Wiskott Aldrich syndrome protein regulates non-selective autophagy and mitochondrial homeostasis in human myeloid cells
Figures
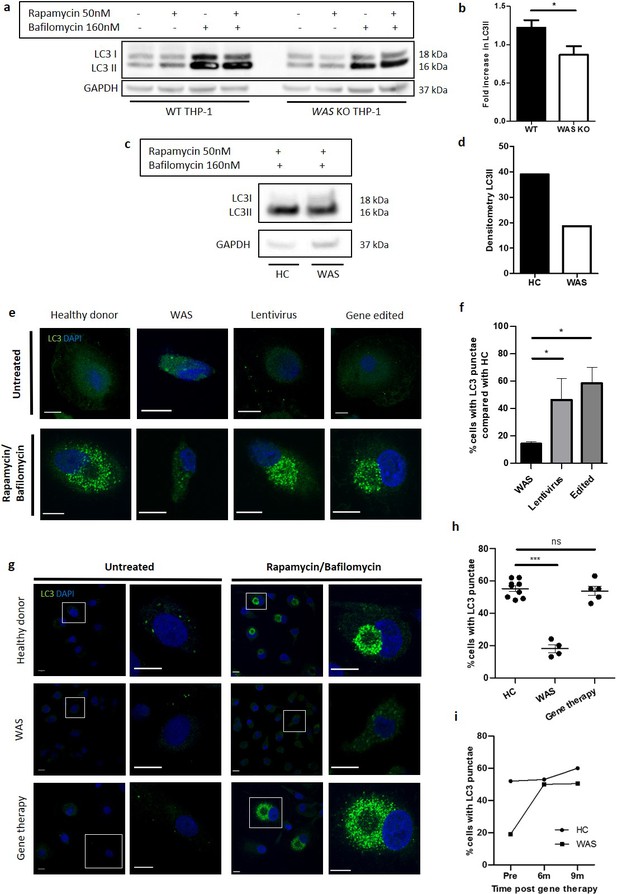
WASp is necessary for autophagosome formation.
(a) WT and WAS KO THP-1 were cultured with rapamycin 50 nM +/- bafilomycin 160 nM for 6 hr and cell lysates immunoblotted for LC3I and LC3II with GAPDH as loading control. Representative blot from five independent experiments. (b) Combined densitometric analysis from (a). Histogram of fold increase in LC3II from baseline control normalised to GAPDH loading control following rapamycin treatment. Bars represent mean +/- SEM (n = 5 in duplicate). Two-tailed t-test *p<0.05 (c). MDMs from a healthy donor and WAS patient cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr and cell lysates immunoblotted for LC3I and LC3II with GAPDH as loading control. (d) Densitometric analysis of (c). Histogram of LC3II densitometry normalised to GAPDH loading control following rapamycin and bafilomycin treatment (n = 1 in duplicate). (e) Representative images of SDMs from a healthy donor, WAS patient and in vitro corrected WAS patient SDMs using lentiviral transduction or gene editing techniques cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x. Scale bar represents 10 µm. Details of the level of WASp expression and gene marking in the experiments summarised in e and f can be found in Figure 1—source data 2. (f) SDMs from healthy donor, WAS patient and in vitro corrected WAS patient SDMs using gene therapy or gene editing techniques cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3. Slides blinded for imaging and analysis. Cells forming significant LC3 punctae were counted from confocal microscopy images taken at 20x. 300–1000 cells per slide analysed from six fields of view. Histogram displays percentage of cells with LC3 punctae compared with their matched healthy donors from the same experiment with bars representing median +/- IQR. Combined analysis from four independent experiments. Mann Whitney *p<0.05. (g) Representative images of MDMs from a healthy donor (top panel), WAS patient (middle panel), and WAS patient after treatment with gene therapy (lower panel) cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x (first column), with further detail of areas highlighted by white boxes shown to the right. Scale bars represent 10 µm. (h) MDMs from healthy donors (n = 5), WAS patients (n = 3) and WAS patients after treatment with gene therapy (n = 2) cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green) and nuclei (DAPI, blue). Slides blinded for imaging and analysis. Cells forming significant LC3 punctae were counted from confocal microscopy images taken at 20x. 300–1000 cells per slide analysed from six fields of view. Dot plot of percentage of cells forming LC3 punctae displayed, with points representing independent experiments. Bars represent mean +/- SEM. Two-tailed t-test ***p<0.0001. (i) MDMs from healthy donors and the same WAS patient before and after gene therapy treatment cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green) and nuclei (DAPI, blue). Slides blinded for imaging and analysis. Cells forming significant LC3 punctae were counted from confocal microscopy images taken at 20x. 300–1000 cells per slide analysed from six fields of view. Percentage of cells forming LC3 punctae displayed. DAPI, 4’,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HC, healthy control; IQR, interquartile range; MDM, monocyte-derived macrophage; ns, not significant; SDM, stem-cell-derived macrophage; SEM, standard error of the mean; WAS, Wiskott Aldrich syndrome.
-
Figure 1—source data 1
Molecular details of patient monocyte-derived macrophages used in experiments.
^Additional variant in WAS gene (c.391G > A, p.Glu1331Lys). ARPC1B, actin-related protein C1B-deficiency; GT, in vivo gene therapy; PBMCs, peripheral blood mononuclear cells; VCN, vector copy number; WAS, Wiskott Aldrich syndrome.
- https://cdn.elifesciences.org/articles/55547/elife-55547-fig1-data1-v2.docx
-
Figure 1—source data 2
Molecular details of patient stem-cell-derived macrophages used for in vitro WAS correction WASp, Wiskott Aldrich syndrome protein.
^Same patient.
- https://cdn.elifesciences.org/articles/55547/elife-55547-fig1-data2-v2.docx
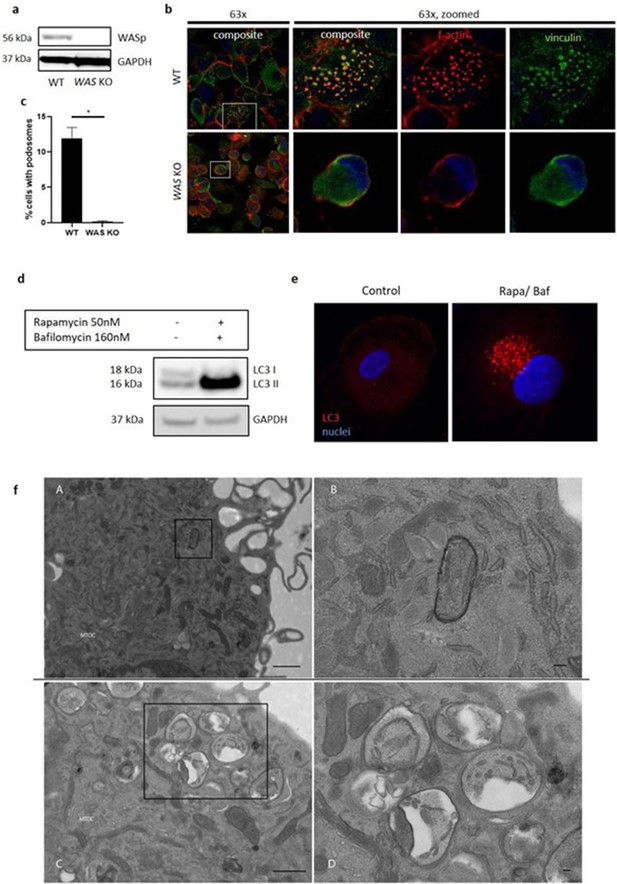
WASp is necessary for autophagosome formation.
(a) WT and WAS KO THP-1 cell lysates immunoblotted for WASp expression with GAPDH as loading control. (b) WT and WAS KO THP-1 cells fixed and stained for vinculin (green) and f-actin (phalloidin, red). Imaged at 63x by confocal microscopy, with higher magnification of white box highlighted to the right. (c) Podosome analysis from (b) taken from at least 100 cells per sample. Two tailed t-test *p<0.05. (d) Healthy donor MDMs were cultured in complete RPMI or exposed to rapamycin 50 nM and bafilomycin 160 nM for 6 hr as indicated and immunoblotted for LC3 expression. (e) MDMs from the same experiment in (d) were fixed and stained for LC3 (red) and nuclei (DAPI, blue) and imaged by confocal microscopy at 63x. (f) Healthy donor MDMs were cultured in complete RPMI (A, enlarged in B) or rapamycin 50 nM/bafilomycin 160 nM (C, enlarged in D) and analysed by electron microscopy. MTOC denotes microtubule organising centre.
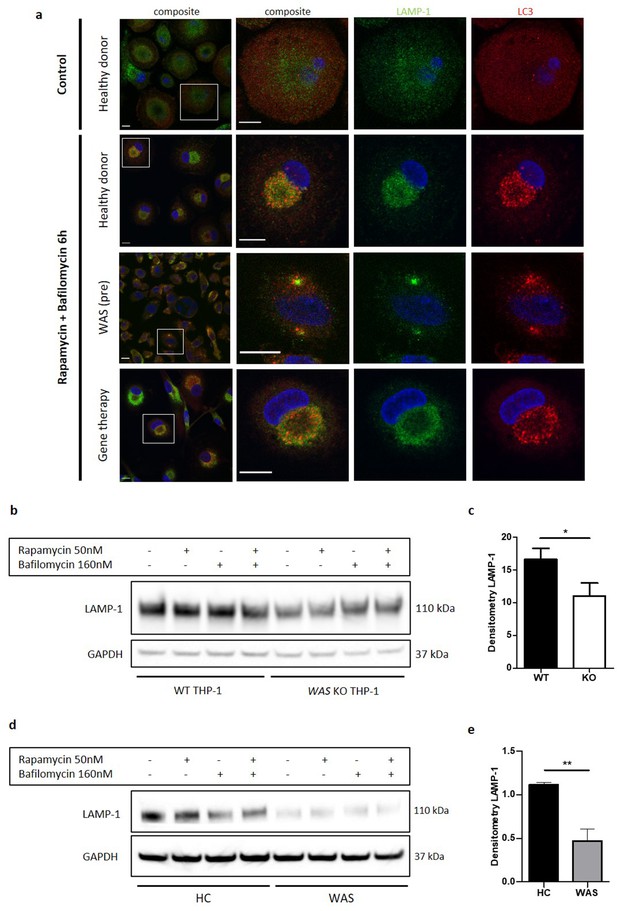
WASp is necessary for delivery of autiophagosomes to lysosomes.
(a) MDMs from a healthy donor, WAS patient and WAS patient following treatment with gene therapy cultured with or without rapamycin 50 nM and bafilomycin 160 nM as indicated for 6 hr. Cells fixed and stained for LC3 (red), LAMP-1 (green), and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x, with further magnification of selected area highlighted by white box. Scale bar 10 µm. Representative images from five healthy donors, two WAS patients and two WAS patients post gene therapy. (b) WT and WAS KO THP-1 cultured with rapamycin 50 nM +/- bafilomycin 160 nM for 6 hr as indicated. Cell lysates immunoblotted for LAMP-1 expression, with GAPDH as loading control. Representative blot from three independent experiments. (c) Combined densitometric analysis from (b). Histogram shows densitometry of LAMP-1 normalised to GAPDH loading control from unstimulated WT and WAS KO THP-1 cells (n = 3 in duplicate). Bars represent mean +/- SEM. Two-tailed t-test *p<0.05. (d) CD14+ PBMCs from healthy donors or WAS patients cultured with rapamycin 50 nM +/- bafilomycin 160 nM for 1.5 hr as indicated. Cell lysates immunoblotted for LAMP-1 expression, with GAPDH as loading control. Representative blot from three independent experiments in duplicate. (e) Combined densitometric analysis from (d). Histogram shows densitometry of LAMP-1 normalised to GAPDH loading control for unstimulated healthy donor and WAS patient MDMs. Bars represent mean +/- SEM (n = 3 in duplicate). Two-tailed t-test **p<0.01 DAPI, 4’,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HC, healthy control; MDM, monocyte-derived macrophage; PBMCs, peripheral blood mononuclear cells; SEM, standard error of the mean; WAS, Wiskott Aldrich syndrome.
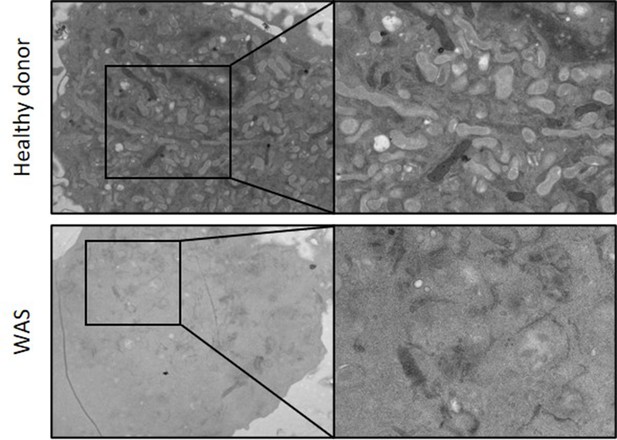
WASp is necessary for autophagosome formation.
Healthy donor and WAS MDMs cultured in complete RPMI and analysed by electron microscopy.
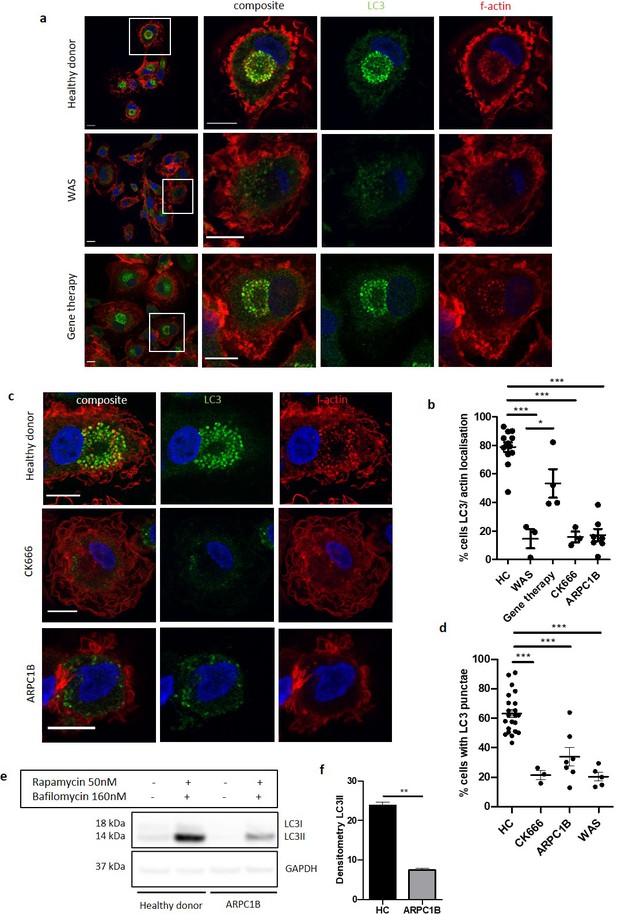
The role of WASp in non-selective autophagy is ARP2/3-dependent.
(a) Representative images of MDMs from a healthy donor, WAS patient and WAS patient post gene therapy cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green), f-actin (phalloidin, red) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x, with further magnification of highlighted area inside white boxes. Scale bar = 10 µm (b) MDMs from healthy donors (n = 7), WAS patients (n = 2), WAS patients post gene therapy (n = 2), healthy donor MDMs treated with CK666 20 µM (n = 2) and ARPC1B (n = 4)-deficient patients cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green), f-actin (phalloidin, red) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x and percentage of cells with LC3 and actin localisation calculated from 50 to 100 cells per slide from at least three fields of view. Dots represent independent experiments and bars represent mean +/- SEM. Two-tailed t-test *p<0.05, ***p<0.0001. (c) Representative images of MDMs from healthy donors, CK666 20µM-treated healthy donors and ARPC1B-deficient patients cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green), f-actin (phalloidin, red), and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x. Scale bar = 10 µm. (d) MDMs from healthy donors (n = 7), CK666 20µM-treated healthy donors (n = 2) and ARPC1B (n = 4)-deficient patients cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3. Imaged by confocal microscopy at 20x and cells forming significant LC3 punctae analysed from 500 to 1000 cells from at least six fields of view per slide. Dots represent independent experiments with bars representing mean +/- SEM. Two-tailed t-test ***p<0.0001. (e) MDMs from two healthy controls and two ARPC1B-deficient patients cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cell lysates immunoblotted for LC3I and LC3II expression, with GAPDH for loading control. Representative blot from two independent experiments in duplicate. (f) Combined densitometric analysis from (e). Histogram shows densitometry of LC3II expression normalised to GAPDH loading control from rapamycin and bafilomycin treated MDMs (n = 2 independent experiments in duplicate). DAPI, 4’,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HC, healthy control; MDM, monocyte-derived macrophage; SEM, standard error of the mean; WAS, Wiskott Aldrich syndrome.
-
Figure 3—source data 1
Summary of LC3-interating regions of human WASp and all seven subunits of the ARP2/3 complex as identified using the web resource developed by Kalvari et al.
Meaningful PSSM scores predictive of possible LIR domains are identified in the range 13–17 (highlighted in grey), with balanced accuracy most optimal at 15 or 16.
- https://cdn.elifesciences.org/articles/55547/elife-55547-fig3-data1-v2.docx
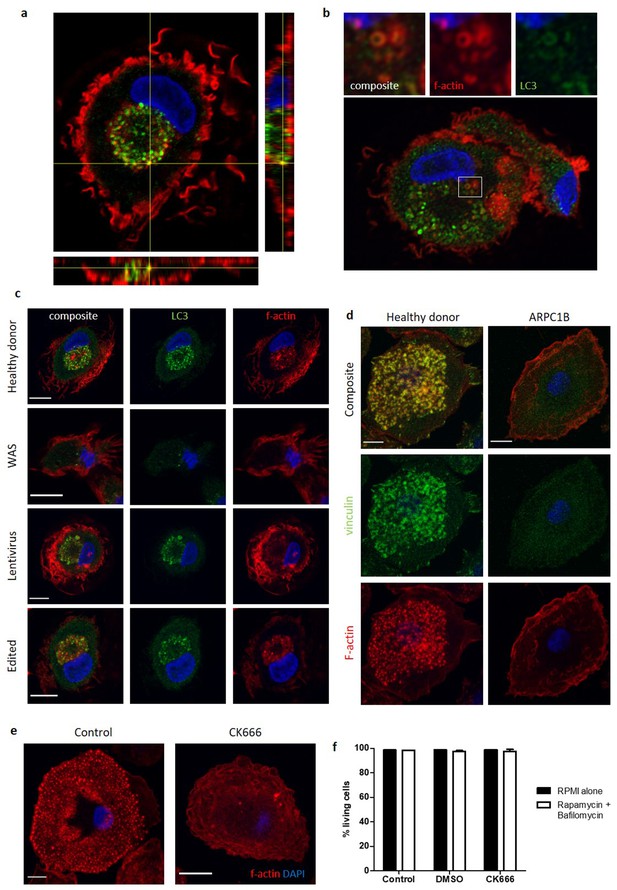
The role of WASp in non-selective autophagy is ARP2/3 dependent.
(a) Representative image of healthy donor MDMs cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr, fixed and stained for f-actin (phalloidin, red), LC3 (green), and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x with z axis with respect to the x and y axes highlighted below and to the right of the image respectively, to provide depth detail of the cell pictured. A highlighted yellow punctum signifies co-localisation of LC3 and f-actin, which through following the yellow lines to the z axis can be seen to be present in the centre of the cell. (b) Deconvolved super resolution confocal microscopy image of healthy donor MDM cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Stained for f-actin (phalloidin, red), LC3 (green) and nuclei (DAPI, blue) and imaged at 63x. Higher magnification of area highlighted by white box detailed above. (c) Healthy donor, WAS patient and in vitro WASp reconstituted SDMs using lentiviral vector or gene editing techniques cultured with rapamycin 50 nM and bafilomycin 160 nM for 6 hr. Cells fixed and stained for LC3 (green), f-actin (phalloidin, red) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x. Scale bar = 10 µm. Representative images shown from n = 4. Details of the level of WASp expression and gene marking in the experiments can be found in Figure 1—source data 2. (d) Healthy donor and ARPC1B-deficient MDMs fixed on cover slips coated in fibronectin and stained for vinculin (green) and f-actin (phalloidin, red). Imaged at 63x by confocal microscopy. Scale bar = 10 µm. Representative images shown from n = 3. (e) Healthy control MDMs with or without ARP2/3 inhibition with CK666 20 µM, fixed and stained for f-actin (phalloidin, red) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x. Scale bar = 10 µm. Representative images from n = 4. (f) Cell death from rapamycin 50 nM, bafilomycin 160 nM, DMSO 0.1%, CK666 20 µM treatment of healthy donor MDMs measured by percentage of cells negative for DAPI staining by flow cytometry. Summary of n = 2 in duplicate. Error bars represent mean +/- SEM. DAPI, 4’,6-diamidino-2-phenylindole; DMSO, dimethylsulfoxide; MDMs, monocyte-derived macrophages; SEM, standard error of the mean; SDMs, stem-cell-derived macrophages; WAS, Wiskott Aldrich syndrome.
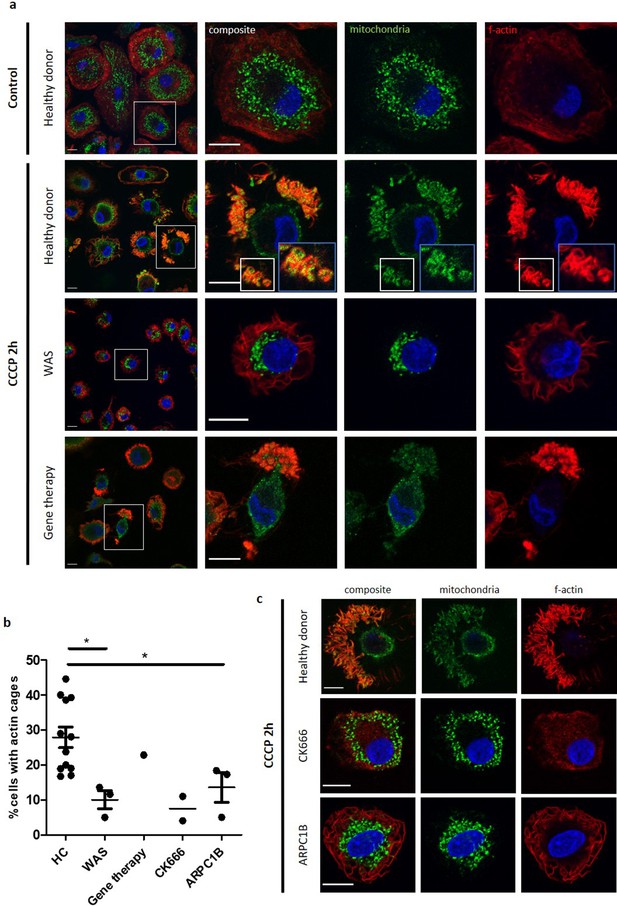
WASp plays an ARP2/3-dependent role in mitophagy.
(a) MDMs from a healthy donor, WAS patient and WAS patient post gene therapy cultured with or without CCCP 10 µM (plus bafilomycin 160 nM) for 2 hr. Cells fixed and stained for mitochondria (biotin, green), f-actin (phalloidin, red) and nuclei (DAPI, blue) and imaged by confocal microscopy. Representative images illustrated at 63x, with higher magnification of areas highlighted by white boxes. Actin cages further magnified from healthy donor, denoted by blue boxes (inset). Scale bars = 10 µm. (b) Healthy donor (n = 7), WAS patient (n = 2), WAS patient post-gene therapy (n = 1), CK666 100µM-treated healthy donor (n = 1) and ARPC1B-deficient patient (n = 3) MDMs cultured with CCCP 10 µM (plus bafilomycin 160 nM) for 2 hr. Cells fixed and stained for mitochondria and f-actin. Slides blinded prior to imaging and analysis. At least six fields of view per slide were imaged at 63x by confocal microscopy. Cells forming actin cages around mitochondria were analysed from at least 100 cells per slide. Dots represent independent experiments, with bars denoting mean +/- SEM. Two-tailed t-test *p<0.05. (c) Representative images of healthy donor, CK666 100µM-treated healthy donor and ARPC1B-deficient MDMs cultured with CCCP 10 µM (plus bafilomycin 160 nM) for 2 hr. Cells fixed and stained for mitochondria (biotin, green), f-actin (phalloidin, red), and nuclei (DAPI, blue) and imaged by confocal microscopy at 63x. Scale bars = 10 µm. CCCP, carbonyl cyanide m-chlorophenylhydrazone; DAPI, 4’,6-diamidino-2-phenylindole; HC, healthy control; MDM, monocyte-derived macrophage; SEM, standard error of the mean; WAS, Wiskott Aldrich syndrome.
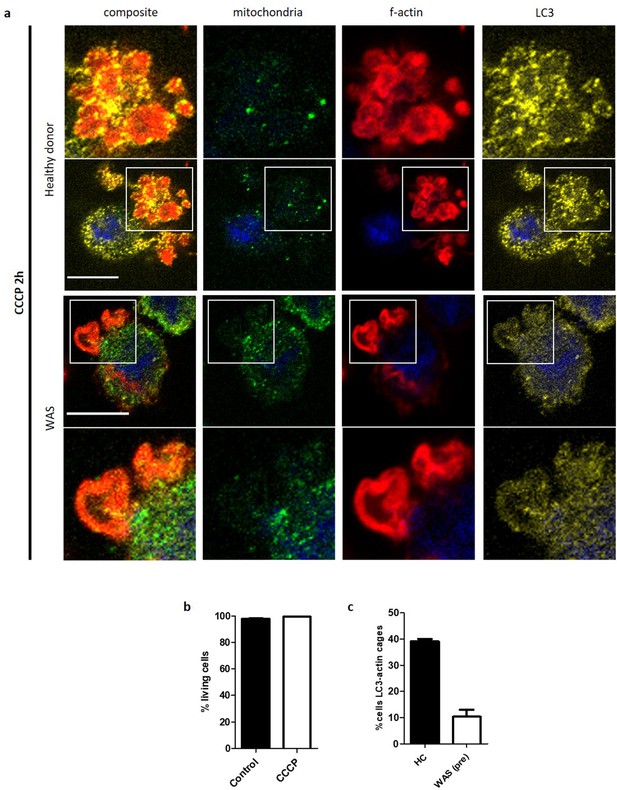
WASp plays an ARP2/3-dependent role in mitophagy.
(a) Healthy donor and WAS patient MDMs cultured with CCCP 10 µM and bafilomycin 160 nM for 2 hr. Fixed and stained for mitochondria (biotin, green), f-actin (phalloidin, red), LC3 (yellow), and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x, with area highlighted by white boxes enlarged either above (for healthy donor) or below (for WAS patient). Scale bar = 10 µm. (b) Percentage of healthy donor or WAS patient MDMs with LC3 associating with actin cages following 2 hr treatment with CCCP 10 µM and bafilomycin 160 nM. Mean +/- SEM from two independent experiments. (c) Cell death from CCCP treatment calculated from healthy donor MDMs treated with CCCP 10 µM for 2 hr and analysed by flow cytometry following DAPI staining. Summary of n = 2 in duplicate. Mean +/- SEM. CCCP, carbonyl cyanide m-chlorophenylhydrazone; DAPI, 4’,6-diamidino-2-phenylindole;HC, healthy control; MDMs, monocyte-derived macrophages; SEM, standard error of the mean; WAS, Wiskott Aldrich syndrome.

WASp is necessary for mitochondrial homeostasis.
(a) Representative images of different mitochondrial morphologies from healthy donor MDMs fixed and stained for mitochondria (biotin, green) and nuclei (DAPI, blue). Imaged by confocal microscopy at 63x. Scale bar = 10 µm. (b) Representative images of healthy donor (n = 7), WAS patient (n = 3) or WAS patient post gene therapy (n = 2) MDMs fixed and stained for mitochondria (biotin, green), f-actin (phalloidin, red) and nuclei (DAPI, blue). Imaged at 63x by confocal microscopy, with higher magnification of area inside white boxes displayed to the right. Scale bar = 10 µm. (c) Combined analysis from (b and d). Slides blinded prior to imaging and analysis. At least 100 cells per slide analysed for mitochondrial morphology and categorised according to appearance as indicated. Bars represent mean +/- SEM. (d) Representative images of healthy donor MDMs (n = 7), CK666 100µM-treated healthy donor MDMs (n = 2), or MDMs from ARPC1B-deficient patients (n = 3) fixed and stained for mitochondria (biotin, green) and nuclei (DAPI, blue). Imaged at 63x by confocal microscopy and higher magnification of area highlighted by white boxes shown to the right. Scale bar = 10 µm. DAPI, 4’,6-diamidino-2-phenylindole; HC, healthy control; MDM, monocyte-derived macrophage; SEM, standard error of mean; WAS, Wiskott Aldrich syndrome.
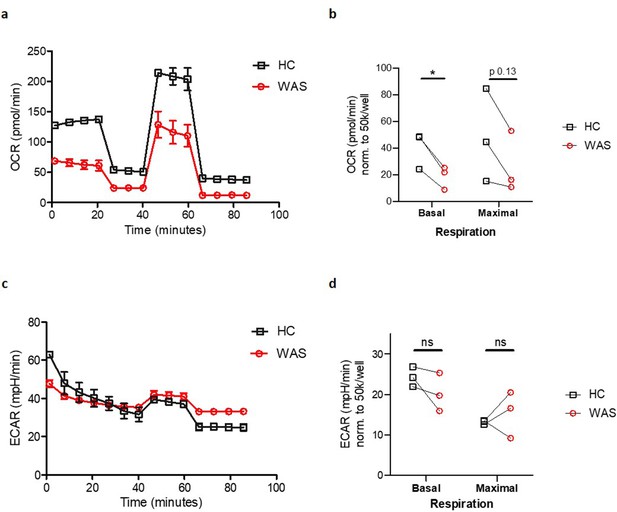
WASp deficiency is associated with impaired mitochondrial function.
(a) Oxygen consumption rate (OCR) of MDMs from healthy donors and WAS patients during Mito Stress test. Representative plot from three independent experiments in triplicate. (b) Summary of OCR from three independent experiments, normalised to 50 × 103 cells/well, with OCR at basal and maximal respiration from WAS patient MDMs paired to their respective healthy controls (HC). Paired t-test *p<0.05, ns = not significant. (c) Extracellular acidification rates (ECAR) of MDMs from healthy donors and WAS patients during Mito Stress test. Representative plot from three independent experiments in triplicate. (d) Summary of ECAR from three independent experiments, normalised to 50 × 103 cells/well, with ECAR at basal and maximal respiration from WAS patient MDMs paired to their respective healthy controls (HC). Paired t-test ns = not significant.
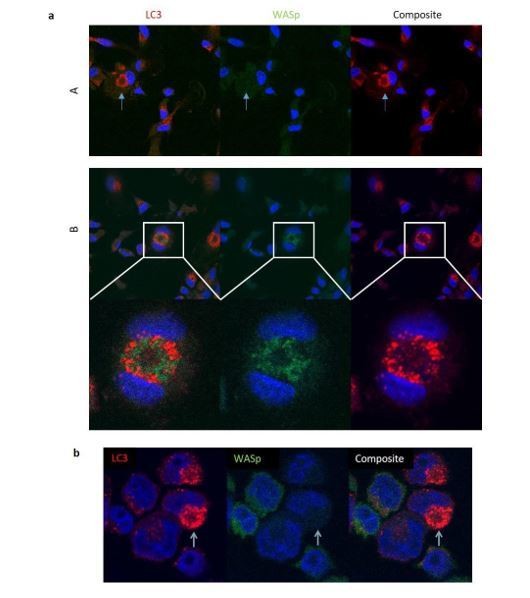
a) Healthy donor MDMs cultured with rapamycin and bafilomycin for 6 hours.
Fixed and stained for LC3 (red), WASp (green) and DAPI (blue). Analysed by confocal microscopy at 63x. Panel A displays an example of LC3 punctae without WASp localisation. Panel B displays an example where LC3 appears to be localising with WASp. b) eGFP-WASp THP-1 cells cultured with rapamycin and bafilomycin for 6 hours. Fixed and stained for LC3 (red), nuclei (DAPI). Analysed by confocal microscopy at 63x.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | WT THP-1 | ATCC | TIB-202 | Male, 1 year infant |
| Cell line (Homo sapiens) | WAS KO THP-1 | doi: 10.1038/s41467-017-01676-0 | Male, 1 year infant | |
| Antibody | Anti-human WASP (Mouse monoclonal) | BD Bioscience | Cat. #: 557773, RRID:AB_396867 | FACS (1:100), WB (1:500) |
| Antibody | Anti-mouse IgG Alexa Fluor 647 (Goat monoclonal) | BioLegend | Cat. #: 405301, RRID:AB_315005 | FACS (1:200) |
| Antibody | Anti-human LAMP-1 (Mouse monoclonal) | BD Bioscience | Cat. #: 17–611043; RRID:AB_3983356 | WB (1 in 1000) |
| Antibody | Anti-human LC3B (Rabbit monoclonal) | Sigma | Cat. #: L7543; RRID:AB_796155 | WB (one in1000) |
| Antibody | Anti-human GAPDH (Mouse monoclonal) | Santa Cruz | Cat. #: sc365062; RRID:AB_10847862 | WB (1 in 1000) |
| Antibody | Anti-mouse IgG HRP (Sheep monoclonal) | GE Healthcare | Cat. #: NA9310-1ML; RRID:AB_772193 | WB (1 in 2000) |
| Antibody | Anti-rabbit IgG HRP (Donkey monoclonal) | GE Healthcare | Cat. #: NA9340-1ML; RRID:AB_772191 | WB (1 in 2000) |
| Antibody | Anti-human LC3 (Rabbit polyclonal) | MBL | Cat. #: PM036; RRID:AB_2274121 | IF (1 in 200) |
| Antibody | Anti-human LAMP-1 (Mouse monoclonal) | CST | Cat. #: 15665; RRID:AB_2798750 | IF (1 in 50) |
| Antibody | Anti-human Vinculin (Mouse monoclonal) | Sigma | Cat. #: V4505; RRID:AB_477617 | IF (1:200) |
| Antibody | Anti-mouse IgG Alexa Fluor 488 (Goat polyclonal) | Molecular Probes | Cat. #: A32723; RRID:AB_2633275 | IF (1 in 500) |
| Antibody | Anti-rabbit IgG Alexa Fluor 546 (Goat polyclonal) | Molecular Probes | Cat. #: A-11035; RRID:AB_143051 | IF (1 in 500) |
| Antibody | Anti-rabbit IgG Alexa Fluor 647 (Goat polyclonal) | Molecular Probes | Cat. #: A27040; RRID:AB_2536101 | IF (1 in 500) |
| peptide, recombinant protein | Fibronectin | R and D system | Cat. #: 4305-FNB-200 | |
| commercial assay or kit | CD14+ Microbead | Miltenyi Biotec | Cat. #: 130-050-201 | |
| Chemical compound, drug | Rapamycin | Calbiochem | Cat. #: 553211 | |
| Chemical compound, drug | Bafilomycin A1 | Sigma | Cat. #: SML1661 | |
| Chemical compound, drug | CK666 | Abcam | Cat. #: ab141231 | |
| Chemical compound, drug | CCCP | Merck | Cat. #: 215911 | |
| Chemical compound, drug | Oligomycin | Sigma | Cat. #: 75351 | |
| Chemical compound, drug | FCCP | Sigma | Cat. #: C2920 | |
| Chemical compound, drug | Rotenone | Sigma | Cat. #: R8875 | |
| Software, algorithm | ImageJ | NIH | RRID:SCR_003070 | |
| Software, algorithm | Prism | GraphPad | Version 8 | |
| Software, algorithm | iLIR | doi: 10.4161/auto.28260 | Version 1 | |
| Other | Prolong Diamond anti-fade mounting solution with DAPI | Molecular probes | Cat. #: P36962 | |
| Other | Phalloidin-633 | Molecular Probes | Cat. #: A22284 |





