Unc-4 acts to promote neuronal identity and development of the take-off circuit in the Drosophila CNS
Figures
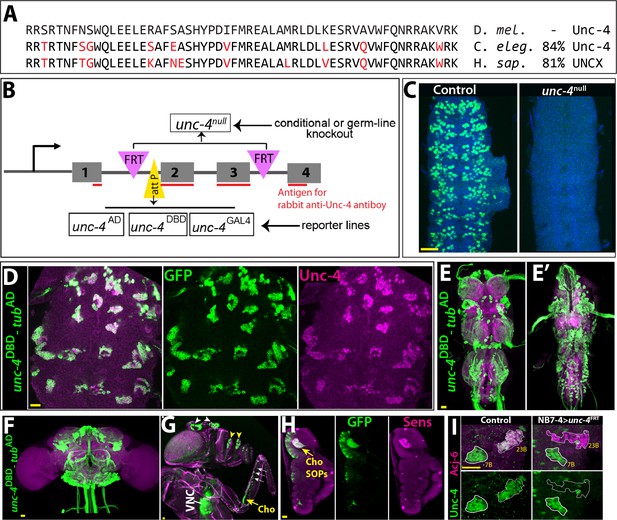
Tools generated to study Unc-4 function.
(A) Homeodomains of Unc-4 protein in D. melanogaster, C. elegans and H. sapiens are shown with the percent identity on the right. Amino acids that differ from Drosophilase quence were highlighted in red. (B) Schematic representation of edited unc-4 locus in unc-4FRT line shown (drawing is not to scale). Two FRT sites (magenta) are present with the same orientation flanking 2nd and 3rd exons (the homeodomain coding exons) and an attP site (yellow) is located 5’ to the second exon. unc-4null fly line was generated by FLP/FRT mediated excision of these exons in the germline. The same FLP/FRT mediated excision in somatic tissues was used for conditional mutant analysis. Trojan exons were inserted into the attP site to generate unc-4AD, unc-4DBD, unc-4GAL4 reporter lines. The region that is recognized by rabbit Unc-4 antibody underlined in red. (C) Dissected VNC samples from control (left) and unc-4null (right) embryos are shown. In the control VNC, Unc-4 protein (green) is expressed in a segmentally-repeated pattern. In the mutant embryo, no Unc-4 expression is detected. (D–H) unc-4DBD-tubAD driven GFP expression (green) shown. (D) Expression of this split GAL4 combination overlaps with Unc-4 protein expression (magenta) in the larval VNC. The clusters of neurons correspond to ventrally located Unc-4+ hemilineages. (E–F) Unc-4 is expressed in clusters of neurons in the adult VNC (E, E’) and brain (F). Ventrally located VNC lineages shown in E; dorsally located VNC lineages shown in E’. Here and in other figures, CadN antibody staining (magenta) used to define the contours of the adult CNS tissues. (G) Anterior body parts of an adult fly shown. Unc-4 reporter marks different types of sensory neurons: chordotonal (cho) organ (yellow arrow) and bristle sensory (white arrows) neurons in the leg, olfactory neurons in maxillary pulps (yellow arrowheads) and antennas (white arrowheads), and Johnston’s organ (not visible in this image). Magenta is cuticle autofluorescence. (H) A leg disc from an early stage pupa shown; anterior is to the left. Unc-4 is expressed in progenitors of chordotonal neurons, also called sensory organ precursors (SOPs), which were marked with Sens expression (magenta). (I) An example for conditional removal of unc-4 shown. Unc-4 (green) and Acj6 (magenta) are co-expressed in 23B neurons in the larval VNC but only Unc-4 is expressed in 7B neurons. NB7-4 specific FLP expression in the unc-4FRT larva (right panel) removes Unc-4 expression from the 23B neurons (NB7-4 progeny) but not the 7B neurons. Anterior is up in all images in this and other figures unless indicated otherwise. Scale bar (yellow line) is 20 microns.
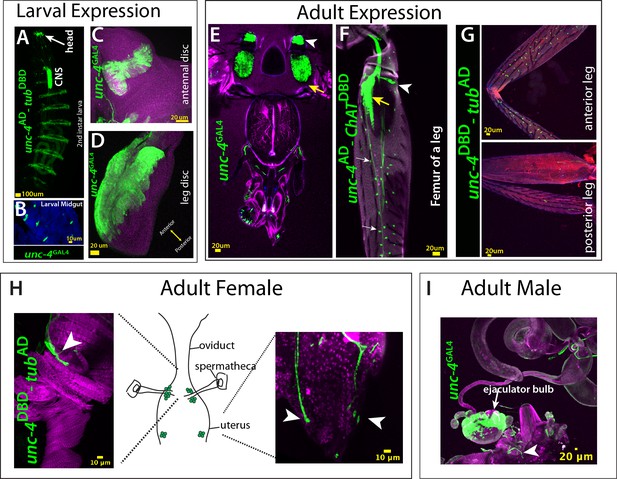
Unc-4 expression pattern in larval and adult tissues.
(A–I) GFP expression (green) driven by unc-4AD, unc-4DBD, unc-4GAL4 reporter lines shown from larval (A–D) and adult tissues (E–I). (A) A whole-mount second instar larval sample shown. Unc-4 is expressed in the CNS, head taste organ, and ventral epidermal cells, which partially overlap with denticle bends. (B) Unc-4 is expressed in the midgut enteroendocrine cells. (C, D) Unc-4 is expressed in antennal (C) and leg (D) imaginal discs. In the leg disc, Unc-4 expression is confined to the anterior region of the disc. (E) Adult head shown. Olfactory neurons (yellow arrowhead) and Johnston’s organ are marked with the Unc-4 reporter. (F–G) Adult legs shown. (F) unc-4AD-ChATDBD driven GFP expression marks chordotonal neurons (yellow arrow), campaniform sensilla neurons (arrowhead), and bristle sensory neurons (white arrows). Unc-4 expression marks bristles sensory neurons in the anterior compartment of the leg but not the ones in the posterior compartment (G). (H–I) Unc-4 is expressed in sensory neurons of female (H) and male (I) reproductive tracts (arrowheads) in addition to ejaculatory bulb of males (arrow). The schematic representation of the female reproductive tract was drawn based on Rezával et al. (2012).
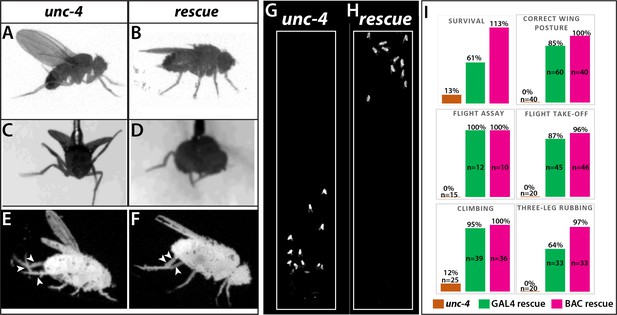
Loss of Unc-4 function results in behavioral defects in adult flies.
(A–H) Still images from recorded videos showing the behavioral defects observed in unc-4 mutants (A, C, E, and G) and the rescue of these defects with uncDBD- tubAD driven UAS-unc-4 transgene (B, D, F, and H). Images from control animals not shown as they were virtually identical to the images from rescued animals. Wings of unc-4 mutants are locked in an erect position (A); they fail flying in a tethered flight assay (C). Restoring Unc-4 function enables mutant animals to rest their wings in tucked position (B) and fly in the flight assay (D). Mutant animals fail to bring their three legs together for rubbing off dust (E) and rescue animals regain this three-leg rubbing behavior (F). (G–H) Restoring Unc-4 expression also improves climbing defects of unc-4 mutants. Positions of mutant and rescued animals after bang-induced climbing shown; the image was reproduced from Video 5. (I) Percentage bar graphs showing the quantification of unc-4 mutant phenotypes (orange bars) and their rescue with uncDBD- tubAD driven UAS-unc-4 (green bars) or a BAC transgene (PBac(DC335)) containing the unc-4 locus (magenta bars). See methods for detailed explanations for the assays quantified here. See also Videos 1–7.
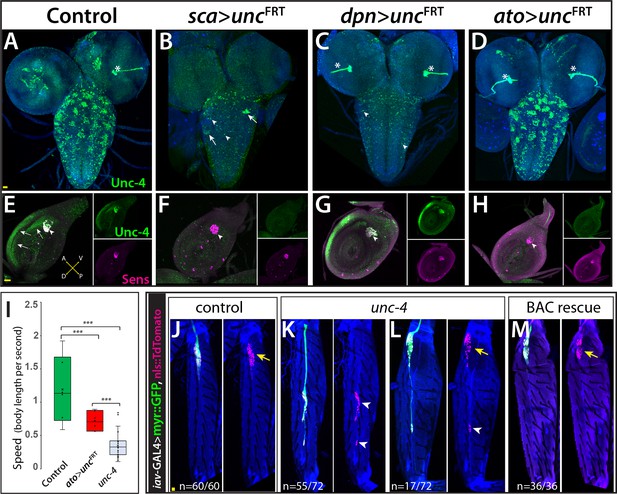
Unc-4 function in the periphery is required for animal coordination and the formation of chordotonal sensory organ.
Unc-4 expression (green) in the CNS (A–D) and leg disc (E–H) of wandering-stage larvae shown. sca-GAL4 (B, F), dpnEE-GAL4 (C, G), ato-GAL4 (D, H) driven FLP expression in the unc-4FRT line used to remove Unc-4 function in a tissue-specific manner. (A) In the control CNS, Unc-4 is expressed in clusters of post-embryonically born neurons (~50 clusters per CNS). (B) Manipulation with sca-GAL4 removed Unc-4 expression in most of the neuronal clusters but spared a few clusters stochastically (arrows; 3.4 clusters per CNS, N = 5). (C) dpnEE-GAL4 removed Unc-4 expression in all clusters without sparing any cluster (N = 5). In both sca-GAL4 and dpnEE-GAL4 manipulations, some dorsally located neurons (presumably embryonic-born) retained Unc-4 expression (arrowheads). (D) ato-GAL4 manipulation did not appear to affect Unc-4 expression in the CNS. Asterisks in (A–D) indicates the bleed-through from the 3xP-DsRed transgene present in unc-4FRT line; thus, they do not represent Unc-4 expression. (E–H) Unc-4 (green) and Sens (magenta) expression in leg discs shown. (E) In the control leg disc, Unc-4 is expressed in precursors of the chordotonal organ (arrowhead, marked with Sens) as well as in epithelial cells in the anterior compartment (arrows). (F) sca-GAL4 removed Unc-4 expression in all SOPs (16/16 leg discs, N = 5 animals), and also from the epithelial cells completely (10/16) or partially (6/16; not shown). (G) dpnEE-GAL4 manipulation did not affect Unc-4 expression in most leg discs (15/23, N = 9 animals) but sometimes removed Unc-4 expression in SOPs partially (3/23; not shown) or completely (5/23; not shown). (H) ato-GAL4 manipulation removed Unc-4 expression in all SOPs (25/25 leg discs; N = 8 animals) but also in epithelial cells completely (20/25) or partially (5/25; not shown). (I) ato-GAL4 mediated removal of Unc-4 resulted in slower walking animals, which were still faster than unc-4 mutant animals (***p<0.0001 student’s t test) (see also Videos 11 and 12). The walking speed was 1.4 +/- 0.6 body length per second (BLPS) for control , 0.76 +/- 0.4 BLPS for ato-GAL4> unc4FRT and 0.35 +/- 0.2 BLPS for unc-4 mutants (n > 27 tracts from 10 animals each). (J–L) The femur of T2 (J, K, M) and T1 (L) legs shown. iav-GAL4 driven myr::GFP (green) and nls::TdTomato (red) used to visualize processes and cell-body location of the chordotonal neurons in the femur. (J) In the control leg, clusters of chordotonal neurons were located proximally and close to the trochanter border in the femur (arrow). (K–L) In the unc-4 mutant animals, organization of the chordotonal neurons was disrupted. All animals showed this phenotype with varying degree of severity. In many legs (55/72; N = 12 animals), all of the iav-GAL4 marked chordotonal neurons were located away from their proximal position (arrowheads in K). In some legs (17/72), the phenotype was milder and only a small number of neurons were misplaced (arrowhead in L). Milder phenotype was observed mostly in T1 legs (15/17). (M) The presence of a BAC transgene containing the unc-4 locus restored the organization of the chordotonal neurons (36/36 legs, N = 6 animals) in the unc-4 mutant background. In E-H, A:anterior; P:posterior; V:Ventral; D; Dorsal Whisker box plots in I: the box delineates the first and third quartiles, the line in the box is median, and whiskers showing minimum and maximum values within 1.5 times the interquartile range. Scale bar is 20 microns.
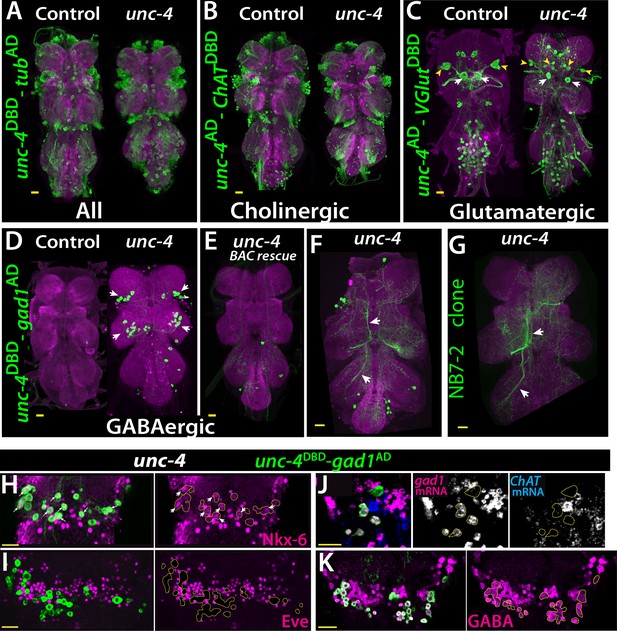
Cholinergic 11A neurons adopt GABAergic identity in unc-4 mutant animals.
(A–D) The adult VNC from control (left) and unc-4 mutant (right) animals shown; images are from maximum Z projection of confocal slices. (A) All Unc-4+ neurons were visualized with unc-4DBD-tubAD driven GFP expression (green). No major difference was observed between the control and mutant. (B) unc-4AD-ChATDBD split-GAL4 combination used to label cholinergic Unc-4+ neurons. Most Unc-4+ neurons were marked with this combination in both the control and mutant VNC and no major difference was seen between two. (C) unc-4AD-VGlutDBD driver marked a number of embryonic-born Unc-4+ glutamatergic motor neurons in the control VNC, which included flight related motor neurons MN1-4 (arrowheads) and MN5 (arrows). In the mutant, these motor neurons were disorganized and some showed reduction in cell size; for example, MN5 neurons. (D) unc-4DBD-gad1AD combination did not consistently show any GABAergic Unc-4+ neuron in the control CNS; however, in the unc-4 mutant background GABAergic interneurons were found in both the T1 (9 +/- 5 neurons; N = 30) and T2 (17 +/- 12 neurons; N = 30) segments. (E) A BAC transgene containing unc-4 locus suppressed the induced GABAergic phenotype and reduced the number of these neurons to 1.5 +/- 1 neurons per T1 segment and 1.3 +/- 1 neurons per T2 segment (N = 11 VNCs each). (F) A partial Z projection showing the main neuronal processes of unc-4DBD-gad1AD marked neurons in the VNC of an unc-4 mutant animal. They extend ipsilateral processes both anteriorly and posteriorly (arrows). (G) NB7-2 intersected reporter immortalization used to label all lineage 11 neurons in unc-4 mutant background and partial Z projection made to show only 11A neuronal processes but not that of 11B. The projections by the mutant cells match those of the 11A neurons (arrows). (H–K) unc-4DBD-gad1AD expression (outlined with dashed yellow lines) in the T2 segment from unc-4 mutants shown. Nkx-6, a marker for 11A neurons, is expressed in a subset of unc-4DBD-gad1AD marked neurons (arrows in H) while Eve, a marker for 11B neurons, is not (I). (J) ChAT (blue) and gad1(magenta) mRNAs were visualized via RNA in situ hybridization. Majority of unc-4DBD-gad1AD marked neurons expressed gad1 mRNA but not ChAT mRNA. (K) unc-4DBD-gad1AD marked neurons were immunopositive for GABA reactivity (magenta). Scale bar is 20 microns.
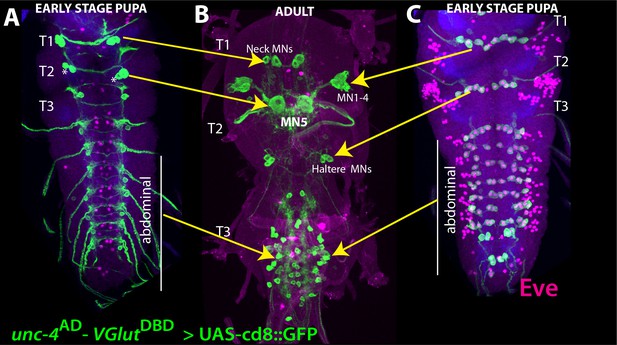
Emryonic-born Unc-4+ neurons become flight motor neurons in the adult VNC.
(A–C) unc-4AD-VGlutDBD driven GFP expression in the larval (A, C) and adult (B) VNC shown. Eve expression (magenta) marks segmentally repeated larval U/CQ motor neurons (C). Based on our tracing experiment, these motor neurons are remodeled during metamorphosis to be adult MN1-4 and haltere motor neurons in thoracic segments, and dorsal body-wall innervating motor neurons in abdominal segments (B). (A) Unc-4 is also expressed in two pairs (in T1 and T2) and one pair (in T3 and abdominal segments) neurons, which are located on the dorsal surface of the pupal VNC. Based on lineage clones we generated (not shown) and previously published work (Rogulja-Ortmann et al., 2008), these neurons are likely embryonic progeny of NB2-4. (B) In T1, these neurons become adult neck motor neurons. In T2, one of these neurons becomes MN5 motor neuron while its cousin is eliminated by apoptosis during pupal development (asterisks). Genotype: unc-4AD/+;VGluTDBD/+;10XUAS-IVS-myr::GFP/+.
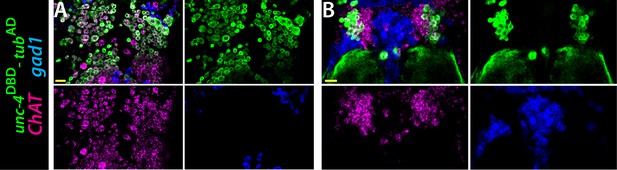
ChAT (magenta) and gad1(blue) mRNAs were visualized via RNA in situ hybridization.
unc-4DBD-tubAD marked Unc-4+ hemilineages (green) express ChAT mRNA but not gad1 mRNA. Single confocal slices from the T2 segment of the dorsal (A) and ventral (B) VNC shown. Scale bar is 20 microns. unc-4DBD/+;tubAD/+;10XUAS-IVS-myr::GFP/+.
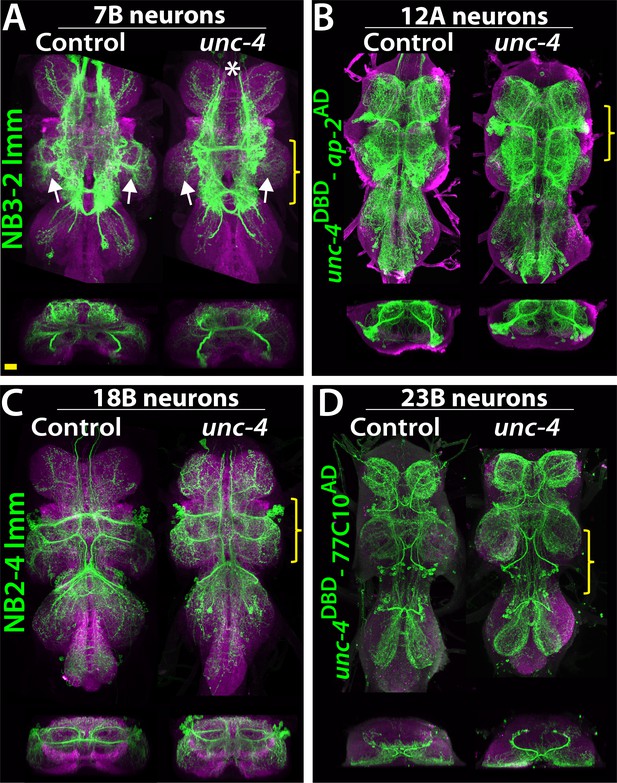
Projections of Unc-4+ lineages in control and unc-4 mutant backgrounds.
(A–D) VNC examples from control (left panels) and unc-4 mutant (right panels) adults. Transverse views from a 3D projection of the region indicated by yellow bracket are shown under the main images. (A) NB3-2 intersected reporter immortalization with ey-AD;dbxDBD driver was used to visualize the 7B neurons. In the unc-4 mutant animal, the 7B neurons did not innervate the T2 leg neuropil (compare the regions indicated by arrows). Note that 7B neurons in the mutant T1 segment were not labeled in this sample (asterisk), but in other mutant samples, projections of 7B neurons in T1 segments were similar to the control (not shown). (B) uncDBD- AP-2AD driver GFP used to mark 12A neurons. No dramatic difference was observed between the control and mutant VNCs. (C) Poxn-GAL4 driven reporter immortalization in NB2-4 used to visualize 18B neurons. Their neuronal projections appeared similar in the control and mutant. (D) unc-4DBD;77C10AD driven GFP expression used to visualize projections of 23B neurons, which did not differ between the control and mutant. Z projection images were made from a subset of confocal slices to visualize projections of all neurons. Scale bar is 20 microns.
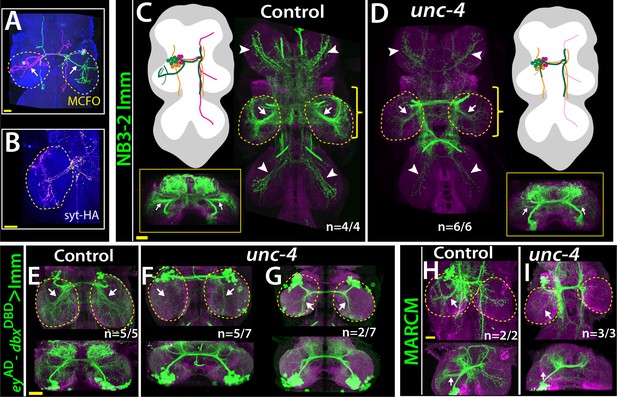
Unc-4 function is necessary in 7B neurons for the leg neuropil innervation .
(A) An adult VNC containing MCFO clones showing two 7B neurons in the T2 segment; both extend projections (arrows) to the ipsilateral leg neuropil. (B) A subset of 7B neurons (magenta) in the T2 segment marked with SS20852-GAL4 driven syt-HA (yellow) showing synaptic outposts in the leg neuropil. (C, D) NB3-2 intersected reporter immortalization via ey-AD;dbxDBD driver used to visualize projections of 7B neurons in the control (C) and mutant (D) animals. Cartoons in the left panels schematized projections of the T2 7B neurons observed in confocal images. Transverse views (corresponding the yellow brackets) shown under the cartoons. In the mutant, ipsilateral (arrows) and contralateral (arrowheads) projections in the leg neuropils were reduced. Note that the numbers of T2 7B neurons marked with the reporter in control and mutant animals was the same (67 + - 6 and 68+/- 9 neurons in control and mutant animals, respectively; N = 7 clones for each). (E–G) A different immortalization technique involving FLP/FRT induced LexA:p65 without NB intersection used to mark 7B neurons in the VNC of control (E) and mutant (F,G) animals. ey-AD;dbxDBD expression was immortalized without NB intersection. Top panels show maximum z projections; bottom panels show the transverse view of the same region. As with the above manipulations, mutant 7B neurons failed to show the normal innervation of the leg neuropil (compare arrows in E and F). In some mutant animals, ipsilateral 7B projections extended posteriorly without innervating leg neuropils (arrows in G). (H–I) 7B MARCM clones in T2 segments of the control (H) and (I) unc-4 mutant animals shown. Both ipsilateral (arrows) and contralateral (arrowheads) projections into leg neuropils were reduced in the mutant clone (I). Control clone has 68 cells; unc-4 clone has 73 cells. (C–H) Maximum z projections were made of a subset of confocal slices to reveal neuronal processes in leg neuropils. n indicates the number of animals showing the phenotype. Leg neuropils are outlined with yellow dashed lines.
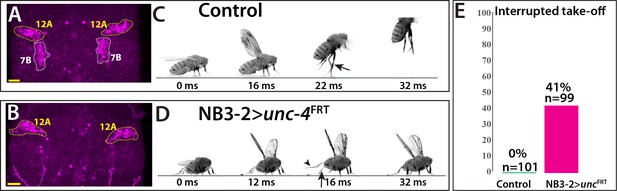
Removal of Unc-4 specifically in 7B neurons impairs take-off behavior.
(A–B) Unc-4 expression (magenta) in the T1/T2 region of the VNC shown. (A) Unc-4 expression in T1 12A and T2 7B neurons are outlined with dashed lines. (B) NB3-2-GAL4 (eyAD;dbxDBD) driven FLP removes Unc-4 expression specifically from 7B neurons. (C–D) Sequences of still images shown from videos recording the response of flies to a looming stimulus. (C) The control animal exhibited a stereotyped take-off behavior. It first raised its wings, and then extended its T2 legs (arrow) to take off. (D) The NB3-2 specific Unc-4 knock-out animal exhibited an interrupted take-off behavior. It responded to the looming stimulus and raised its wings. It attempted to jump by lifting its hindleg (arrowhead) but did not extend its T2 legs (arrow), thus failed to take off. See also Videos 14 and 15. (E) The percentage of animals that showed interrupted take-off behavior shown. See Materials and methods section for more information. The time under each image is in milliseconds and indicates the elapsed time after the visual stimulus. Scale bar is 20 microns.
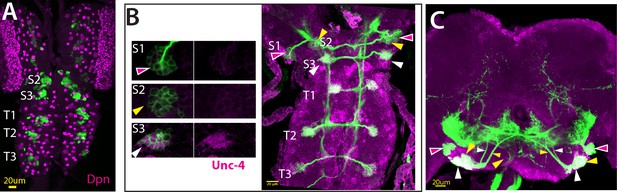
eyAD;dbxDBD marks NB3-2 in the VNC.
(A) eyAD;dbxDBD driven GFP expression specifically marks NB3-2 in all thoracic segments (T1, T2, T3) in addition to first and second abdominal segments (not visible in this projection) in the larval CNS. The same driver also marks a single NB in gnathal segments (S1, S2, S3; S1 expression is not visible in projection). No NB is marked in the central brain. (B–C) NB3-2 intersected reporter immortalization used to visualize progeny of eyAD;dbxDBD marked NBs in the larval (B) and adult CNS (C). From S3 through A2, reporter immortalization marked Unc-4+ 7B neurons. Marked neurons in S1 and S2 segments do not express Unc-4; in the S2 segment, marked neurons extend axons in a similar manner to 7B neurons. Red, yellow, and white arrowheads the neurons in S1, S2, and S3 segments, respectively.
Videos
unc-4 mutant males (unc-4DBD /y) in open field.
Climbing activity of individual flies: left, unc-4 mutant males (unc-4DBD /y); right, rescued males (uncDBD/y; tubAD/10XUAS-IVS-myr::GFP; UAS-unc-4).
Climbing activity of group of flies: left, unc-4 mutant males (unc-4DBD /y); right, rescued males (uncDBD/y; tubAD/10XUAS-IVS-myr::GFP; UAS-unc-4).
Three-leg rubbing activity of individual flies; 2X slower: left, unc-4 mutant males (unc-4DBD /y); right, rescued males (uncDBD/y; tubAD/10XUAS-IVS-myr::GFP; UAS-unc-4).
Tethered flight assay, 10X slower: left, unc-4 mutant males (unc-4DBD /y); right, rescued males (uncDBD/y; tubAD/10XUAS-IVS-myr::GFP; UAS-unc-4).
Rescue of the flight take-off activity by GAL4/UAS system (uncDBD/y; tubAD/10XUAS-IVS-myr::GFP; UAS-unc-4).
Rescue of the flight take-off activity by a BAC transgene (uncDBD/y;; PBac(DC335) /+).
Climbing activity of group of flies with the CNS and PNS specific deletion of unc-4: left, control (unc-4FRT /+; sca-GAL4/UAS-FLP.D); right, sca >unc-4FRT (unc-4FRT /y; sca-GAL4/UAS-FLP.D).
Climbing activity of group of flies with the CNS specific deletion of unc-4: left, control (unc-4FRT /+; dpnEE-GAL4/UAS-FLP); right, dpnEE >unc-4FRT (unc-4FRT /y; dpnEE-GAL4/UAS-FLP).
Three-leg rubbing activity of individual flies with the chordotonal organ lineage specific deletion of unc-4: left, control (unc-4FRT /+; ato-GAL43.6/UAS-FLP); right, ato >unc-4FRT (unc-4FRT /y; ato-GAL43.6/UAS-FLP).
Walking activity of individual flies in small-diameter tubes: left, control (unc-4FRT /+; ato-GAL43.6/UAS-FLP); middle, ato >unc-4FRT (unc-4FRT /y; ato-GAL43.6/UAS-FLP); right, unc-4 mutant (unc-4DBD /y).
Magnified version of Video 11.
Single fly shown for each gentype.
The flight take-off behavior of flies with the 7B hemilineage specific deletion of unc-4 in response to banging the vial; 2X slower: left, control (5XUAS-FLP::PEST, unc-4FRT/+; dbxDBD/+; eyAD/+); right, NB-3–2-GAL4 > unc-4FRT (5XUAS-FLP::PEST, unc-4FRT/y; dbxDBD/+; eyAD/+).
The flight take-off behavior of flies with the 7B hemilineage specific deletion of unc-4 in response to visual looming stimuli; 200X slower: left, control (5XUAS-FLP::PEST, unc-4FRT/+; dbxDBD/+; eyAD/+); right, NB-3–2-GAL4 > unc-4FRT (5XUAS-FLP::PEST, unc-4FRT/y; dbxDBD/+; eyAD/+).
Flies with 7B specific unc-4 deletion initiating flight without take-off (5XUAS-FLP::PEST, unc-4FRT/y; dbxDBD/+; eyAD/+).
The flight take-off behavior of flies with the ablated 7B hemilineage in response to banging the vial; 2X slower (UAS-hidAla5/+;; dbxDBD/+; eyAD/+).
Tables
Three-leg rubbing and climbing behavior in response to tissue specific removal of Unc-4.
| Success in three-leg rubbing | Success in climbing, time to climb§ (in seconds) | ||||
|---|---|---|---|---|---|
| Driver | Target cell types | Control | unc-4 removal | Control | unc-4 removal |
| sca-GAL4 | All CNS and PNS neurons | 88.9%, N = 18 | 0%*, N = 20 | 100%, 17.7 s. +/- 8, N = 22 | 9.1%*, N/A¶, N = 22 |
| dpn-GAL4 | All CNS neurons | 81.8%, N = 22 | 28.5%*, N = 21 | 90%, 20.1 s. +/- 15, N = 30 | 80%†, 27.6 s.‡+/- 20.5, N = 28 |
| ato-Gal4 | Chordotonal neurons | 90%, N = 33 | 14.8%*, N = 27 | N/A** | N/A** |
-
*Significant change, Fisher exact test P value < 0.001.
†Non-significant change, Fisher exact test p value>0.1.
-
‡Non-significant change, Mann-Whitney U Test p value>0.1.
§failed climbing attempts not included for quantification.
-
¶ not assessed since most animals failed to climb.
**ato-GAL4 >FLP expression impaired the negative geotaxis in both control and experimental animals, thus climbing assay was not performed.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Guinea pig anti-Sens polyclonal | PMID:10975525 | 1:1000 dilution | |
| Antibody | Rabbit anti- GABA polyclonal | Sigma | A2052 | 1:1000 dilution |
| Antibody | Chicken anti-GFP polyclonal | Life Tech. | A10262 | 1:1000 dilution |
| Antibody | Rabbit anti-HA polyclonal | Cell sig. | 3724S | 1:500 dilution |
| Antibody | Rabbit anti-GFP polyclonal | Thermo Fisher S. | A11122 | 1:1000 dilution |
| Antibody | Rabbit anti-Unc-4 polyclonal | PMID:24550109 | 1:1000 dilution | |
| Antibody | Rabbit Eve | PMID:2884106 | 1:2000 dilution | |
| Antibody | Mouse anti-Acj6 monoclonal | DSHB | Acj6 | 1:100 dilution |
| Antibody | Rat anti-Cadherin monoclonal | DSHB | DN-Ex#8 | 1:25 dilution |
| Antibody | goat anti-rabbit Alexa Fluor 488 | Life Technologies | A-11034 | 1:500 dilution |
| Antibody | goat anti-rabbit Alexa Fluor 568 | Life Technologies | A-11011 | 1:500 dilution |
| Antibody | goat anti- rabbit Alexa Fluor 633 | Life Technologies | A-21070 | 1:500 dilution |
| Antibody | goat anti-chicken Alexa Fluor 488 | Life Technologies | A-11039 | 1:500 dilution |
| Antibody | goat anti-mouse Alexa Fluor 568 | Life Technologies | A-11004 | 1:500 dilution |
| Antibody | goat anti-mouse Alexa Fluor 633 | Life Technologies | A-21050 | 1:500 dilution |
| Antibody | goat anti-rat Alexa Fluor568 | Life Technologies | A-11077 | 1:500 dilution |
| Antibody | goat anti-rat Alexa Fluor633 | Life Technologies | A-21094 | 1:500 dilution |
| Genetic reagent (D. melanogaster) | MKRS, P{ry[+t7.2]=hsFLP}86E/TM6B, P{w[+mC]=Crew}DH2, Tb[1] | BloomingtonDrosophila Stock Center | RRID:BDSC_1501 | |
| Genetic reagent (D. melanogaster) | P{ry[+t7.2]=hsFLP}1, w[1118]; Adv[1]/CyO | BloomingtonDrosophila Stock Center | RRID:BDSC_6 | |
| Genetic reagent (D. melanogaster) | y1 w*; P{UAS-FLP.D}JD1 | BloomingtonDrosophila Stock Center | RRID:BDSC_4539 | |
| Genetic reagent (D. melanogaster) | w*; P{UAS-p35.H}BH1 | BloomingtonDrosophila Stock Center | RRID:BDSC_5072 | |
| Genetic reagent (D. melanogaster) | P{Tub-dVP16AD.D} | BloomingtonDrosophila Stock Center | RRID:BDSC_60295 | |
| Genetic reagent (D. melanogaster) | P{Tub-GAL4DBD.D} | Bloomington Drosophila Stock Center | RRID:BDSC_60298 | |
| Genetic reagent (D. melanogaster) | w[*]; P{w[+mC]=iav-GAL4.K}3 | BloomingtonDrosophila Stock Center | RRID:BDSC_52273 | |
| Genetic reagent (D. melanogaster) | y[1] w[*]; Mi{y[+mDint2]=MIC}TfAP-2[MI04611]/TM3, Sb[1] Ser[1] | BloomingtonDrosophila Stock Center | RRID:BDSC_37965 | |
| Genetic reagent (D. melanogaster) | 13XLexAop2-IVS-myr::GFP in attP40 | BloomingtonDrosophila Stock Center | RRID:BDSC_32210 | |
| Genetic reagent (D. melanogaster) | w[*]; P{w[+mC]=tubP-GAL80[ts]}2/TM2 | BloomingtonDrosophilaStock Center | RRID:BDSC_7017 | |
| Genetic reagent (D. melanogaster) | hsFlp2::PEST;; HA_V5_FLAG-MCFO | PMID:25964354 | RRID:BDSC_9494 | |
| Genetic reagent (D. melanogaster) | y1 w*; P{ato-GAL4.3.6}10 | PMID:10774724 | ||
| Genetic reagent (D. melanogaster) | Dpn-EE-GAL4-attp16;T3/T6b | PMID:26700685 | ||
| Genetic reagent (D. melanogaster) | pJFRC29-10XUAS-IVS-myr::GFP-p10 in attP40 | PMID:22493255 | ||
| Genetic reagent (D. melanogaster) | pJFRC105-10XUAS-IVS-nlstdTomatoi n VK00040 | PMID:22493255 | ||
| Genetic reagent (D. melanogaster) | Poxn-GAL4.13 | PMID:12421707 | ||
| Genetic reagent (D. melanogaster) | y[1] w[*]; Mi{y[+mDint2]=MIC}TfAP-2[MI04611]/TM3, Sb[1] Ser[1] | BloomingtonDrosophilaStock Center | RRID:BDSC_37965 | |
| Genetic reagent (D. melanogaster) | UAS-hidAla5 | PMID:9814704 | ||
| Genetic reagent (D. melanogaster) | Actin5Cp4.6>dsFRT > nlsLexAp65 | G Rubin | ||
| Genetic reagent (D. melanogaster) | pJFRC51-3XUAS-IVS-Syt::smGFP-HA in attP40 | G Rubin | ||
| Genetic reagent (D. melanogaster) | pJFRC12-10XUAS-IVS-myr::GFP in attP18 | G Rubin | ||
| Genetic reagent (D. melanogaster) | unc-4-GAL4 | this study | FBgn0024184 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | unc-4-AD | this study | FBgn0024184 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | unc-4-DBD | this study | FBgn0024184 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | unc-4-FRT | this study | FBgn0024184 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | unc-4-null | this study | FBgn0024184 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | TfAP-2-AD | this study | FBgn0261953 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | TfAP-2-GAL4 | this study | FBgn0261953 | built by H. Lacin |
| Genetic reagent (D. melanogaster) | w*;sca-GAL4 | PMID:2125959 | ||
| Chemical compound, drug | Paraformaldehyde | EMS | 15713 | |
| Chemical compound, drug | Vectashield | Vector | H-1000 | |
| Chemical compound, drug | Prolong Diamond | Molecular Probes | P36961 | |
| Recombinant DNA reagent | pHD-DsRed-attP | Addgene | 51019 | |
| Recombinant DNA reagent | pCFD4-U6:1_U6:3tandemgRNAs | Addgene | 49411 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(1)-T2A-Gal4-Hsp70 | Addgene | 62897 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(1)-T2A-Gal4DBD-Hsp70 | Addgene | 62903 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(1)-T2A-p65AD-Hsp70 | Addgene | 62914 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(0)-T2A-Gal4-Hsp70 | Addgene | 62896 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(0)- T2A-Gal4DBD-Hsp70 | Addgene | 62902 | |
| Recombinant DNA reagent | pBS-KS-attB2-SA(0)-T2A-p65AD-Hsp70 | Addgene | 62912 |
Additional files
-
Supplementary file 1
Genotypes of fly lines used in each experiment.
- https://cdn.elifesciences.org/articles/55007/elife-55007-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55007/elife-55007-transrepform-v2.docx





