Mouse brain transcriptome responses to inhaled nanoparticulate matter differed by sex and APOE in Nrf2-Nfkb interactions
Abstract
The neurotoxicity of air pollution is undefined for sex and APOE alleles. These major risk factors of Alzheimer’s disease (AD) were examined in mice given chronic exposure to nPM, a nano-sized subfraction of urban air pollution. In the cerebral cortex, female mice had two-fold more genes responding to nPM than males. Transcriptomic responses to nPM had sex-APOE interactions in AD-relevant pathways. Only APOE3 mice responded to nPM in genes related to Abeta deposition and clearance (Vav2, Vav3, S1009a). Other responding genes included axonal guidance, inflammation (AMPK, NFKB, APK/JNK signaling), and antioxidant signaling (NRF2, HIF1A). Genes downstream of NFKB and NRF2 responded in opposite directions to nPM. Nrf2 knockdown in microglia augmented NFKB responses to nPM, suggesting a critical role of NRF2 in air pollution neurotoxicity. These findings give a rationale for epidemiologic studies of air pollution to consider sex interactions with APOE alleles and other AD-risk genes.
Introduction
Air pollution is a major global environmental risk factor of morbidity and mortality across the human lifespan (Landrigan et al., 2018; Shaffer et al., 2019; Finch, 2018). Air pollution exposure is also associated with neurodegeneration, accelerated cognitive decline of aging and increased risk of Alzheimer’s disease (AD) (Kulick et al., 2020; Calderón-Garcidueñas et al., 2020). However, little is known of interaction of air pollution neurotoxicity for sex and APOE alleles and other AD risk factors (Finch and Kulminski, 2019).
Epidemiological studies of air pollution neurotoxicity have not identified interactions of gender by APOE alleles. Findings are typically ‘adjusted or controlled’ for gender differences (Clifford et al., 2016; Chen and Schwartz, 2009; Ailshire and Clarke, 2015; Gatto et al., 2014). In the WHIMS cohort of elderly women, APOE4 homozygotes had a greater risk of dementia and accelerated cognitive decline (Cacciottolo et al., 2017). The APOE4 vulnerability for accelerated cognitive aging was recently extended to ozone, as well as PM10 and PM2.5 in a large sample of both sexes from New York City (Kulick et al., 2020). A recent study from China suggested greater male vulnerability to air pollution for verbal test deficits (Zhang et al., 2018). Sex-APOE interactions for air pollution neurotoxicity remain undefined. In a small sample from polluted Mexico City, APOE4 heterozygous females with high BMI had higher risk of severe cognitive deficit than other groups (Calderón-Garcidueñas and de la Monte, 2017). Developmental air pollution exposure has received greater attention for gender because of the consistent male excess vulnerability for behavioral and cognitive deficits in the pre-adolescent and young adult (Chiu et al., 2013; Sunyer et al., 2015).
Mouse models have not addressed sex and APOE in responses to air pollution. Our initial study examined female EFAD (APOE-TR/5xFAD-Tg+/-) mice carrying transgenes for familial AD genes (5xFAD) together with human APOE alleles by targeted replacement (APOE-TR), which had APOE-e3+/+ (E3; APOE3) or APOE-e4+/+ (E4; APOE4) genotype. Consistent with WHIMS findings, E4FAD female mice accumulated more brain amyloid in response to nPM than the E3FAD (Cacciottolo et al., 2017). However, for ozone exposure, male APOE-TR showed the converse of greater behavioral impairments in APOE3 than APOE4 (Jiang et al., 2019). For further study of both sexes, we examined the cerebral cortex transcriptomic responses of APOE-TR and wildtype mice (C57BL/6J) by RNAseq for the main regulators of air pollution toxicity in AD pathways.
We focused on genomic pathways mediated by NRF2 and NFKB, which responded to air pollution in our prior studies (Zhang et al., 2012; Woodward et al., 2017a). These redox-sensitive transcription factors control hundreds of genes that mediate cellular responses to oxidative stress and immunity. They respond to oxidative stress, tobacco smoke, traumatic brain injury, and ischemic stroke, and are altered by aging and AD (Sivandzade et al., 2019). NRF2 downstream genes include antioxidants (e.g. glutathione, thioredoxin), anti-inflammatory cytokines (e.g. IL10), phase I and II xenobiotic detoxifying enzymes (e.g. CYP450) and free radical scavengers (Sandberg et al., 2014). The NFKB family transcriptionally regulates the expression of immune related proteins including cytokines (e.g. TNFA, IL1A, IL1B), antigen presentation proteins (e.g. MHCI, β2-microglobulin), chemokines (e.g. MCP1, MIP1), adhesion molecules (e.g. ICAM1, VCAM1), inducible nitric oxide synthase (INOS), and proapoptotic (e.g. BIM, BAX) or antiapoptotic proteins (e.g. XIAP, BCL2) (Hayden and Ghosh, 2012). The complex interplay of NRF2 and NFKB signaling pathways can alter the balance of anti-oxidative or inflammatory responses, depending on the type of stress, and target cell or tissue (Sivandzade et al., 2019).
Sex and APOE alleles can also alter NRF2 and NFKB activities, as shown for the larger response of female mice for hepatic NRF2 activation by phenobarbital and oxazepam and other xenotoxins (Rooney et al., 2018a). NRF2 downstream genes including Gsta2, Ho1, and Nqo1 showed lower hepatic expression in APOE4-TR than APOE3-TR mice (Graeser et al., 2011). We therefore examined both sex and APOE allele for interactions with NRF2/NFKB responses of air pollution neurotoxicity.
Results
To define brain transcriptional responses of air pollution and interactions with sex and APOE alleles, we examined responses of adult C57BL/6J (wild type, ‘B6’) and B6 mice carrying human APOE3 and APOE4 alleles by targeted replacement (APOE-TR) to nPM, a subfraction of ultrafine PM (PM0.2). Three independent exposures used different batches of nPM at specified durations of exposure (details on sample collection and chemical composition in Figure 1—figure supplement 1 and Zhang et al., 2019). In vitro studies on BV2 microglia examined the role of NRF2 and NFKB in responding inflammatory pathways.
Cerebral cortex transcriptome responses to nPM
Differentially expressed genes (DEGs) were analyzed by RNAseq for nPM responses. Stratification by APOE and sex was done subsequently to establish general effects. The multivariate model of combined B6 and APOE-TR data was adjusted statistically for sex, APOE genotype, and different nPM batches of the two exposures. For p=0.005, there were 140 DEGs (118 increased, 22 decreased) responses to nPM (Figure 1A). Ingenuity pathway analysis (IPA) of responding pathways included synapse function (e.g. axonal guidance, calcium signaling, endocannabinoid neuronal synapse), inflammation (e.g. AMPK, SAPK/JNK), circadian rhythm, NRF2 mediated antioxidant response, and hypoxia-inducible factor 1-α (HIF1A) signaling (Figure 1B). The top DEGs include Grin1 (+20%) and Rap1a (−20%) (Figure 1C).
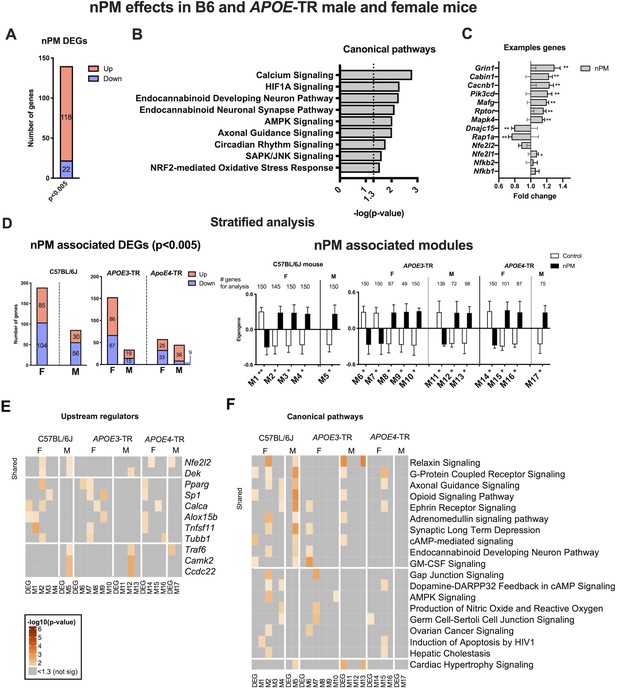
Cerebral cortex transcriptome responses to nPM in B6 and APOE-TR mice.
(A) Multivariate differential expression analysis of nPM responses in combined data from the independent exposures of C57BL/6J (B6) and APOE-TR. Covariates included sex, APOE genotype, and nPM. DEGs identified at p-value, 0.005. (B) Canonical pathways associated with nPM DEGs. (C) Examples of nPM associated DEGs. (D) Sex- and APOE-stratified DE and WGCNA modules associated with nPM responses. Male, M; Female, F. The top 150 genes of modules (kME inter-module connectivity) were used for IPA analysis. Significance was calculated from the Pearson correlation of eigengene of the modules with nPM. (E) Upstream regulators and F) canonical pathways associated with nPM transcriptome responses in B6 and APOE-TR mice. Solid horizontal lines separate responses that are shared and sex-specific. Heatmaps were sorted by the sum of -log10 (p-values) in each row. p-values<10−6 were converted to 10−6 for better visualization; grey, not significant. RNAseq sample size was 4/genotype/sex/treatment.
RNAseq data were stratified to identify sex- and APOE-specific nPM responses by linear models and by weighted gene co-expression (WGCNA). Females had more DEG than males for both B6 and APOE-TR mice, by up to two-fold (Figure 1D). Females of APOE3 and B6 had the most nPM responding genes (153 vs 189, respectively). Gene modules identified by WGCNA also had more female responses to nPM for B6 and APOE-TR (Figure 1D). Modules were constrained to a maximum of 150 hub genes, based on connectivity (Materials and methods). Both analyses (DE, WGCNA) showed more nPM-responding gene responses for APOE3 than APOE4.
Upstream regulators and canonical pathways were identified by IPA for sex-specific and shared nPM responses. The top upstream candidate was Nfe2l2 (Nrf2), a regulator of Phase II detoxification (Figure 1E, Figure 1—figure supplements 2–3), which had the strongest associations for B6 and APOE4. Sexes differed in immune-related upstream regulators of gene responses. Female-specific responses included Pparg (peroxisome proliferator activated receptor gamma), Sp1 (specificity protein1 transcription factor), and Tnfsf11 (TNF superfamily 11). Male-specific responses included Traf6 (TNF associated receptor factor 6), Camk2 (regulator of synaptic plasticity and AMPA glutamate receptors), and Ccdc22 (regulator of NFKB signaling by interaction with COMMD proteins). These results paralleled the enrichment of NRF2 and immune response pathways in the combined multivariate model above.
Stratified analysis by APOE and sex for canonical pathways showed nPM responses of neuronal pathways; for example G-protein-coupled receptors, axonal guidance, ephrin receptors, synaptic long-term depression, and endocannabinoid development neuron pathway (Figure 1F). Other nPM responding genes were related to relaxin, GM-CSF, and c-AMP-mediated signaling. Female-specific responses include genes associated with the following pathways: AMPK, dopamine-PARPP32 feedback in cAMP, gap junction signaling, and nitric oxide production. Male-specific responses in both mouse strains were enriched for ‘cardiac hypertrophy’ signaling; for example, Elk1 (transcription factor) and Hsp27 (Figure 1F). APOE3 and APOE4 of both sexes had different inflammatory responses for NFKB, IL6, CREB, and IL22 pathways (Figure 1—figure supplement 3). Cell-type deconvolution analysis of RNAseq also showed APOE and sex-specificity for microglial and astrocyte responses to nPM (Figure 1—figure supplement 3B).
Baseline effects of APOE4 allele and the overlap with nPM responses
Baseline differences by APOE in non-exposed controls were analyzed by sex in two steps. The combined multivariate model showed 133 DEGs differed in baseline APOE allele effect (5% FDR) (Figure 2A). These DEGs were enriched for immune-related pathways including rheumatoid arthritis, granulocyte adhesion, IL10, and NFKB signaling. APOE4 baseline differences included pathways of glutamate metabolism, and production of nitric oxide, superoxide and hydrogen peroxide, the LXL/RXR pathway of cholesterol efflux, and atherosclerosis.
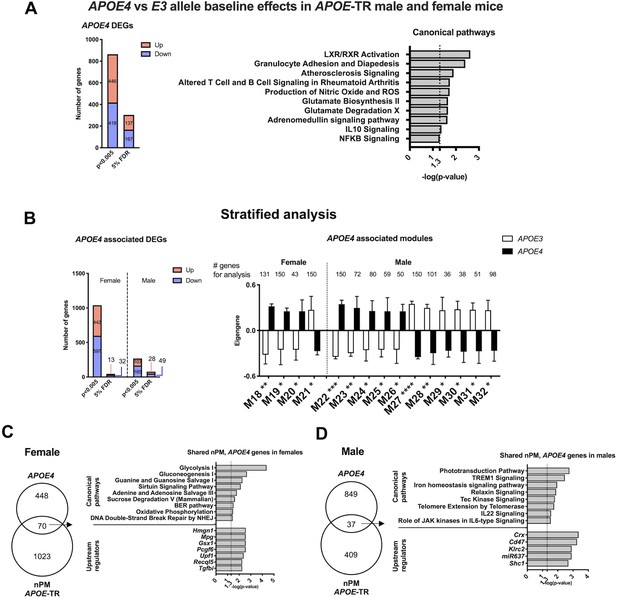
APOE allele baseline differences of RNA in cerebral cortex.
(A) Differential expression analysis of APOE4- vs APOE3-TR, at 5% FDR and p-value, 0.005. (B) WGCNA modules associated with APOE4 allele. IPA of the top 150 genes of the modules identified by kME (inter-module connectivity). Significance was calculated from the Pearson correlation of eigengenes for modules with APOE4 allele. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. IPA analysis of overlapped genes between baseline differences by APOE allele and nPM response in females (C) and males (D). The genes in each group are a combination of identified genes based on DE and WGCNA. RNAseq sample size was 4/genotype/sex/treatment. Detailed IPA analysis of APOE allele baseline DEGs (Figure 2—figure supplement 1).
In stratified analysis, males had 60% more DEGs differing by APOE alleles (male, 75 genes; female, 45 genes (Figure 2B, Figure 2—figure supplement 1). For WCGNA modules differing by APOE alleles, males had 3-fold excess (male 12 modules; female, four modules). Subsets of DEG (females, 70 DEGs; males, 37) had 7% overlap with nPM responding genes (Figure 2C–D), suggesting convergent effects of APOE4 allele and nPM. For females, the shared responding genes involved metabolic pathways (glycolysis, oxidative phosphorylation) and DNA repair (HMGN1). The male overlap involved a different set of genes related to iron homeostasis, telomere extension, and immune response (TREM1, IL11, JAK signaling).
AD-related pathways differed by sex and APOE alleles for nPM responses (Figure 3A–B). Only female APOE3 responded to nPM in five AD pathway genes for amyloid precursor protein (APP) processing and for tau: App, Bace1, Psen1 (Figure 3A); Tau and Gsk3b (Figure 3B). In contrast to mRNA changes in amyloidogenesis genes, the levels of amyloid peptides (Aβ40 and Aβ42) were not affected by nPM exposure in the cerebral cortex of APOE-TR mice (Figure 3C). However, Aβ40 peptide had a 50% lower baseline in males than females of both APOE3 and APOE4-TR mice (Figure 3C). Aβ40 peptide also had a significant negative correlation with mRNA levels of amyloidogenesis pathway including App (r, −0.65) and Psen1 (r,−0.51 to −0.67) for both APOE3 and APOE4-TR cerebral cortex (Figure 3D).
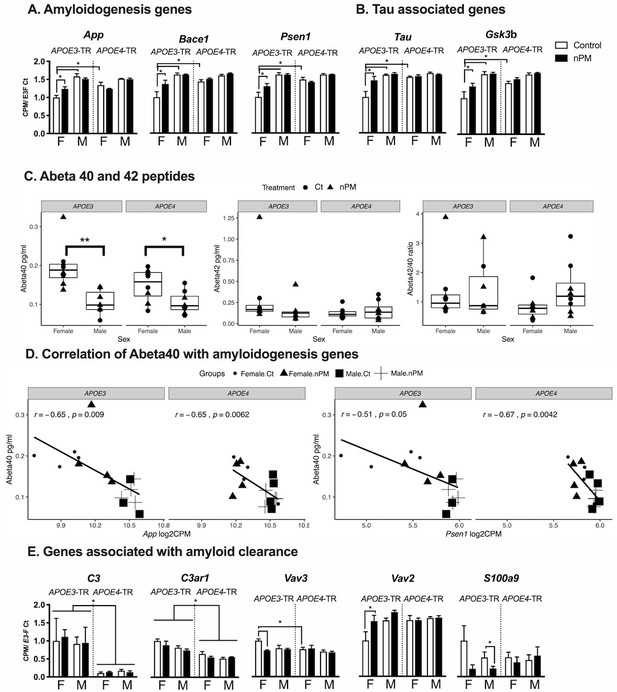
AD-associated gene responses to nPM in cerebral cortex.
(A) Amyloidogenic pathway responses. Female APOE3 had largest nPM response. (B) Tau and its kinase, Gsk3b. (C) Levels of Aβ40 and Aβ42 peptides did not respond to nPM. However, females had higher Aβ40 (pg/ml) of both APOE3 and APOE4 mice. **p<0.01, *p<0.05 in t-test. (D) Aβ40 peptide was negatively correlated with amyloidogenesis gene expression including App (r = −0.65), and Psen1 (r = −0.51 to −0.67). The expression was reported as Log2 count per million (cpm). (E) Aβ-amyloid clearance pathway responses to nPM. A small subset (10%, 5/46) of amyloid clearance genes differed by APOE or nPM (genes identified in the IPA database for phagocytosis, proteolysis, degradation, deposition). Only APOE3 responded to nPM. Mean ± SEM. ANOVA; FDR multiple test correction. * Adj. p-value, 0.05. Sample size: 4/genotype/sex/treatment.
About 10% of genes related to amyloid clearance (5/46) differed by APOE or nPM. For amyloid clearance genes, only APOE3 carriers in both sexes responded to nPM (Figure 3E). Three genes responded to nPM with sex differences only in APOE3. For APOE3 females, Vav2 (+50%); Vav3 (decreased −50%); the Vav guanine nucleotide exchange factors mediate phagocytosis of fibrillar Aβ (Wilkinson et al., 2006). For APOE3 males, S100a9 (−60%), also known as Mrp14, regulates microglial phagocytosis of fibrillar Aβ (Kummer et al., 2012). Baseline expression of two complement genes was higher in APOE3 than APOE4: C3 (10-fold), C3ar1 (+50%).
Sex- and APOE- specific nPM-mediated NFKB responses
Next, we examined genes of the NFKB pathway, which regulate pro-inflammatory responses to nPM, as shown for responses of wildtype mice (B6 males) in hippocampus to nPM (Woodward et al., 2017a). In cerebral cortex, APOE-TR mice responded to nPM with a subset of genes downstream of NFKB (13%, 8/133) that differed by sex and APOE (Figure 4A). Two clusters of nPM responses were identified by Principal Component Analysis for these 133 NFKB downstream genes: Principal component (PC) 2 (20% of variance, nPM: sex interaction) and PC4 (2.5% of variance nPM:APOE4 interaction) (Figure 4B).
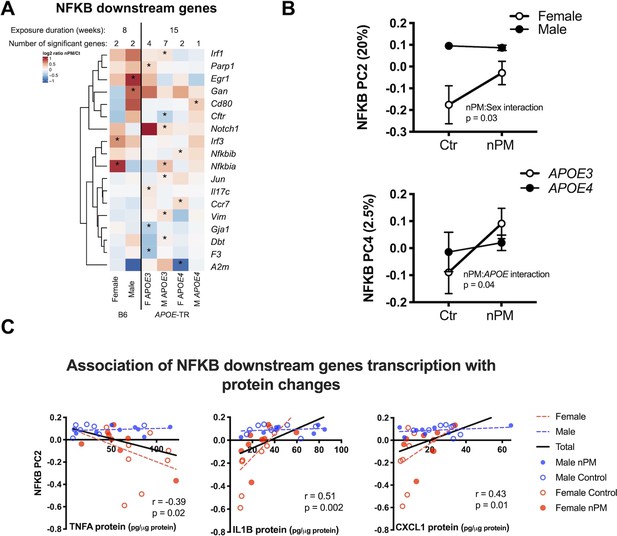
nPM induced inflammatory responses with sex- and APOE specificity.
(A) Stratified analysis of NFKB downstream genes responses to nPM. The combined IPA datasets included 133 NFKB downstream genes. (B) Principal component analysis of 133 NFKB downstream genes in APOE-TR: PC2 (20% of total variance) was associated with nPM: sex interaction; PC4 (2.5% of variance), associated with nPM:APOE interaction. (C) Protein levels of genes downstream of NFKB were correlated with PC2: positive correlations for CXCL1 and IL1B; inverse correlation with TNFA. Only females responded to nPM. Sample size of 4/genotype/sex/treatment.
Cytokine protein levels were examined for association with these PCs in cerebral cortex. PC2, but not PC4, was correlated with TNFA (r = −0.39, p=0.02), IL1B (r = 0.51, p=0.002), and CXCL1 (r = 0.43, p=0.01) proteins (Figure 4C). Only females responded to nPM for these cytokines. These RNA and protein responses are notable for consistent sex-specific inflammatory responses to nPM.
Sex- and APOE allele-specific NRF2 responses
NRF2 downstream responses to nPM differed by sex and APOE (Figure 5A). The 60 responding genes included Gpx3, and Gstp1, Jun, Nfe2l1 (Nrf1), and several Maf family transcription factors. A subset of gene responses was validated by qPCR, for example Nrf1 (Figure 5B; Figure 1—figure supplement 4). Female B6 and APOE-TR had 2-fold or more Nrf2 downstream genes responding to nPM. PC2 is associated with nPM for interactions with sex (p=0.01) and APOE (p=0.02), 6.4% of the variance, mainly associated with APOE3 females (Figure 5C). The strong inverse correlation of Nrf2 PC2 with Nfkb PC2 (Figure 5E, r = - 0.95, p=0.0001) suggests crosstalk between these transcriptional factors during exposure to nPM.
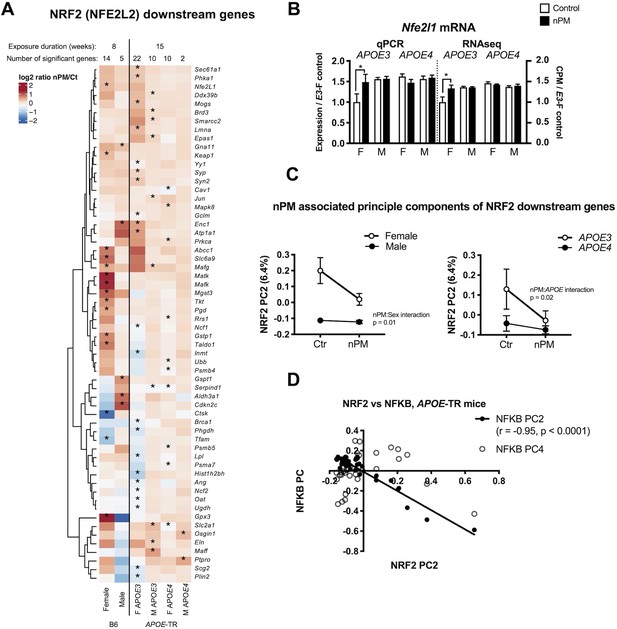
Nrf2 responses to nPM in B6 and APOE-TR mice.
(A) Heatmap of log2 fold changes of nPM responses, showing altered expression of at least 60 genes downstream of Nrf2, differing by sex or APOE genotype. (B) Validation by qPCR of Nfe2l1 (Nrf1) changes in RNAseq. (C) Principal component analysis of 513 Nrf2 downstream genes in APOE-TR: Only PC2 (6.4% of variance) had nPM-sex interaction (p=0.01) and APOE (p=0.02). APOE3 females had the highest nPM response. (D) PC2 of Nrf2 downstream genes varied inversely (R2 = 0.91, p=0.0001) with the PC2 of Nfkb downstream genes. Sample size of 4/genotype/sex/treatment.
Inhibitory effects of NRF2 on NFKB response to nPM
The relationship of NRF2 and NFKB responses of nPM was further explored in an independent dose-response experiment. The duration of inhalation exposure was only 3 weeks to assess the initial responses of nPM by male C57BL/6 mice. After 3-week exposure to 300 μg/m3 nPM, the cerebral cortex had nuclear translocation of NRF2 protein (+50%) and increased cytosolic NFKBP65 (+25%) (Figure 6A). Downstream of NRF2, the rate limiting enzyme of glutathione synthetase (GCLC) had dose-dependent increase correlated with Nrf2 mRNA (r, 0.6, p=0.005) (Figure 6B). Nrf2 and Nfkb responses of B6 male mice at three nPM doses had opposing changes of increased Nrf2 mRNA, but decreased Nfkbp65, Nfkbp50 mRNA and IL2 protein, all with dose-dependence (Figure 6C).
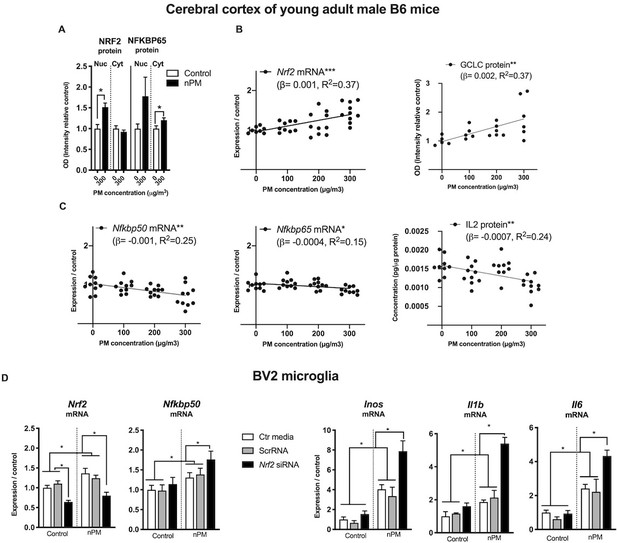
NRF2 and NFKB interact with nPM toxicity in cerebral cortex of male C57BL/6 mice and in mouse microglia (BV2 cells, in vitro).
(A) Increased nuclear translocation of NRF2 and cytosolic NFKBP65 of B6 mice exposed to 300 μg/m3 nPM for three wks. (B) nPM exposure dose-dependent increase of Nrf2 mRNA and positive correlation with increase of GCLC protein. (C) nPM dose-dependent decrease of Nfkbp65 and Nfkbp50 mRNA, and IL2 protein levels. Inhalation exposure to nPM at 100, 200, and 300 μg/m3 nPM (in vivo sample size, 10/group; exposure, 5 hr/d, 3 d/wk, 3 wks. **p=0.001, ***p=0.0001. (D) BV2 microglia in vitro response to nPM at 5 μg/ml nPM for 6 hr after partial knockdown of Nrf2 (sample size, 6/group; two independent biological replicates). Nrf2 mRNA knockdown was >60% at time 0. ANOVA with FDR multiple test correction. Mean ± SEM. *Adj. p=0.05. nPM chemical characterization (Figure 1—figure supplement 1).
Nrf2 and Nfkb interactions were examined in BV2 microglia in vitro during acute (6 hr) exposure to nPM. Partial knockdown of Nrf2 (−40%) by siRNA increased the Nfkb mediated responses of nPM (Figure 6D), with 30% higher Nfkbp50 mRNA, and 200% higher mRNA of Inos, Il1b and Il6.
NRF2 and NFKB are potential regulators of the nPM responses in the cerebral cortex and mixed glial culture
The top canonical pathways related to nPM exposure in the cerebral cortex of adult mice comprised of calcium signaling, HIF1α signaling, circadian rhythm pathway, AMPK signaling, SAPK/JNK signaling, endocannabinoid pathways and NRF2 oxidative stress responses (Figure 1A). These highly interconnected pathways comprise a larger network of oxidative and inflammatory responses (Figure 7). Thus, the hub regulators of nPM responses could broadly affect these pathways. Using IPA analysis, we built two networks from the NRF2 and NFKB downstream genes in the identified canonical pathways. Exposure to nPM caused expression changes in four of these genes in the cerebral cortex of adult mice: Smarca4 (+25%), Cftr (+25%), Hdac1 (−65%), and Vegfc (−65%).
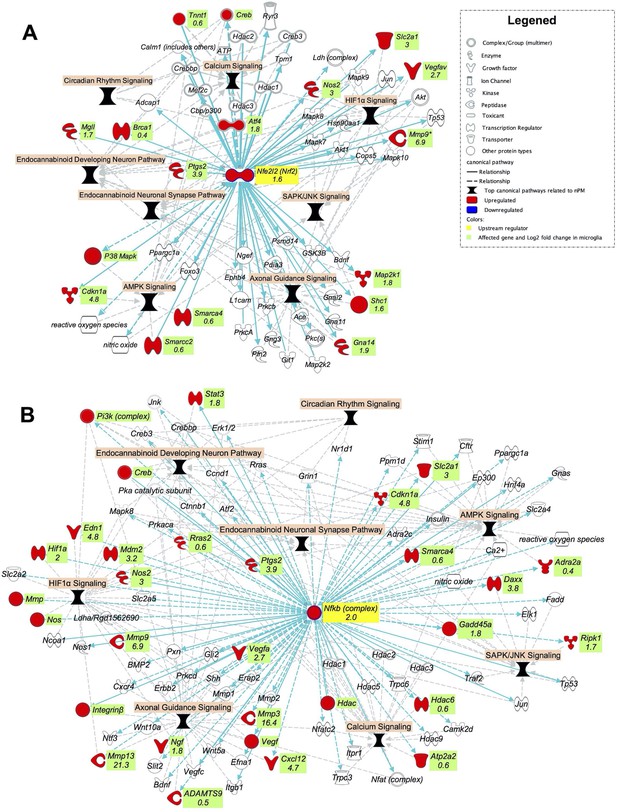
NRF2 and NFKB are potential upstream regulators of the top canonical pathways related to nPM effects in the cerebral cortex of adult mice.
Gene networks of (A) NRF2 and (B) NFKB downstream genes in the top nPM related canonical pathway (Figure 1A). The network was made by IPA software. The networks were overlaid with the significant responses to nPM in mixed glial culture (Woodward et al., 2017a). The numbers indicate log2 fold-changes of gene expression in Affymetrix microarray. Dataset from prior studies (Woodward et al., 2017a). In vitro sample size, 4/group.
We hypothesized that these networks are among the initial responses to nPM exposure and contributed to chronic damage at later stages. The transcriptional responses of these networks were examined in mixed glial culture after 24 hr exposure to 10 μg/ml nPM (Dataset from prior nPM studies Woodward et al., 2017a). Major increases of both Nrf2 (300%) and Nfkb (400%) mRNA were induced by acute nPM exposure (Figure 7). A large portion of genes from the constructed networks was also upregulated by nPM: 17 NRF2 and 30 NFKB DEGs from these downstream networks. These genes were related to all selected signaling pathways: HIF1A, calcium, AMPK, SAPK/JNK, and the endocannabinoid system. Several of these genes are also transcriptional regulators that could initiate further downstream changes, body-wide. Some of these genes include Brca1 (+30%), Atf4 (350%), Smarca4 (50%), Hif1a (400%), and Stat3 (350%) (Figure 7).
Discussion
These finding address the gap in how sex and APOE allele interactions may alter neurodegenerative responses to air pollution. Cerebral cortex genomic responses to nPM, were examined in C57BL/6 mice (B6, wildtype) and B6 mice carrying human transgenes for APOE3 and APOE4 alleles. Female mice had two-fold more genes that responded to nPM, further enhanced for APOE3. Responding genes included neuronal pathways (e.g. axonal guidance; glutamate synapse genes), inflammation (e.g. AMPK, and APK/JNK signaling), and antioxidant and hypoxic signaling (e.g. NRF2, HIF1A signaling). Genes in pathways downstream of NFKB and NRF2 responded oppositely to nPM. These interactions of NRF2 and NFKB may modulate sex and APOE risk for AD and accelerated cognitive aging during air pollution exposure.
The nPM responding genes may help to identify GxE in neurodegenerative risks from air pollution and cigarette exposure. For example, a combination of air pollution and a specific IL1B variant increased the risk of Parkinson disease (Lee et al., 2016). Our findings extend microarray analysis of frontal cortex of children and young adults from Mexican cities differing in air pollution levels: the 134 responding genes include inflammation (e.g. NFKB) and antioxidant responses (e.g. GPX2, GPX3) (Calderón-Garcidueñas et al., 2012). Microarray analysis of rat brain chronically exposed to PM0.2 also overlapped with our results: S100a9, calcium channels (e.g. Cacna1i), and glutamate receptors (Ljubimova et al., 2013).
These glutamatergic gene responses extend findings that hippocampal neurites are selectively sensitive to nPM (Morgan et al., 2011; Woodward et al., 2017b; Fonken et al., 2011). The increased levels of the ionotropic receptor NMDA type subunit 1 (Grin1) is notable: a GRIN polymorphism is associated with the risks of Parkinson (Wu et al., 2010), schizophrenia (Demontis et al., 2011), and also interacts with APOE alleles for earlier AD onset (de Oliveira et al., 2016), GRIN variants might also alter air pollution neurodegenerative responses, for example mutations in GRIN2A and GRIN2B increased the risk of cognitive impairments for lead poisoning of children (Rooney et al., 2018b). The nPM responses of Grin2a and Grin2b mRNA, while modest (p=0.05–0.06), merit further study among xenobiotics.
AD-associated genes responded differently to nPM by sex and APOE allele. APOE3 females had the lowest baseline and the highest nPM response in genes associated with amyloidogenesis and TAU. The smaller responses of APOE4 mice to nPM may be a ceiling effect, because about 7% of the nPM responsive genes also had APOE4 baseline differences in both sexes. For non-exposed controls (baseline), APOE4 vs APOE3 differed in 300 genes related to known APOE pathways including LXR/RXR (Courtney and Landreth, 2016), Atherosclerosis (Mahley et al., 2009), and Rheumatoid arthritis (Toms et al., 2012). A similar APOE allele specificity was found in responses to ozone (O3), which impaired memory in APOE3, but not APOE4-TR mice (Jiang et al., 2019). This pattern may be compared with ceiling effects of aging on responses to air pollution which we observed in middle-aged B6 mice. Both sexes at age 18 months have elevated baseline levels of antioxidant and inflammatory gene mRNA and protein, which did not respond to nPM or O3, unlike young mice (Jiang et al., 2019; Woodward et al., 2017b; Zhang et al., 2017a).
Brain amyloid must also be considered in the complex interactions of APOE genotype and sex with air pollution. The current study of APOE-TR mice response to nPM and a study of response to ozone (Jiang et al., 2019) showed that neither air pollutant altered brain Abeta40 and Abeta42 peptide levels for mice of ages 3 to 18 months (young to middle-age). The APOE-TR mice have the normal murine wildtype amyloid peptide Abeta42, which differs from the human in three amino acids and aggregates less avidly (Otvos et al., 1993). Thus, our study in APOE-TR mice do not include additional stress due to amyloid aggregates present in the FAD transgenic models. Unlike APOE-TR, nPM exposure increased the amyloid plaque loads in EFAD (APOE-TR plus five mutation in amyloidogenic genes) and J20 AD mouse models (Cacciottolo et al., 2017; Cacciottolo et al., 2020). In EFAD mice, nPM caused greater amyloid plaque load in E4FAD than E3FAD. Even in that study, nPM increased levels of Abeta oligomers, but not plaque load in E3FAD more than for E4FAD mice. These differences suggest the hypothesis that APOE alleles cause differences in amyloid clearance, phagocytosis, and proteolysis. As expected, some nPM-responding genes that mediate amyloid clearance differed by the APOE allele. The complement factor C3, which mediates amyloid clearance (Maier et al., 2008), had a higher baseline in APOE3, but lacked nPM response, whereas Vav and S100 calcium-binding protein A9 (S100a9) responses to nPM were restricted to APOE3. Vav and S100a9 regulate phagocytosis of fibrillary Abeta by microglia (Wilkinson et al., 2006; Kummer et al., 2012). The APOE3 and 4 proteins, and Abeta compete differentially for uptake by astrocytes via the lipoprotein receptor-related protein 1 (LRP1)-dependent pathway (Verghese et al., 2013). Moreover, APOE4 astrocytes have less efficient Abeta clearance, attributed to acidification of endosomes (Verghese et al., 2013; Prasad and Rao, 2018). Possibly, an efficient clearing system could compensate even for a greater amyloidogenic effect of nPM in APOE3 than APOE4 mice. This hypothesis will be tested further in vitro and in vivo.
Similar to the amyloid pathway, APOE4 had higher baseline level of inflammation, while their response to nPM was less than the APOE3. This allele specificity was also shown for responses to ozone (Jiang et al., 2019). In contrast, responses to LPS endotoxin by intra-peritoneal injection were greater in APOE4 male mice for microglial activation and systemic inflammatory cytokines (Zhu et al., 2012). In vitro, APOE4 macrophages also had higher induction of NFKB, TNFA, and heme oxygenase one in response to LPS (Jofre-Monseny et al., 2007). While air pollution can include endotoxins, the comparisons with injected LPS for sex and APOE are limited because the systemic responses are downstream of the lung, which receives most inhalants.
Female mice had greater response to nPM than males for immune and antioxidant pathways, for wildtype and APOE-TR mice. Sex hormones and early life gonadal programming during brain development could underly these differences. Ex-vivo microglial cultures from male and female brains had divergent inflammatory response to estradiol (E2) and LPS (Loram et al., 2012). A gene expression microglial developmental index showed a sex difference in maturation and immune reactivity, which correlated with the risk of AD and autism spectrum disorders (Hanamsagar et al., 2017). In SH-SY5Y neuroblastoma cells, E2 increased cell survival and NRF2 antioxidant defense against homocysteine (Chen et al., 2013). Similarly, E2 treatment of postnatal rats ameliorated acute ethanol-induced oxidative stress, neuroinflammation, and neuronal cell death through increase of sirt1, P53 acetylation inhibition, and reduction in phospho-NFKB nuclear localization (Khan et al., 2018). Higher adaptive genomic response by females might favor faster detoxification or recovery from air pollution. Metformin mediated NRF2 activation could ameliorate tight junction proteins, blood–brain barrier (BBB) integrity, reduce inflammation and oxidative stress, and also normalize the levels of BBB glucose transporter GLUT1 protein after cigarette smoke exposure in mice (Prasad et al., 2017).
As opposed to potential protective effects of estrogen hormones against air pollution, female mice had higher baseline levels of Abeta40 than males. This results parallels with female excess amyloid plaque load of aged AD mouse models (Callahan et al., 2001; Hirata-Fukae et al., 2008). Neonatally demasculinized or defeminized 3xTg-AD or other AD transgenic mice shows the major role of sex steroids in determining adult sex differences in Abeta accumulation (Carroll et al., 2010; Pike et al., 2009). Thus, sex steroids can act as a double-edged sword for amyloidogenic responses to air pollution and other environmental neurotoxins. Resolving the role of steroid hormones in this intricate relationship of background biology and air pollution requires studies on the recovery after nPM exposure in gonadectomized and older mice. We plan further studies of the complex interface of sex-APOE allele and nPM in mixed glial cultures derived from male and female APOE3 and APOE4-TR mice.
NFKB and NRF2 had opposite responses to nPM, that included downstream genes in wildtype B6 and APOE-TR. This divergence was also shown in a short term (3 weeks) exposure of B6 male mice. The NRF2 and NFKB crosstalk was validated in BV2 microglia. This is the first evidence for NRF2 and NFKB interactions in response to air pollution of both in vivo and in vitro models. These results parallel the LPS responses of monocytes, which showed redox-mediated transcriptional cross-talk between NRF2 and NFKB responses to LPS (Zhang et al., 2017b). Concurrent increase of nuclear NRF2 and cytosolic NFKBP65 in cerebral cortex after nPM exposure suggest that NRF2 activation can attenuate NFKB nuclear localization. We hypothesize involvement of KEAP1, the NRF2 repressing protein, which can mediate IKKB degradation and inhibit NFKB nuclear localization (Kim et al., 2010; Lee et al., 2009). Other mechanisms could be mediated by direct protein-protein interaction, and by secondary messengers. For example, NRF2 can inhibit NFKB through reduction of reactive species and suppress RAC1-mediated NFKB activation (Sanlioglu et al., 2001; Cuadrado et al., 2014). In contrast, NFKB can also inhibit NRF2 activity through enhancing the recruitment of histone deacetylase (HDAC3) to ARE region (Wakabayashi et al., 2010), or competing with NRF2 for binding to CH1-KIX domain of CBP protein inside the nucleus (Liu et al., 2008). These interactions are also shown for thein nematode C. elegans: the NRF2 homologue skn-1 and the antibacterial factor 2 (abf-2) responded rapidly to nPM, with persisting developmental effects (Haghani et al., 2019).
Since immortalized BV2 microglial cells have limited comparability with in vivo microglial cells, we further corroborated the nPM responses of adult brain in primary mixed glial culture. In adult brain, nPM affected a network of genes from a set of highly interconnected canonical pathways including NRF2 oxidative stress response, HIF1A, AMPK, circadian rhythm, and endocannabinoid related signaling pathways. Interestingly, numerous genes from this network including Nrf2 and Nfkb mRNA were upregulated in mixed glial culture after 24 hr acute exposure to nPM. A large portion of this gene network were considered as downstream of NRF2 and NFKB transcriptional factors. Thus, these two molecules are potentially the hub upstream regulators of long-term nPM neurotoxic effects. This hypothesis remained to be tested in further in vitro and in vivo studies.
The statistical power to identify all responding genes is intrinsically limited by the high dimensionality of RNAseq data with >20,000 genes. Thus, p=0.05 threshold needs to be adjusted for multiple testing to identify the real changes. Unfortunately, the small effect size of nPM exposure together with the necessarily small sample size of animals led to lack of enogh power to detect the changes at 5% FDR rate. To minimize the rate of false positives, we reported the changes at a nominal significance of p<0.005. Nonetheless, there could be small changes in some genes that we could not detect but still critical for air pollution toxicity. For example, while RNAseq analysis did not detect responses of Nrf2, Nfkb, and Gclc mRNA at p<0.005, the nPM dose-response experiment showed a 50% dose-dependent change in Nrf2, Nfkb mRNA, and GCLC protein, which confirmed findings on the cerebellum (Zhang et al., 2012). Thus, RNAseq and other high dimensional data are inherent with potential false-positive and false-negative results.
Another limitation of air pollution studies is the heterogeneity in the chemical composition and toxic activity PM. Recently, we identified physical and chemical characteristics of nPM that altered in vitro and in vivo toxicity of nPM (Zhang et al., 2019; Haghani et al., 2020). Regardless, NRF2 and NFKB responses were consistent in all the nPM batches used in these different experiments.
In summary, air pollution neurotoxicity was shown to have sex- and APOE allele-specificity, which are main risk factors for AD. These findings give a rationale for including APOE-gender interactions in epidemiological studies of cognitive aging and dementias.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | APOE3-TR+/+ APOE4-TR+/+ | PMID:8980023 | ||
| Strain, strain background (M. musculus) | C57BL/6J (B6) | Jackson laboratory | 000664; RRID:IMSR_JAX:000664 | |
| Cell line (M. musculus) | BV2 microglia | ATCC | EOC 20 (ATCC CRL-2469); RRID:CVCL_5745 | Female originated |
| Other | Mixed glia (microglia and astrocyte) | R. norvegicus | Postnatal days 3–5, mixed sexes | |
| Transfected construct (M. musculus) | Nfe2l2 siRNA | Thermofisher Scientific | 156499 | |
| Other | Lipofectamine RNAiMAX reagent | Thermofisher Scientific | 13778500 | |
| Antibody | anti-NRF2 (rabbit polyclonal) | Abcam | ab137550; RRID:AB_2687540 | WB, 1:1000 |
| Antibody | anti-H3 (rabbit polyclonal) | Cell Signaling Technology | D1H2; RRID:AB_10544537 | WB, 1:1000 |
| Antibody | anti-GAPDH (Mouse monoclonal) | Santa Cruz Biotechnology | sc-32233; RRID:AB_627679 | WB, 1:500 |
| Antibody | anti-NFKBP65 (Rabbit polyclonal) | Cell Signaling Technology | D14E12; RRID:AB_10859369 | WB, 1:750 |
| Antibody | anti-mouse IRDye 800CW | LICOR | 926–32210; RRID:AB_621842 | WB, 1:20,000 |
| Antibody | anti-rabbit IRDye 680RD | LICOR | 926–68070; RRID:AB_10956588 | WB, 1:20,000 |
| Commercial assay or kit | RNAeasy Mini Kit | Qiagen | 74104 | |
| Commercial assay or kit | TRUseq Stranded mRNA Kit | Illumina | 20020594 | |
| Commercial assay or kit | qScript cDNA Supermix | Quantabio | ||
| Commercial assay or kit | Taq master mix | Biopioneer | MAT-2.1–10 | |
| Commercial assay or kit | 12–230 kDa Jess or Wes Separation Module | Protein Simple | SM-W004 | |
| Commercial assay or kit | V-PLEX proinflammatory panel one immunoassay | Mesoscale Diagnostics, Rockville, MD | K15048D | |
| Commercial assay or kit | V-PLEX Aβ Peptide Panel 1 (4G8) Kit | Mesoscale Diagnostics, Rockville, MD | K15199E | |
| Chemical compound, drug | TRIzol | Invitrogen | 15596026 | |
| Software, algorithm | Rstudio | Packages: LIMMA, WGCNA, BRETIGEA | ||
| Software, algorithm | Ingenuity pathway analysis | Qiagen | ||
| Software, algorithm | GraphPad Prism | Version 8 |
Animals
Husbandry and experimental procedures were approved by the USC Institutional Animal Care and Use Committee (approval numbers: APOE-TR experiment, 20417; B6 experiments and mixed glial culture, 11233). The C57BL/6J and APOE-TR (Xu et al., 1996) mice were aged 2 months at exposure onset (Cacciottolo et al., 2016; Youmans et al., 2012). For long-term nPM exposures (8–15 weeks), four mice for each sex, genotype (C57BL/6J, APOE3-TR, APOE4-TR), and exposure (48 mice total) were randomly assigned to nPM exposure or control. The dose-response experiment was done with 10 male C57BL/6J mice per group. After exposure, mice were euthanized by isoflurane anesthesia and perfused transcardially with phosphate-buffered saline. Brains were hemisected at midline; total cerebral cortex was frozen on dry ice and stored at −80°C. Investigators were blinded to exposure groups during data measurement and analyses.
Air pollution nPM collection and exposure
Request a detailed protocolMice were exposed to nPM, a nano-sized subfraction of airP particulate matter of 2.5 microns diameter (PM2.5) collected from a local urban freeway corridor (Woodward et al., 2017b; Haghani et al., 2020). Briefly, PM0.2 samples were collected by a High-Volume Ultrafine Particle (HVUP) Sampler (Misra et al., 2002) at 400 L/min flow rate on an 8 × 10 inch-Zeflour PTFE filter (Pall Life Sciences, Ann Arbor, MI). The Particle Instrumentation Unit of University of Southern California is located within 150 m downwind of a major freeway (I-110). Chemical composition and size distribution of re-aerosolized nPM is characterized by high-resolution mass spectrometry (SF-ICPMS) and Sievers 900 Total Organic Carbon Analyzer as described before (Morgan et al., 2011; Haghani et al., 2020). Chemical characterization of the nPM batches in this study is in Supplementary data (Figure 1—figure supplement 4). Re-aerosolized nPM or filtered air (control) was delivered to the sealed exposure chambers at approximately 300 μg/m3 concentration to model chronic exposure: 5 h/day, 3 days/week, for 8 (C57BL/6J) or 15 weeks (APOE-TR). For dose-response experiment, 8 weeks male C57BL/6J mice were exposed to approximately 100, 200, and 300 μg/m3 for 3 weeks. The duration and nPM dosages were based on brain responses in prior studies (Haghani et al., 2020; Cheng et al., 2016). The 3 weeks of intermittent exposure (5 h/day, 3 days/week) to 300 μg/m3 nPM yields an average hourly exposure of 27 μg/m3, as experienced in many cities.
RNA sequence (RNAseq) analysis of mouse cerebral cortex
Request a detailed protocolRNA was extracted with TRIzol (Invitrogen) and RNAeasy Mini Kit (Qiagen) with DNase digestion. Libraries were made with the TRUseq Stranded mRNA Kit (Illumina) with 1 mg of RNA. For Illumina NextSeq500sequencing, a single end-sequencing length of >50 nt was used. Reads were aligned and quantified to the mouse reference genome RefSeq mm10 with Tophat2 (v2.0.8b), restricted to uniquely mapping reads with three possible mismatches using the Partek flow software platform (Trapnell et al., 2012).
Differential gene expression was calculated by linear modeling (Limma package in R). Significance was calculated at 5% FDR rate or p-value, 0.005. Gene responses were analyzed by Qiagen Ingenuity Pathway Analysis (IPA) software. In combining the datasets generated from B6 and APOE-TR experiments, the models were adjusted by the COMBAT method to control for unknown variance (Johnson et al., 2007). Additional downstream analysis and plotting were done in Rstudio and GraphPad Prism. Cell type deconvolution analysis was done using BRETIGEA (BRain cEll Type specIfic Gene Expression Analysis) R package (McKenzie et al., 2018), which uses single-cell RNAseq data to identify cell-type-specific gene-signatures to predict the proportion of each cell type in bulk RNAseq data.
Weighted gene co-expression network analysis (WGCNA)
Request a detailed protocolThe co-expression network, based on WGCNA, was constructed from RNAseq data. WGCNA is an unsupervised clustering approach, which assigns groups of genes with shared expression patterns into modules (Langfelder and Horvath, 2008). Briefly, the adjacency matrix (correlations between genes) was converted to a scale-free network using soft threshold power (tuned in each group) of the signed matrix. The result was converted to a topological overlap matrix (TOM). Hierarchical clustering used 1-TOM distance measure (dissimilarity). A dynamic tree-cut algorithm was used to assign modules containing at least 30 genes. Module eigengenes (MEs) were calculated as the maximum amount of the variance of the model for each module, based on the Singular Value Decomposition method. Linear regression models estimated association of nPM or APOE with the MEs. The top 150 hub genes of the modules were selected for IPA analysis by the highest eigengene connectivity (kME). In total, 32 nPM associated gene modules were identified based on different analyses. Thus, the modules were renamed to M1-M32 to distinguish each analysis.
Cell culture and Nrf2 siRNA
Request a detailed protocolBV2 microglia (mouse-derived) were grown in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Cellgro, Mediatech, Herndon, VA) containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine (Woodward et al., 2017a). These cells were authenticated by expression of microglial markers, cell morphology, phagocytic activity, and comparison of nPM or LPS responses with mixed glial culture and other literature. The cells were not tested for mycoplasma. Nfe2l2 (Nrf2) siRNA (156499, Thermofisher Scientific) was delivered by Lipofectamine RNAiMAX reagent (Thermofisher Scientific).
Quantitative real-time PCR
Request a detailed protocolExtracted RNA was reverse transcribed to cDNA using qScript cDNA Supermix (Quantabio). qPCR used Taq master mix (Biopioneer) and gene-specific primers (supplementary file 1; Figure 1—figure supplement 4).
Protein extraction
Request a detailed protocolFrontal cerebral cortex (anterior to Bregma, excluding olfactory bulbs) was homogenized (20 mg in 0.2 ml) in 1x RIPA buffer supplemented with 1 mM Na3VO2, 1 mM phenylmethane sulfonyl fluoride (PMSF), 10 mM NaF, phosphatase inhibitor cocktail (Sigma), and Complete Mini EDTA-free Protease Inhibitor Cocktail Tablet (Roche). Supernatants were obtained by centrifugation at 12,000 g/15 min.
NRF2 subcellular localization
Request a detailed protocolNuclear and cytosolic fractions were separated after tissue homogenization in sucrose-Tris-MgCl2 (STM) buffer with phosphatase and protease inhibitors and centrifuged 800g × 15 min (Dimauro et al., 2012). After removing supernatant, the nuclear pellet was washed in STM buffer, resuspended in HEPES pH 7.9 buffer (20 mM HEPES 1.5 mM MgCl2, 0.5 M NaCl, 0.2 mM EDTA, 20% glycerol, 1% Triton-X-100, protease and phosphatase inhibitors) and sonicated. Cell fraction purity was validated by immunoblotting for nuclear histone 3 (H3) and cytosolic glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Protein analysis
Request a detailed protocolNRF2 was detected by Western blot using anti-NRF2 primary antibody (1:1000, rabbit polyclonal, ab137550). Proteins (20 μg) were electrophoresed on Criterion 4–15% TGX gels (Biorad) and transferred to PVDF membranes. After washing with TBS+0.05% Tween-20 (PBST), membranes were blocked (LiCOR) 1 hr/ambient, then incubated with primary antibody overnight at 4°C: anti-NRF2 (1:1000, rabbit polyclonal, ab137550), anti-NFKBP65 (1:750, Rabbit polyclonal, Cell Signaling Technology, D14E12), anti-H3 (1:1000, Rabbit polyclonal, Cell Signaling Technology, D1H2), and anti-GAPDH (1:500, Mouse monoclonal, Santa Cruz Biotechnology, sc-32233). Bands were identified by incubation with 1:20,000 fluorochome-conjugated LICOR-antibodies (anti-mouse IRDye 800CW or anti-rabbit IRDye 680RD); band intensity was analyzed by ImageJ. GCLC was assayed by capillary electrophoresis (12–230 kDa range, Jess ProteinSimple, California, USA). Total lysate 1 μg/μl was electrophoresed and treated with anti-GCLC (1:100) and HRP-labeled secondary antibody. Results were normalized to total protein (PN module, ProteinSimple). IL2 was assayed by V-PLEX proinflammatory panel one immunoassay (Mesoscale Diagnostics, Rockville, MD). Abeta 40 and 42 peptides were assayed by 4G8 Kit VPLEX (Peptide Panel 1, Meso Scale Discovery, Rockville, MD).
Data availability
All raw data have been deposited in GEO under accession code GSE142066.
-
NCBI Gene Expression OmnibusID GSE142066. Mouse brain transcriptome responses to inhaled nanoparticulate matter differed by sex and APOE in Nrf2-Nfkb interactions.
References
-
Fine particulate matter air pollution and cognitive function among U.S. older adultsThe Journals of Gerontology Series B: Psychological Sciences and Social Sciences 70:322–328.https://doi.org/10.1093/geronb/gbu064
-
Rust on the brain from microbleeds and its relevance to alzheimer studies: invited commentary on cacciottolo neurobiology of aging, 2016Journal of Alzheimer's Disease & Parkinsonism 6:287.https://doi.org/10.4172/2161-0460.1000287
-
Traffic-related air pollutants (TRAP-PM) promote neuronal amyloidogenesis through oxidative damage to lipid raftsFree Radical Biology and Medicine 147:242–251.https://doi.org/10.1016/j.freeradbiomed.2019.12.023
-
Augmented senile plaque load in aged female beta-amyloid precursor protein-transgenic miceThe American Journal of Pathology 158:1173–1177.https://doi.org/10.1016/S0002-9440(10)64064-3
-
Nanoscale particulate matter from urban traffic rapidly induces oxidative stress and inflammation in olfactory epithelium with concomitant effects on brainEnvironmental Health Perspectives 124:1537–1546.https://doi.org/10.1289/EHP134
-
Associations between traffic-related black carbon exposure and attention in a prospective birth cohort of urban childrenEnvironmental Health Perspectives 121:859–864.https://doi.org/10.1289/ehp.1205940
-
LXR regulation of brain cholesterol: from development to diseaseTrends in Endocrinology & Metabolism 27:404–414.https://doi.org/10.1016/j.tem.2016.03.018
-
Transcription factors NRF2 and NF-κB are coordinated effectors of the rho family, GTP-binding protein RAC1 during inflammationJournal of Biological Chemistry 289:15244–15258.https://doi.org/10.1074/jbc.M113.540633
-
Association of GRIN1 and GRIN2A-D with schizophrenia and genetic interaction with maternal herpes simplex virus-2 infection affecting disease riskAmerican Journal of Medical Genetics Part B: Neuropsychiatric Genetics 156:913–922.https://doi.org/10.1002/ajmg.b.31234
-
The Alzheimer's Disease ExposomeAlzheimer's & Dementia 15:1123–1132.https://doi.org/10.1016/j.jalz.2019.06.3914
-
Nrf2-dependent gene expression is affected by the proatherogenic apoE4 genotype-studies in targeted gene replacement miceJournal of Molecular Medicine 89:1027–1035.https://doi.org/10.1007/s00109-011-0771-1
-
Air pollution alters Caenorhabditis elegans development and lifespan: responses to Traffic-Related nanoparticulate matterThe Journals of Gerontology: Series A 74:1189–1197.https://doi.org/10.1093/gerona/glz063
-
NF-κB, the first quarter-century: remarkable progress and outstanding questionsGenes & Development 26:203–234.https://doi.org/10.1101/gad.183434.111
-
Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expressionBiochemical and Biophysical Research Communications 357:319–324.https://doi.org/10.1016/j.bbrc.2007.03.150
-
The lancet commission on pollution and healthThe Lancet 391:462–512.https://doi.org/10.1016/S0140-6736(17)32345-0
-
Gene-environment interactions linking air pollution and inflammation in Parkinson's diseaseEnvironmental Research 151:713–720.https://doi.org/10.1016/j.envres.2016.09.006
-
NF-κB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafKBiochimica Et Biophysica Acta (BBA) - Molecular Cell Research 1783:713–727.https://doi.org/10.1016/j.bbamcr.2008.01.002
-
Gene expression changes in rat brain after short and long exposures to particulate matter in Los Angeles Basin air: comparison with human brain tumorsExperimental and Toxicologic Pathology 65:1063–1071.https://doi.org/10.1016/j.etp.2013.04.002
-
Sex and estradiol influence glial pro-inflammatory responses to lipopolysaccharide in ratsPsychoneuroendocrinology 37:1688–1699.https://doi.org/10.1016/j.psyneuen.2012.02.018
-
Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDSJournal of Lipid Research 50:S183–S188.https://doi.org/10.1194/jlr.R800069-JLR200
-
Glutamatergic neurons in rodent models respond to nanoscale particulate urban air pollutants in vivo and in vitroEnvironmental Health Perspectives 119:1003–1009.https://doi.org/10.1289/ehp.1002973
-
Human and rodent alzheimer beta-amyloid peptides acquire distinct conformations in membrane-mimicking solventsEuropean Journal of Biochemistry 211:249–257.https://doi.org/10.1111/j.1432-1033.1993.tb19893.x
-
Protective actions of sex steroid hormones in Alzheimer’s diseaseFrontiers in Neuroendocrinology 30:239–258.https://doi.org/10.1016/j.yfrne.2009.04.015
-
Improving and expanding estimates of the global burden of disease due to environmental health risk factorsEnvironmental Health Perspectives 127:105001.https://doi.org/10.1289/EHP5496
-
Apolipoprotein E gene polymorphisms are strong predictors of inflammation and dyslipidemia in rheumatoid arthritisThe Journal of Rheumatology 39:218–225.https://doi.org/10.3899/jrheum.110683
-
When NRF2 talks, who's listening?Antioxidants & Redox Signaling 13:1649–1663.https://doi.org/10.1089/ars.2010.3216
-
Toll-like receptor 4 in glial inflammatory responses to air pollution in vitro and in vivoJournal of Neuroinflammation 14:84.https://doi.org/10.1186/s12974-017-0858-x
-
APOE4-specific changes in aβ accumulation in a new transgenic mouse model of alzheimer diseaseJournal of Biological Chemistry 287:41774–41786.https://doi.org/10.1074/jbc.M112.407957
-
Nrf2-regulated phase II enzymes are induced by chronic ambient nanoparticle exposure in young mice with age-related impairmentsFree Radical Biology and Medicine 52:2038–2046.https://doi.org/10.1016/j.freeradbiomed.2012.02.042
-
Delayed Nrf2-regulated antioxidant gene induction in response to silica nanoparticlesFree Radical Biology and Medicine 108:311–319.https://doi.org/10.1016/j.freeradbiomed.2017.04.002
-
Temporal changes in glutathione biosynthesis during the lipopolysaccharide-induced inflammatory response of THP-1 macrophagesFree Radical Biology and Medicine 113:304–310.https://doi.org/10.1016/j.freeradbiomed.2017.10.010
-
Cell-based assays that predict in vivo neurotoxicity of urban ambient nano-sized particulate matterFree Radical Biology and Medicine 145:33–41.https://doi.org/10.1016/j.freeradbiomed.2019.09.016
Article and author information
Author details
Funding
Cure Alzheimer's Fund
- Caleb Finch
National Institute on Aging (R01-AG051521)
- Caleb Finch
National Institute on Aging (P50-AG05142)
- Caleb Finch
National Institute on Aging (P01-AG055367)
- Caleb Finch
National Institute on Aging (T32- AG052374)
- Amin Haghani
National Institute on Aging (T32-AG000037)
- Max Thorwald
National Institute on Aging (1RF1AG053982-01A1)
- Terrence Town
National Institute on Aging (1R01AG057912-01)
- Morgan Levine
National Institute on Aging (4R00AG052604-02)
- Morgan Levine
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We are grateful to Drs. Arian Saffari and Farimah Shirmohammadi (USC Viterbi School of Engineering) for their contribution to nPM collection and animal exposures.
Ethics
Animal experimentation: All husbandry and experimental procedures were approved by the USC Institutional Animal Care and Use Committee. Approval numbers: APOE-TR experiment, 20417; B6 experiments and mixed glial culture, 11233.
Copyright
© 2020, Haghani et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,022
- views
-
- 202
- downloads
-
- 35
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 35
- citations for umbrella DOI https://doi.org/10.7554/eLife.54822






