s; 72°C, 30 s, 34 cycles; 72°C, 5 min | WT – 245 bp Floxed– 336 bp |
| Lox-gtR | |||
| Frt-gtF | |||
| Frt-gtR | |||
| B-OE | Rosa-gtF | 94°C, 3 min; 94°C, 30 s; 61°C, 30 s; 72°C, 30 s, 30 cycles; 72°C, 5 min | WT – 469 bp Mutant – 188 bp |
| Rosa-gtR | |||
| Mut-R |





