Histone deacetylase knockouts modify transcription, CAG instability and nuclear pathology in Huntington disease mice
Figures
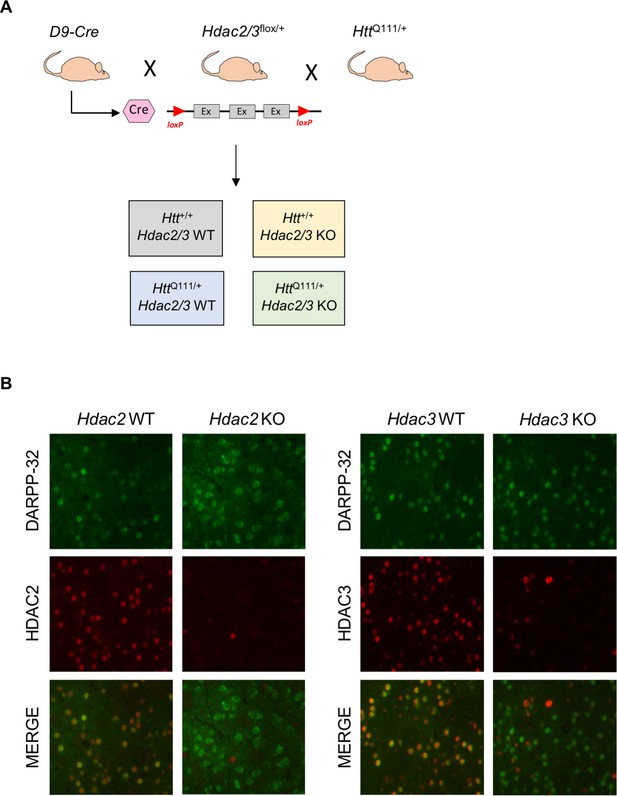
MSN-specific deletion of Hdac2 or Hdac3.
(A) D9-Cre mice, expressing Cre recombinase from the Ppp1r1b promoter from 5 to 6 weeks of age, were crossed with floxed Hdac2 or Hdac3 mice (schematic illustrates exons flanked by loxP sites), and with HttQ111/+ mice to obtain mice with or without a single HttQ111 allele either expressing HDAC2/3 (WT) or with Hdac2 or Hdac3 allele deleted in MSNs (KO). See Figure 1—figure supplement 1 for a detailed breeding scheme. (B) Fluorescent micrographs of 10 week HttQ111/+ Hdac2 KO (left) and HttQ111/+ Hdac3 KO (right) striata co-stained with DARPP-32/HDAC2 and DARPP-32/HDAC3 antibodies, respectively.
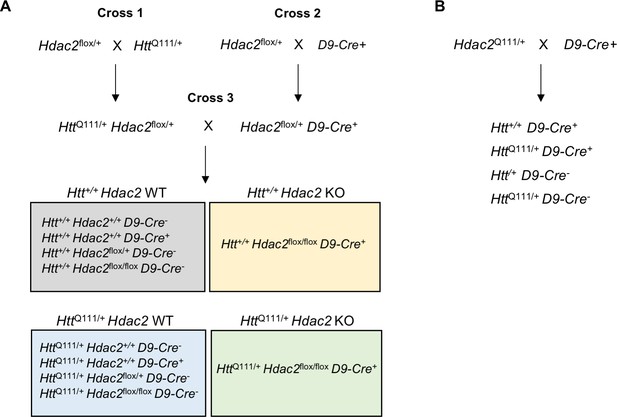
Detailed mating scheme.
(A) The scheme is shown for Hdac2 but the same scheme was implemented for Hdac3. Mice carrying conditional floxed Hdac2 or Hdac3 alleles were crossed with HttQ111 mice (cross 1) and, in a separate cross, with D9-Cre mice (cross 2). Progeny from the two crosses were bred together (cross 3) to obtain Htt+/+ and HttQ111/+ mice with or without the Hdac2 or Hdac3 floxed allele and/or Cre transgene. Mice harboring either no floxed Hdac alleles in the presence of Cre or one, two, or no floxed alleles in the absence of Cre were wild-type with respect to HDAC expression and are referred to as Hdac2 WT or Hdac3 WT. Mice homozygous for the floxed Hdac2 or Hdac3 alleles and expressing Cre are referred to as Hdac2 ΚO or Hdac3 ΚO. In these mice, the Hdac2 or Hdac3 alleles are deleted in MSNs and they do not express HDAC2 or HDAC3 in MSNs. (B). In a separate cross, HttQ111/+ mice were crossed with D9-Cre mice to generate Htt+/+ and HttQ111/+ mice with or without the Cre transgene, with the purpose of testing the effect of Cre expression alone.
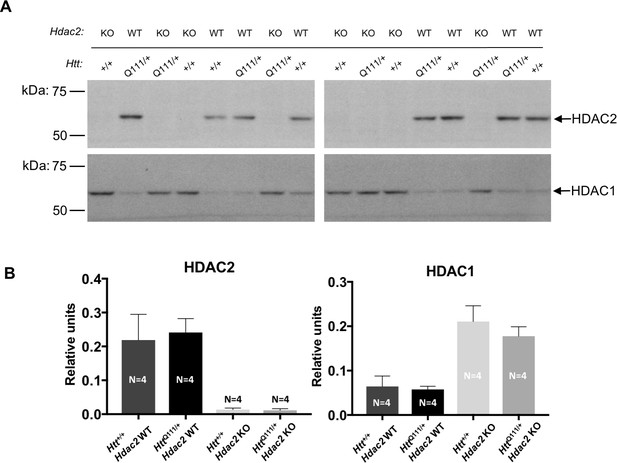
Upregulation of HDAC1 in Hdac2 KO striata.
(A) Total lysates of contralateral striata of a subset of 5-month Hdac2 KO mice from the RNAseq cohort (N = 4 for each of the four compared genotypes, as indicated on the bar graphs), probed with anti-HDAC2 antibody (top) and subsequently with anti-HDAC1 antibody (bottom). HDAC2 and HDAC1 bands are indicated by arrows. (B) Quantification of HDAC2 and HDAC1 levels as detected by western blot, normalized by total protein detected by Novex membrane protein stain. ‘Relative units’ represent the density of HDAC2 or HDAC1 band divided by the density of the total protein stain in the corresponding lane, quantified for each mouse and averaged over the number of mice in each genotype group. Error bars show SD.
-
Figure 1—figure supplement 2—source data 1
HDAC2 and HDAC1 protein levels in Hdac2 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig1-figsupp2-data1-v2.xlsx
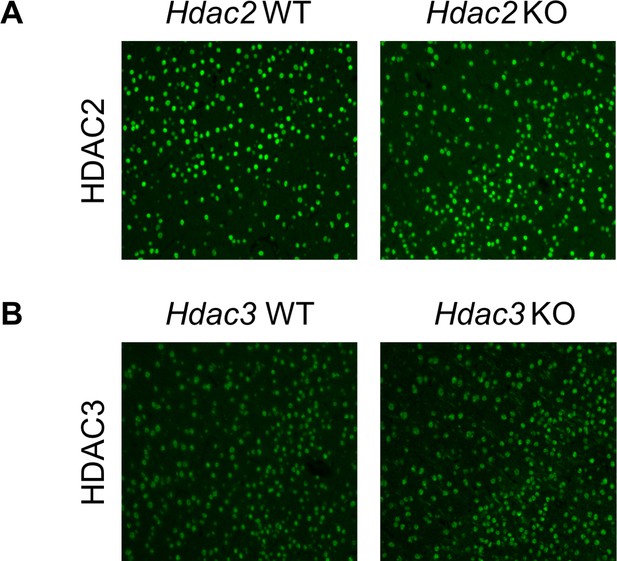
Hdac2 or Hdac3 knockout in striatal MSNs does not affect HDAC2 or HDAC3 levels in cortex.
Shown are fluorescent micrographs of somatosensory cortex of 10-week-old Hdac2 KO (A) and Hdac3 KO (B) mice immunostained with anti-HDAC2 and anti-HDAC3 antibody, respectively.
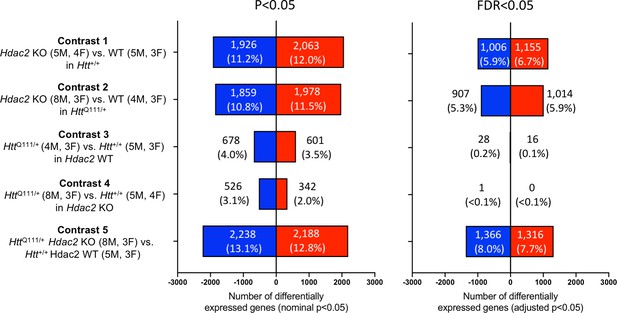
RNA-seq analyses in HttQ111 striata with MSN-specific Hdac2 knockout.
Results of differential gene expression analyses showing the number of differentially expressed genes (DEGs) for each of five contrasts. Number and % of DEGs at nominal p<0.05 or FDR adjusted p<0.05 are indicated. Total number of genes analyzed: contrast 1 = 17,132; contrast 2 = 17,175; contrast 3 = 17,139; contrast 4 = 17,116; contrast 5 = 17,094. The number of male (M) and female (F) mice in each group is indicated.
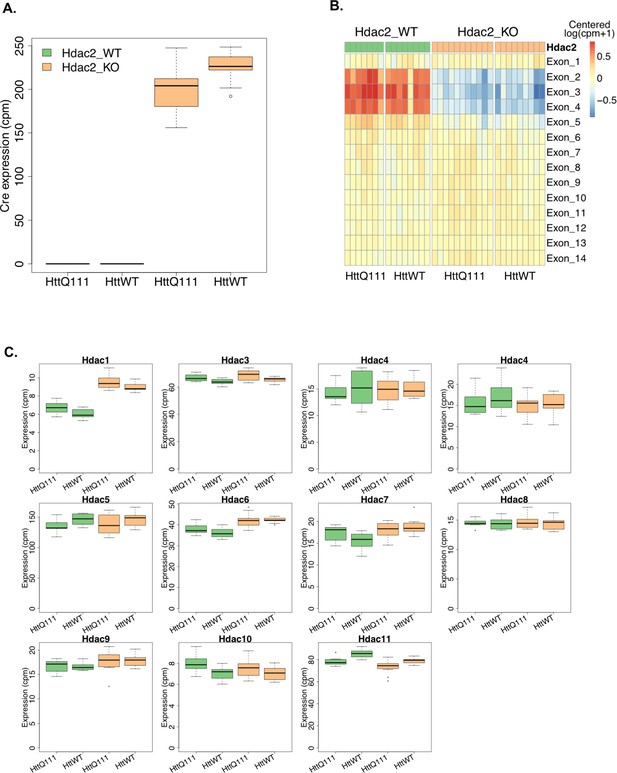
Expression of Hdac genes and Cre recombinase.
(A) Cre expression is increased in Hdac2 KO mice that also express the DARPP-32-Cre transgene. (B) Expression level of each Hdac2 exon showing reduced expression of exons 2–4 in Hdac2 KO mice, as predicted from the floxed Hdac2 allele with loxP sites spanning these exons (Montgomery et al., 2007). (C) Expression levels of other Hdac genes. Hdac1 expression is upregulated in Hdac2 KO mice. Note that mouse reference genome we used in this study, GRCm38 v. 75 had two Ensembl gene identifiers for Hdac4, ENSMUSG00000073617 (third panel on the first row) and ENSMUSG00000026313 (fourth panel on the first row).
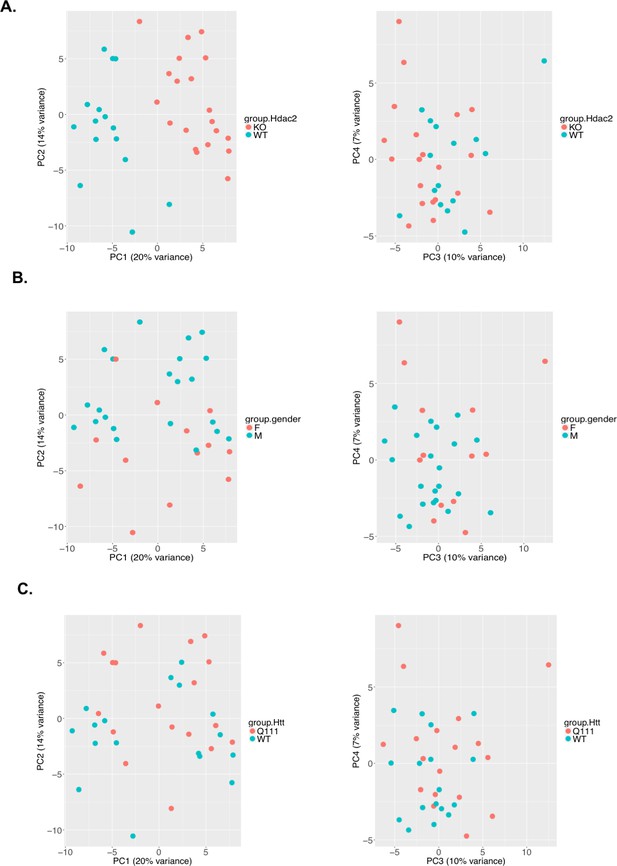
Principal Components Analyses (PCA).
PC1 separates gene expression by Hdac2 genotype (A), while PC2 separates gene expression largely by sex (B). Gene expression data does not clearly separate the samples by HttQ111 genotype (C).
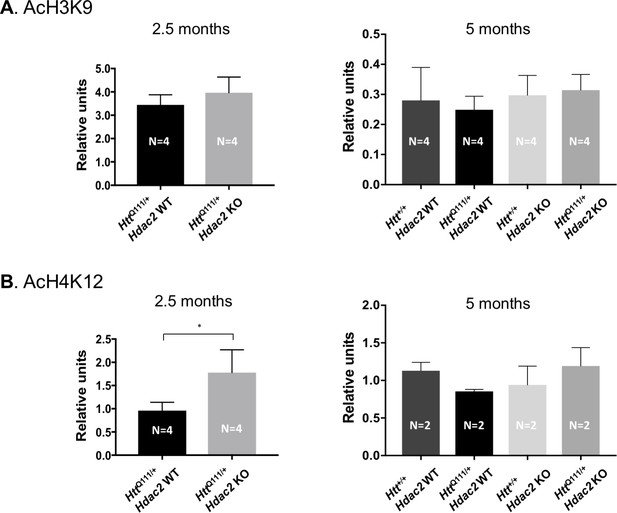
Global H3K9 and H4K12 acetylation levels.
(A) Quantification of acetylated Lys9 on histone H3, normalized by total H3 level, as detected by western blot. ‘Relative units’ represent the density of AcH3K9 band divided by the density of the corresponding histone H3 band, quantified for each mouse and averaged over the number of mice in each genotype group. (B) Quantification of acetylated Lys12 on histone H4 as detected by western blot, normalized by total protein detected by Novex membrane protein stain. ‘Relative units’ represent the density of AcH4K12 band divided by the density of the total protein stain in the corresponding lane, quantified for each mouse and averaged over the number of mice in each genotype group. Error bars show standard deviation. *Hdac2 WT: mean 0.96, 95% CI [0.68,1.24]; Hdac2 KO: mean 1.78, 95% CI [0.99, 2.56]; 2-tailed unpaired t-test p=0.021 in H4K12 acetylation. Numbers of mice for each group are indicated on the bar graphs.
-
Figure 2—figure supplement 3—source data 1
AcH3K9 and AcH4K12 levels in Hdac2 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig2-figsupp3-data1-v2.xlsx
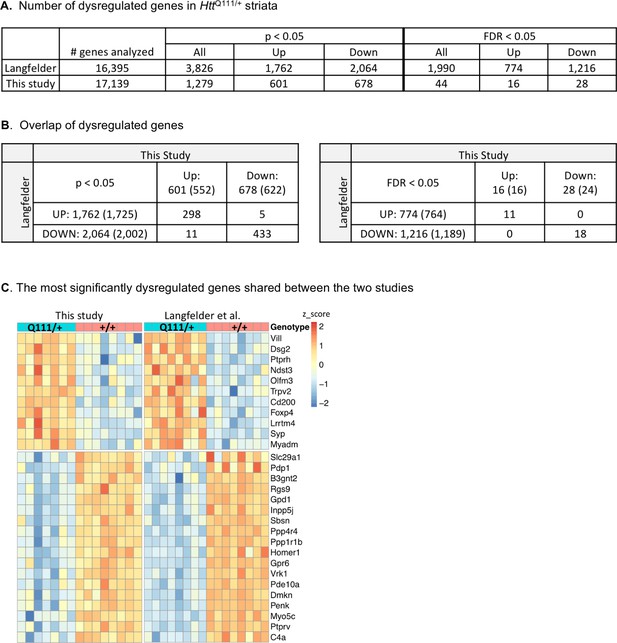
Cross-study comparison of HttQ111-differentially expressed genes.
RNA-seq data from striata of B6J.HttQ111/+ and Htt+/+ mice described in Langfelder et al., 2016 were analyzed in the same way as the current study for comparison. (A) The number of dysregulated genes (all, up- or down-regulated) in each study at two different statistical significance thresholds. (B) The number of overlapping up- or down-regulated genes between the two studies. Numbers in parentheses show the number of shared genes in each study. (C) Heat map of the 29 most significantly up or down-regulated genes shared between the two studies (FDR adjusted p<0.05).
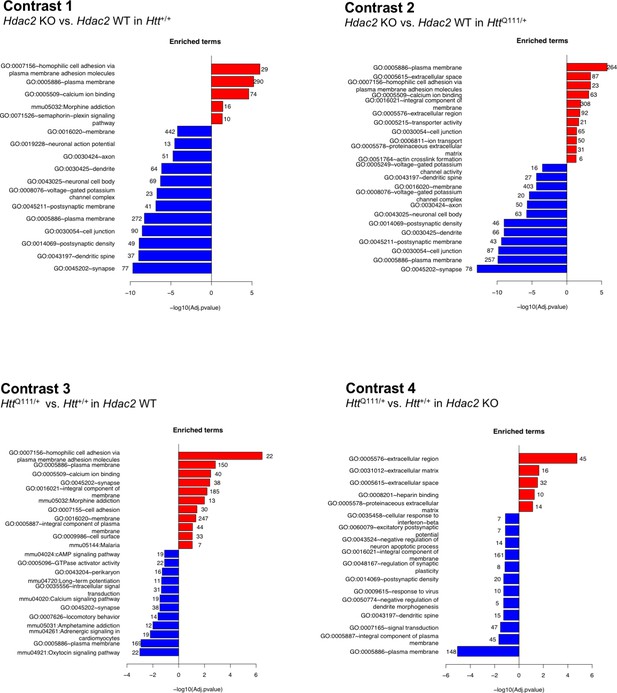
Pathway enrichment of differentially expressed genes.
The most significantly enriched pathways that are up- or down-regulated in each of four differential gene expression contrasts are shown. Numbers at the end of each bar show the number of genes represented in each pathway. For contrasts 1 and 2, DEGs used in the pathway analyses met the FDR < 0.05 threshold; for contrasts 3 and 4 DEG genes had a nominal p<0.05. The full set of pathways analyses is provided in Source data 2.
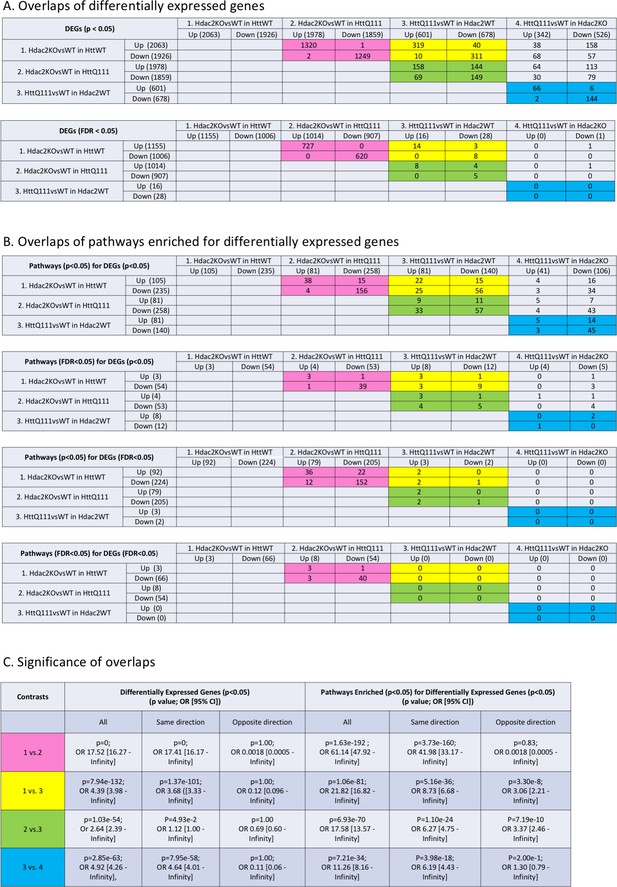
Overlaps of differentially expressed genes and enriched pathways in each of the four contrasts.
Numbers of DEGs genes (A) and enriched pathways (B) that overlap between the four contrasts at different significance threshold cut-offs. (C) The significance, determined using a one-tailed Fisher Exact test, of the enrichment of overlapping genes/pathways, for differentially expressed genes (p<0.05), or pathway enrichment (p<0.05 based on DEGs at p<0.05). The colors highlight overlaps between specific contrasts referred to in the text.
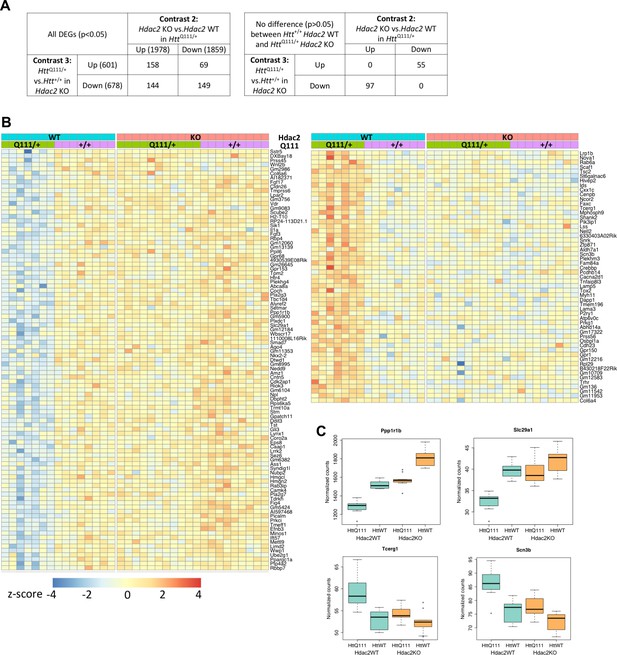
Rescue by Hdac2 KO of gene expression levels of a subset of genes dysregulated by the HttQ111 allele.
(A) To identify genes dysregulated by the HttQ111 allele and by Hdac2 KO in HttQ111/+ mice, the overlapping genes in contrasts 3 and 2 were identified (left table). This shows the numbers of genes (p<0.05) up-regulated by HttQ111 and either further up-regulated, or down-regulated by Hdac2 KO in HttQ111/+ mice, and the numbers of genes down-regulated by HttQ111 and either further down-regulated, or up-regulated by Hdac2 KO in HttQ111/+ mice. To identify HttQ111-dysregulated genes whose expression level was normalized by Hdac2 KO, genes from the contrast 3/2 overlap whose expression level did not differ significantly (p>0.05) between Htt+/+ Hdac2 WT and HttQ111/+ Hdac2 KO striata were identified (right table). (B) Heat map of the 97 genes down-regulated by HttQ111 and up-regulated by Hdac2 KO in HttQ111/+ striata and of the 55 genes up-regulated by HttQ111 and down-regulated by Hdac2 KO in HttQ111/+ striata, whose expression levels did not differ significantly between Htt+/+ Hdac2 WT and HttQ111/+ Hdac2 KO. (C) Examples of four such genes, two down-regulated by HttQ111 and two up-regulated by HttQ111 are displayed as box-plots.
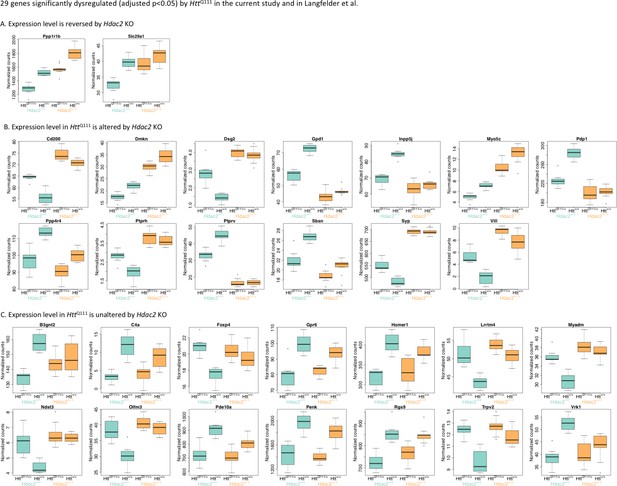
Impact of Hdac2 KO on the most significantly dysregulated genes common to this study and Langfelder et al.
Box-plots showing expression levels of the 29 genes significantly dysregulated in HttQ111/+ striata (FDR-adjusted p<0.05) in both the current study and in Langfelder et al., 2016 across all four genotypes. (A) Two genes had expression levels that did not differ significantly between Htt+/+ Hdac2 WT and HttQ111/+ Hdac2 KO striata; thus the down-regulation of these genes is considered to be rescued by Hdac2 KO. (B) 13 genes had expression levels that were changed by Hdac2 KO in HttQ111 mice; for most of these, Hdac2 KO further dysregulated gene expression in the same direction as the HttQ111/+ allele. (C) For 14 genes there was no change in gene expression in HttQ111/+ Hdac2 KO compared to HttQ111/+ Hdac2 WT striata.
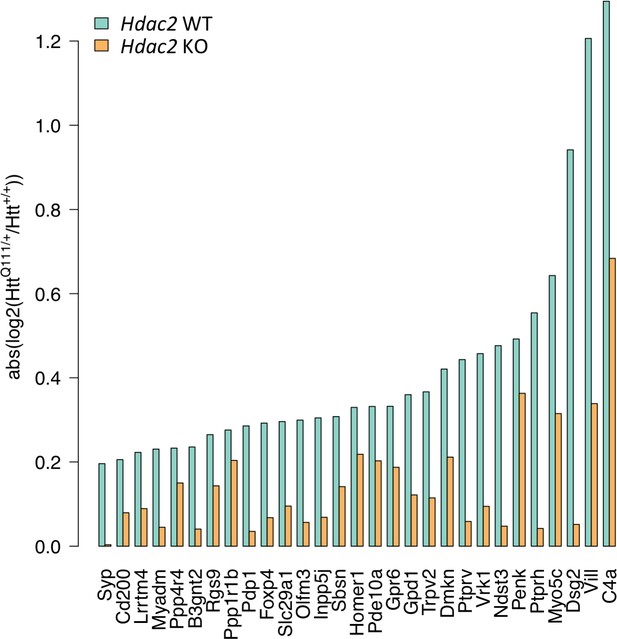
Hdac2 KO reduces the relative impact of the HttQ111 allele.
Absolute log2 fold-change of gene expression differences between HttQ111/+ and Htt+/+ were determined in Hdac2 WT mice and in Hdac2 KO mice as a measure of gene expression change regardless of direction of effect. x-axis indicates the set of 29 genes significantly dysregulated by HttQ111 (FDR adjusted p<0.05) in both this study and in Langfelder et al., 2016; Figure 2—figure supplement 4; Figure 4—figure supplement 1.
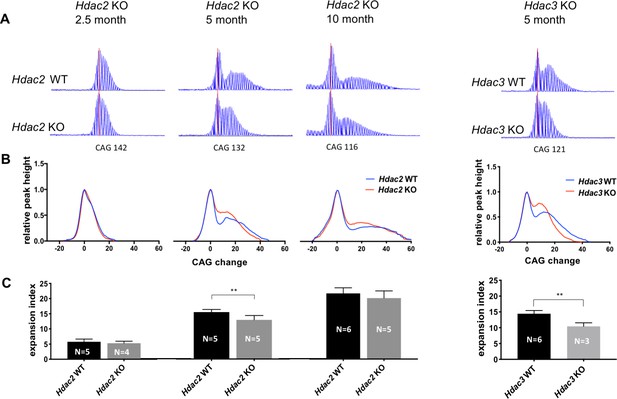
Deletion of Hdac2 or Hdac3 in striatal MSNs decreases striatal HTT CAG repeat expansions.
(A) Representative GeneMapper traces of PCR-amplified striatal HTT CAG repeats from individual mice matched by main allele repeat length within each age group (Source data 1). Main allele is marked with a vertical red line and the number of CAG repeats is indicated below. Peaks to the right of the main allele represent CAG repeat expansions. (B) Genotype-averaged GeneMapper-derived data showing peak heights normalized by the main allele peak height and CAG change relative to the main allele. (C) Mean striatal expansion indices per genotype. Error bars represent standard deviation. **p<0.01 (two-tailed unpaired t-test). *p<0.05, **p<0.01; ***p<0.005 (2-tailed unpaired t-test). Numbers of mice for each group are indicated on the bar graphs.
-
Figure 6—source data 1
Expansion Indices in Hdac2 and Hdac3 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig6-data1-v2.xlsx
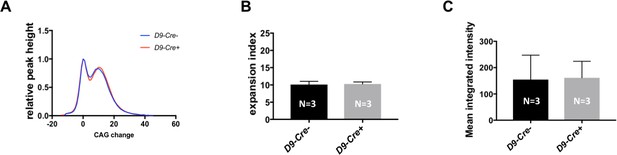
Expression of Cre transgene does not affect HTT CAG repeat instability and nuclear huntingtin accumulation in MSNs.
CAG instability and nuclear mutant huntingtin were measured in a cohort of 5 month HttQ111/+ mice harboring wild-type Hdac2 and Hdac3 alleles, either with or without the DARPP-32 Cre transgene (D9-Cre). (A) Genotype-averaged GeneMapper-derived data showing peak heights normalized by the main allele peak height and CAG change relative to the main allele. (B) Mean striatal expansion indices. (C) Mean integrated intensity of striatal nuclear mutant huntingtin immunostaining normalized by the total number of H3-positive nuclei. Error bars show SD. Numbers of mice for each group are indicated on the bar graphs.
-
Figure 6—figure supplement 1—source data 1
Expansion Indices and mAb5374 nuclear huntingtin immunostaining intensities in Cre-expressing HttQ111 mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig6-figsupp1-data1-v2.xlsx
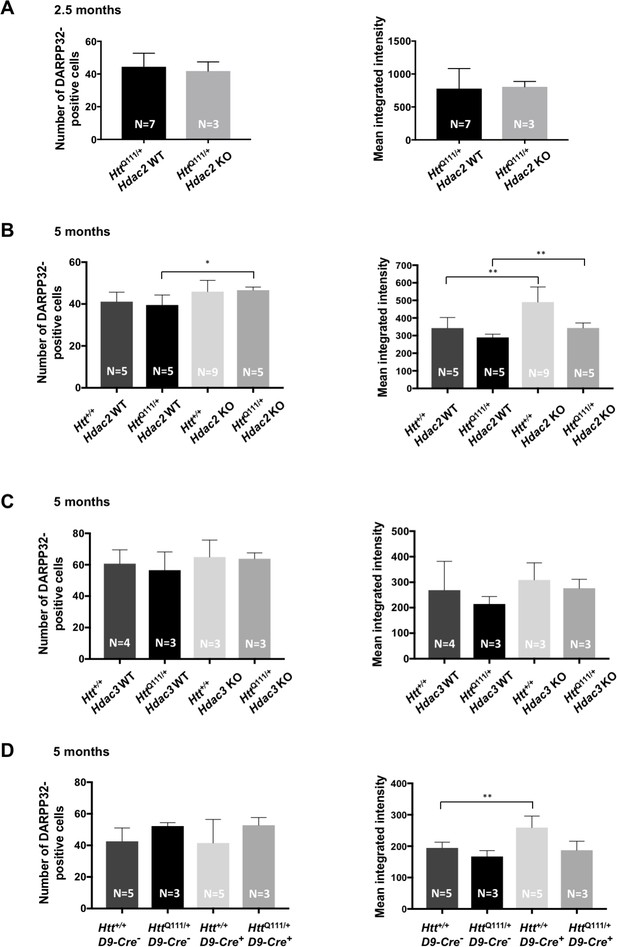
Effect of Hdac2 or Hdac3 deletion in striatal MSNs on number of DARPP-32-positive cells and DARPP-32 levels.
DARPP-32 levels (mean integrated intensity; right panels) are quantified as integrated intensity of DARPP-32 immunostaining, normalized by the number of DARPP-32-positive cells (left panels). Refer to Source data 1 for the cohorts of mice used. (A) 2.5 month Hdac2 KO mice (‘immunohistochemistry (IHC) cohort’). There were no Htt+/+ mice in this group. (B) Five-month Hdac2 KO mice (‘instability/IHC cohort’). (C) 5 month Hdac3 KO mice (‘IHC cohort’). (D) Five-month DARPP-32 Cre (D9-Cre) expression control mice. Error bars show SD. Numbers of mice for each group are indicated on the bar graphs. *p<0.05, **p<0.01 (2-tailed unpaired t-tests).
-
Figure 6—figure supplement 2—source data 1
DARPP32-positive cells and DARP32 levels in Hdac2 and Hdac3 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig6-figsupp2-data1-v2.xlsx
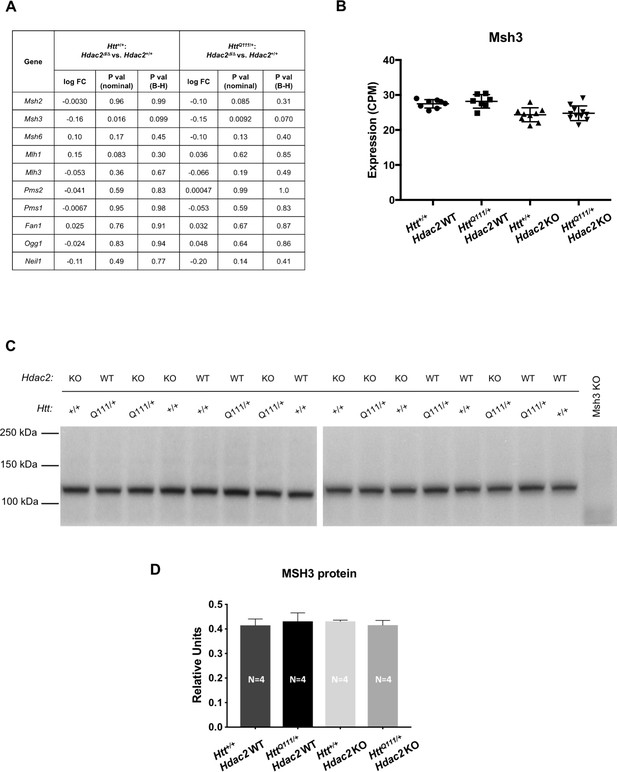
DNA repair gene expression.
(A) Differential expression analysis of DNA repair genes from RNA-seq data in 5-month mice. (B) Effect of Hdac2 KO on levels of Msh3 mRNA from RNA-seq data in 5-month mice. (C) Total lysates of contralateral striata of a subset of mice from the RNA-seq cohort (N = 4 for each of the four genotypes), probed with anti-MSH3 antibody. Msh3 KO lane is striatal lysate from an Msh3-/- knockout mouse, loaded as negative control. (D) Quantification of MSH3 protein levels as detected by western blot, normalized by total protein detected by Novex membrane protein stain. ‘Relative units’ represent the density of MSH3 band divided by the density of the total protein stain in the same lane, quantified for each mouse and averaged over the number of mice in each genotype group. Error bars show S.D. Numbers of mice for each group in the western blot are indicated on the bar graph.
-
Figure 6—figure supplement 3—source data 1
MSH3 protein levels in Hdac2 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig6-figsupp3-data1-v2.xlsx
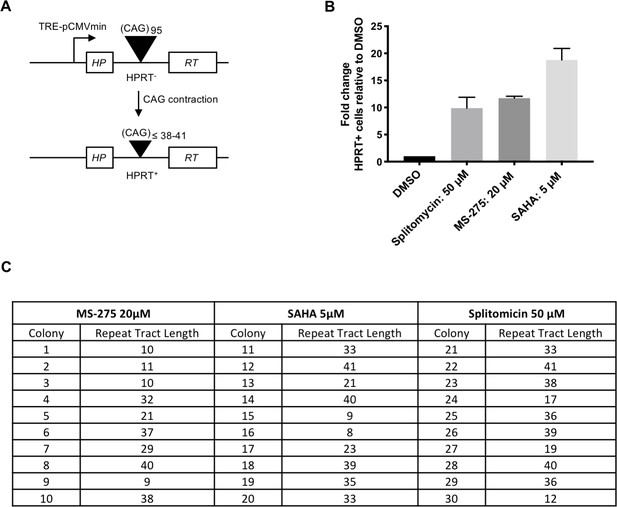
HDAC inhibitors increase instability in selectable cell-based assays for CAG repeat contraction.
(A) HPRT mini-gene reporter construct for contraction assay. (B) Fold-change in the number of HAT-resistant FLAH25 cells (HPRT+) when treated with HDAC inhibitors relative inhibitors relative to the number of resistant colonies in DMSO-treated cells. Bars show mean +/- SD of bars fold-change relative to DMSO in four biological replicates (independent wells treated with drug). (C) CAG repeat lengths in HPRT+ colonies following HDAC inhibitor treatment. DNA was extracted from HPRT+ colonies, and the repeat tract PCR-amplified and sequenced to determine repeat length.
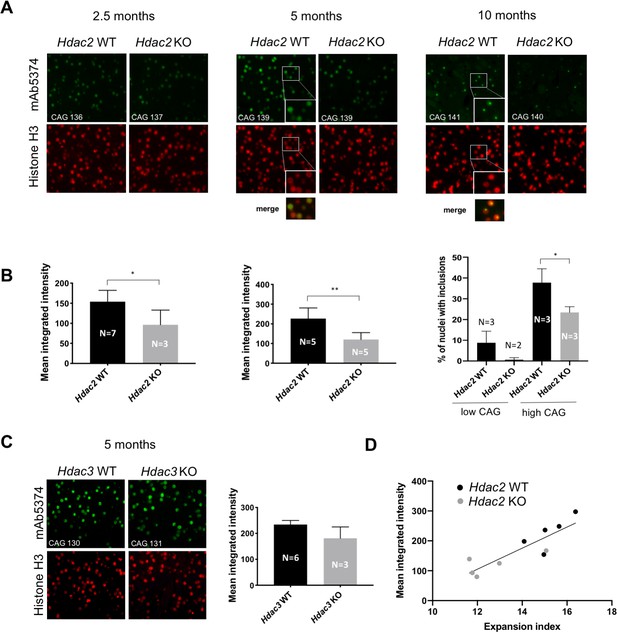
Impact of MSN-specific Hdac2 or Hdac3 knockout on striatal nuclear mutant huntingtin immunostaining phenotypes.
(A) Fluorescent micrographs of HttQ111/+ Hdac2 KO striata immunostained with anti-huntingtin mAb5374 and histone H3 antibodies. Mice were matched by CAG repeat length within each age group. Images of 2.5-month and 5-month striata were taken at 40x; images of 10-month striata taken at 20x with 2x digital zoom. The inserts in the 5-month and 10-month Hdac2 WT mAb5374 images and merged images below highlight the overlap of nuclear huntingtin immunostaining over the entire H3-positive nucleus in the 5-month mice, in contrast to the discrete nuclear huntingtin inclusion immunostaining intensity within the nucleus at 10 months. (B) Mean integrated intensity of mAb5374 staining (integrated intensity normalized by the total number of nuclei as determined by the number of H3-positive nuclei) is shown for 2.5-month and 5-month cohorts of HttQ111/+ Hdac2 KO mice. For 10-month mice, the number of nuclei containing an inclusion (we have not observed more than one inclusion per nucleus) was counted as a percentage of the total number of nuclei, determined by histone H3 immunostaining. The 10-month group of mice was divided into two subgroups based on CAG repeat length: low CAG repeat subgroup represented by the first two columns in the graph (Hdac2 WT CAG 117, 119, 120; Hdac2 KO CAG 110, 116) and high CAG repeat subgroup represented by the last two columns in the graph (Hdac2 WT CAG 138, 141, 146; Hdac2 KO CAG 132, 140, 140). (C) Fluorescent micrographs (20x) of 5 month Hdac3 KO from CAG length-matched mice stained with mAb5374 and histone H3 antibodies (left) and quantified mean integrated intensity of mAb5374 staining (right). 5-months. Numbers of mice for each group are indicated on the bar graphs.
-
Figure 8—source data 1
mAb5474 nulcear intensity and number of nuclear inclusions in Hdac2 and Hdaac3 KO mice.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig8-data1-v2.xlsx
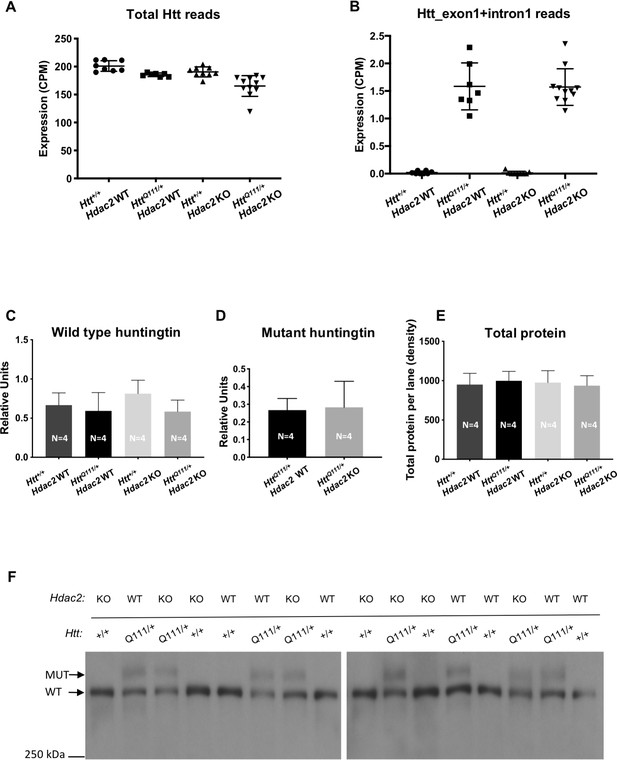
Huntingtin expression.
(A) Effect of Hdac2 KO on levels of total huntingtin mRNA (A) and on aberrant huntingtin transcripts spanning exon 1 + intron 1 (B) from RNA-seq data in 5-month mice. In panel B, reads were aligned to knock-in allele sequence comprising human exon one and part of human intron 1 (93). (C–F) Huntingtin western blot analyses showing quantification of wild type (C) and mutant (D) huntingtin as detected by western blot using mAb2166 (F). Huntingtin protein bands were normalized by total protein detected by Novex membrane protein stain. (E) Average total protein levels as detected by Novex membrane protein stain. ‘Relative units’ represent the density of wild type or mutant huntingtin band divided by the density of the total protein stain in the same lane, quantified for each mouse and averaged over the number of mice in each genotype group (N = 4 for each genotype, as indicated in the bar graphs). Error bars show S.D.
-
Figure 8—figure supplement 1—source data 1
Zip file: RNAseq_codes_and_files.
- https://cdn.elifesciences.org/articles/55911/elife-55911-fig8-figsupp1-data1-v2.xlsx
Additional files
-
Source code 1
RNA-seq codes and data files.
- https://cdn.elifesciences.org/articles/55911/elife-55911-code1-v2.zip
-
Source data 1
CAG lengths of mouse cohorts.
- https://cdn.elifesciences.org/articles/55911/elife-55911-data1-v2.docx
-
Source data 2
Differentially expressed genes.
- https://cdn.elifesciences.org/articles/55911/elife-55911-data2-v2.xlsx
-
Source data 3
Pathway analyses.
- https://cdn.elifesciences.org/articles/55911/elife-55911-data3-v2.xlsx
-
Source data 4
Rescue analysis.
- https://cdn.elifesciences.org/articles/55911/elife-55911-data4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55911/elife-55911-transrepform-v2.docx





