KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage
Figures
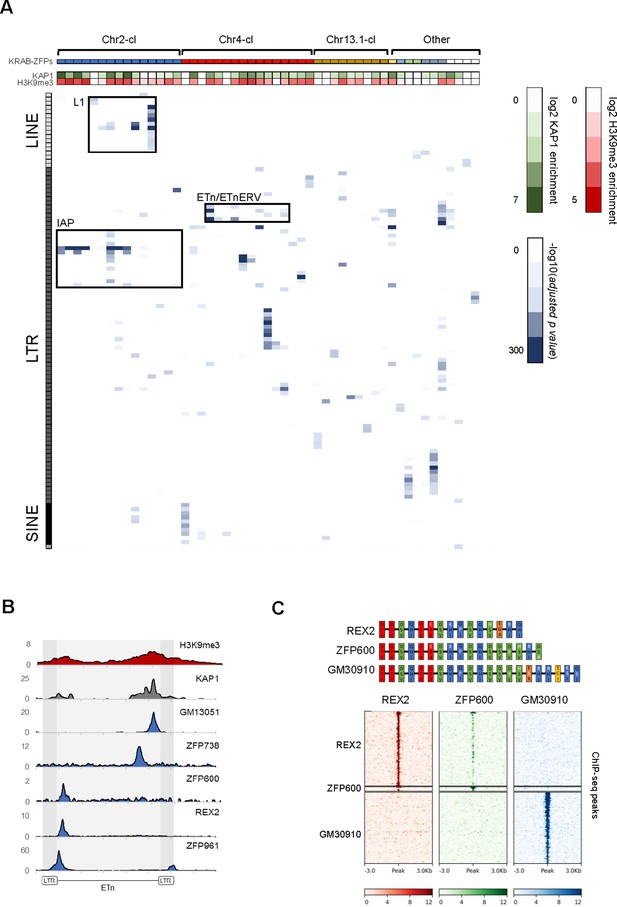
Genome-wide binding patterns of mouse KRAB-ZFPs.
(A) Probability heatmap of KRAB-ZFP binding to TEs. Blue color intensity (main field) corresponds to -log10 (adjusted p-value) enrichment of ChIP-seq peak overlap with TE groups (Fisher’s exact test). The green/red color intensity (top panel) represents mean KAP1 (GEO accession: GSM1406445) and H3K9me3 (GEO accession: GSM1327148) enrichment (respectively) at peaks overlapping significantly targeted TEs (adjusted p-value<1e-5) in WT ES cells. (B) Summarized ChIP-seq signal for indicated KRAB-ZFPs and previously published KAP1 and H3K9me3 in WT ES cells across 127 intact ETn elements. (C) Heatmaps of KRAB-ZFP ChIP-seq signal at ChIP-seq peaks. For better comparison, peaks for all three KRAB-ZFPs were called with the same parameters (p<1e-10, peak enrichment >20). The top panel shows a schematic of the arrangement of the contact amino acid composition of each zinc finger. Zinc fingers are grouped and colored according to similarity, with amino acid differences relative to the five consensus fingers highlighted in white.
-
Figure 1—source data 1
KRAB-ZFP expression in 40 mouse tissues and cell lines (ENCODE).
Mean values of replicates are shown as log2 transcripts per million.
- https://cdn.elifesciences.org/articles/56337/elife-56337-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Probability heatmap of KRAB-ZFP binding to TEs.
Values corresponds to -log10 (adjusted p-value) enrichment of ChIP-seq peak overlap with TE groups (Fisher’s exact test).
- https://cdn.elifesciences.org/articles/56337/elife-56337-fig1-data2-v2.xlsx
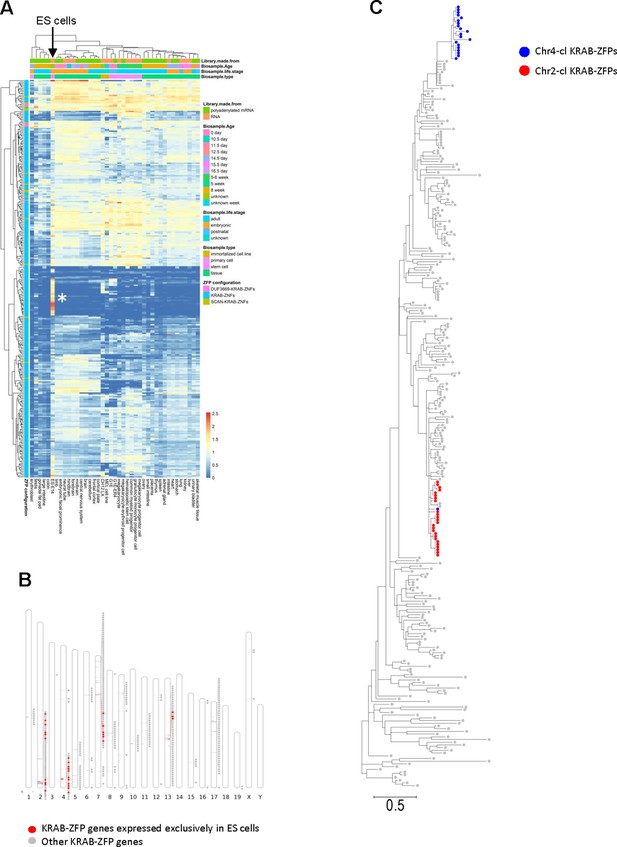
ES cell-specific expression of KRAB-ZFP gene clusters.
(A) Heatmap showing expression patterns of mouse KRAB-ZFPs in 40 mouse tissues and cell lines (ENCODE). Heatmap colors indicate gene expression levels in log2 transcripts per million (TPM). The asterisk indicates a group of 30 KRAB-ZFPs that are exclusively expressed in ES cells. (B) Physical location of the genes encoding for the 30 KRAB-ZFPs that are exclusively expressed in ES cells. (C) Phylogenetic (Maximum likelihood) tree of the KRAB domains of mouse KRAB-ZFPs. KRAB-ZFPs encoded on the gene clusters on chromosome 2 and 4 are highlighted. The scale bar at the bottom indicates amino acid substitutions per site.
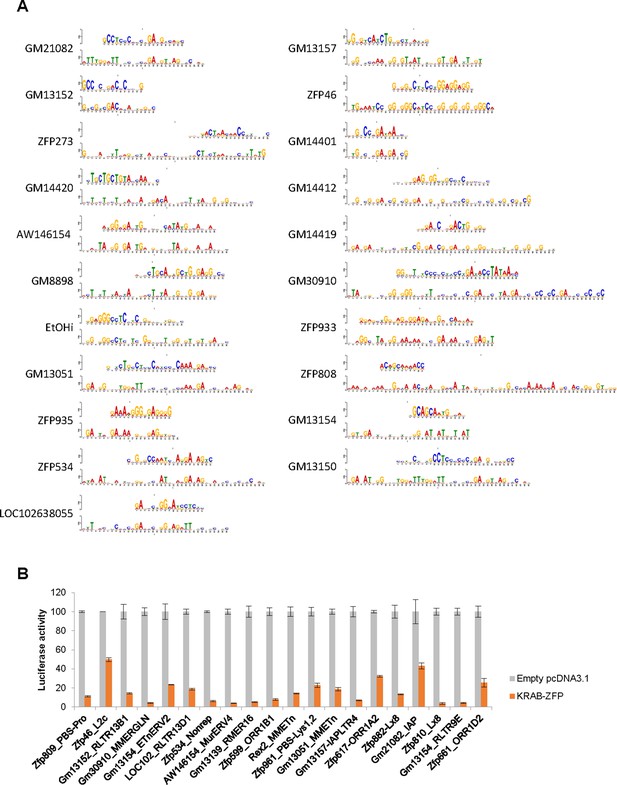
KRAB-ZFP binding motifs and their repression activity.
(A) Comparison of computationally predicted (bottom) and experimentally determined (top) KRAB-ZFP binding motifs. Only significant pairs are shown (FDR < 0.1). (B) Luciferase reporter assays to confirm KRAB-ZFP repression of the identified target sites. Bars show the luciferase activity (normalized to Renilla luciferase) of reporter plasmids containing the indicated target sites cloned upstream of the SV40 promoter. Reporter plasmids were co-transfected into 293 T cells with a Renilla luciferase plasmid for normalization and plasmids expressing the targeting KRAB-ZFP. Normalized mean luciferase activity (from three replicates) is shown relative to luciferase activity of the reporter plasmid co-transfected with an empty pcDNA3.1 vector.
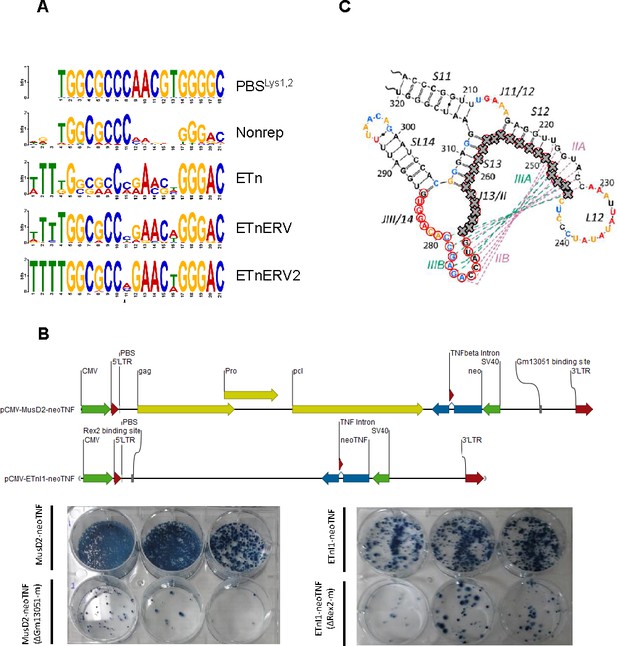
KRAB-ZFP binding to ETn retrotransposons.
(A) Comparison of the PBSLys1,2 sequence with Zfp961 binding motifs in nonrepetitive peaks (Nonrep) and peaks at ETn elements. (B) Retrotransposition assays of original (ETnI1-neoTNF and MusD2-neoTNF Ribet et al., 2004) and modified reporter vectors where the Rex2 or Gm13051 binding motifs where removed. Schematic of reporter vectors are displayed at the top. HeLa cells were transfected as described in the Materials and Methods section and neo-resistant colonies, indicating retrotransposition events, were selected and stained. (C) Stem-loop structure of the ETn RNA export signal, the Gm13051 motif on the corresponding DNA is marked with red circles, the part of the motif that was deleted is indicated with grey crosses (adapted from Legiewicz et al., 2010).
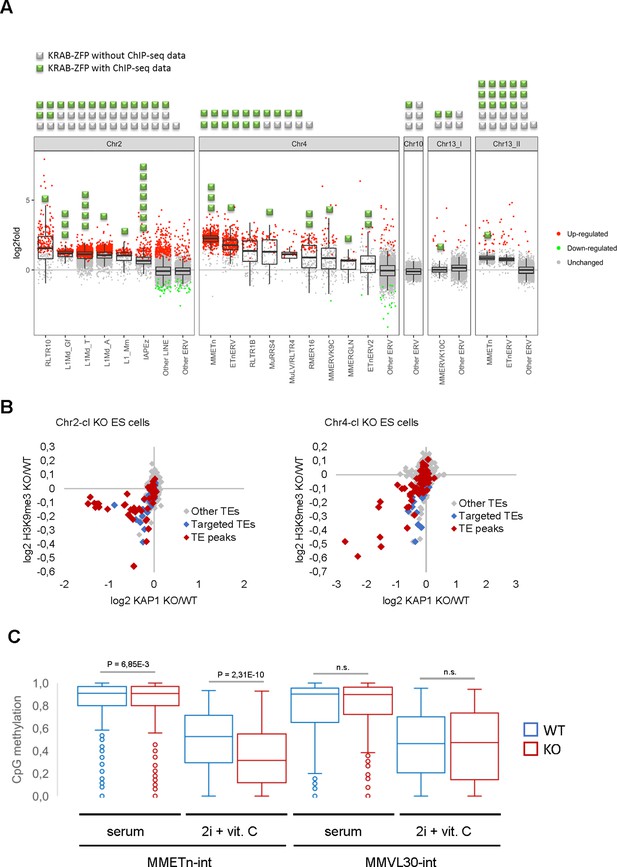
Retrotransposon reactivation in KRAB-ZFP cluster KO ES cells.
(A) RNA-seq analysis of TE expression in five KRAB-ZFP cluster KO ES cells. Green and grey squares on top of the panel represent KRAB-ZFPs with or without ChIP-seq data, respectively, within each deleted gene cluster. Reactivated TEs that are bound by one or several KRAB-ZFPs are indicated by green squares in the panel. Significantly up- and downregulated elements (adjusted p-value<0.05) are highlighted in red and green, respectively. (B) Differential KAP1 binding and H3K9me3 enrichment at TE groups (summarized across all insertions) in Chr2-cl and Chr4-cl KO ES cells. TE groups targeted by one or several KRAB-ZFPs encoded within the deleted clusters are highlighted in blue (differential enrichment over the entire TE sequences) and red (differential enrichment at TE regions that overlap with KRAB-ZFP ChIP-seq peaks). (C) DNA methylation status of CpG sites at indicated TE groups in WT and Chr4-cl KO ES cells grown in serum containing media or in hypomethylation-inducing media (2i + Vitamin C). P-values were calculated using paired t-test.
-
Figure 2—source data 1
Differential H3K9me3 and KAP1 distribution in WT and KRAB-ZFP cluster KO ES cells at TE families and KRAB-ZFP bound TE insertions.
Differential read counts and statistical testing were determined by DESeq2.
- https://cdn.elifesciences.org/articles/56337/elife-56337-fig2-data1-v2.xlsx

Epigenetic changes at TEs and TE-borne enhancers in KRAB-ZFP cluster KO ES cells.
(A) Differential analysis of summative (all individual insertions combined) H3K9me3 enrichment at TE groups in Chr10-cl, Chr13.1-cl and Chr13.2-cl KO ES cells. TE groups targeted by one or several KRAB-ZFPs encoded within the deleted clusters are highlighted in orange (differential enrichment over the entire TE sequences) and red (differential enrichment at TE regions that overlap with KRAB-ZFP ChIP-seq peaks). (B) Top: Schematic view of the Cd59a/Cd59b locus with a 5’ truncated ETn insertion. ChIP-seq (Input subtracted from ChIP) data for overexpressed epitope-tagged Gm13051 (a Chr4-cl KRAB-ZFP) in F9 EC cells, and re-mapped KAP1 (GEO accession: GSM1406445) and H3K9me3 (GEO accession: GSM1327148) in WT ES cells are shown together with RNA-seq data from Chr4-cl WT and KO ES cells (mapped using Bowtie (-a -m 1 --strata -v 2) to exclude reads that cannot be uniquely mapped). Bottom: Transcriptional activity of a 5 kb fragment with or without fragments of the ETn insertion was tested by luciferase reporter assay in Chr4-cl WT and KO ES cells.
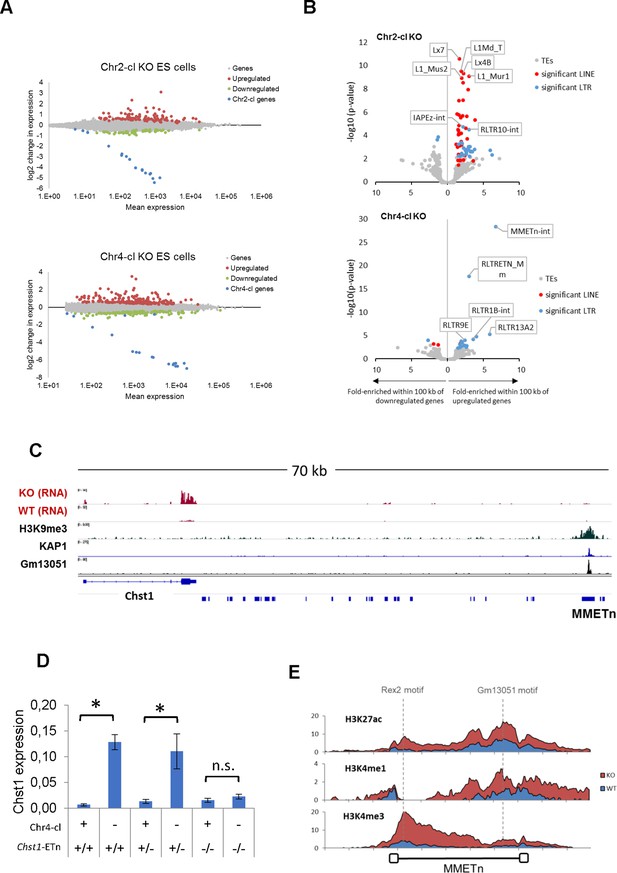
TE-dependent gene activation in KRAB-ZFP cluster KO ES cells.
(A) Differential gene expression in Chr2-cl and Chr4-cl KO ES cells. Significantly up- and downregulated genes (adjusted p-value<0.05) are highlighted in red and green, respectively, KRAB-ZFP genes within the deleted clusters are shown in blue. (B) Correlation of TEs and gene deregulation. Plots show enrichment of TE groups within 100 kb of up- and downregulated genes relative to all genes. Significantly overrepresented LTR and LINE groups (adjusted p-value<0.1) are highlighted in blue and red, respectively. (C) Schematic view of the downstream region of Chst1 where a 5’ truncated ETn insertion is located. ChIP-seq (Input subtracted from ChIP) data for overexpressed epitope-tagged Gm13051 (a Chr4-cl KRAB-ZFP) in F9 EC cells, and re-mapped KAP1 (GEO accession: GSM1406445) and H3K9me3 (GEO accession: GSM1327148) in WT ES cells are shown together with RNA-seq data from Chr4-cl WT and KO ES cells (mapped using Bowtie (-a -m 1 --strata -v 2) to exclude reads that cannot be uniquely mapped). (D) RT-qPCR analysis of Chst1 mRNA expression in Chr4-cl WT and KO ES cells with or without the CRISPR/Cas9 deleted ETn insertion near Chst1. Values represent mean expression (normalized to Gapdh) from three biological replicates per sample (each performed in three technical replicates) in arbitrary units. Error bars represent standard deviation and asterisks indicate significance (p<0.01, Student’s t-test). n.s.: not significant. (E) Mean coverage of ChIP-seq data (Input subtracted from ChIP) in Chr4-cl WT and KO ES cells over 127 full-length ETn insertions. The binding sites of the Chr4-cl KRAB-ZFPs Rex2 and Gm13051 are indicated by dashed lines.
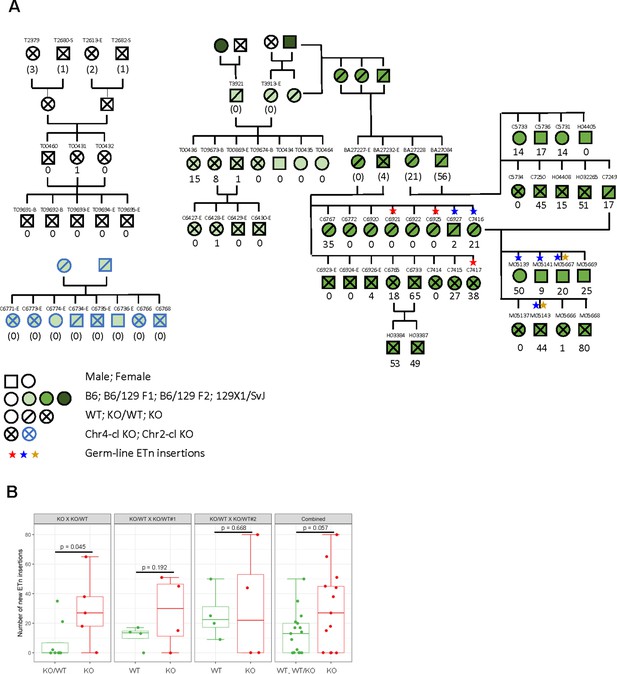
ETn retrotransposition in Chr4-cl KO mice.
(A) Pedigree of mice used for transposon insertion screening by capture-seq in mice of different strain backgrounds. The number of novel ETn insertions (only present in one animal) are indicated. For animals whose direct ancestors have not been screened, the ETn insertions are shown in parentheses since parental inheritance cannot be excluded in that case. Germ line insertions are indicated by asterisks. All DNA samples were prepared from tail tissues unless noted (-S: spleen, -E: ear, -B:Blood) (B) Statistical analysis of ETn insertion frequency in tail tissue from 30 Chr4-cl KO, KO/WT and WT mice that were derived from one Chr4-c KO x KO/WT and two Chr4-cl KO/WT x KO/WT matings. Only DNA samples that were collected from juvenile tails were considered for this analysis. P-values were calculated using one-sided Wilcoxon Rank Sum Test. In the last panel, KO, WT and KO/WT mice derived from all matings were combined for the statistical analysis.
-
Figure 4—source data 1
Coordinates of identified novel ETn insertions and supporting capture-seq read counts.
Genomic regions indicate cluster of supporting reads.
- https://cdn.elifesciences.org/articles/56337/elife-56337-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Sequences of capture-seq probes used to enrich genomic DNA for ETn and MuLV (RLTR4) insertions.
- https://cdn.elifesciences.org/articles/56337/elife-56337-fig4-data2-v2.txt

Birth statistics of KRAB-ZFP cluster KO mice and TE reactivation in adult tissues.
(A) Birth statistics of Chr4- and Chr2-cl mice derived from KO/WT x KO/WT matings in different strain backgrounds. (B) RNA-seq analysis of TE expression in Chr2- (left) and Chr4-cl (right) KO tissues. TE groups with the highest reactivation phenotype in ES cells are shown separately. Significantly up- and downregulated elements (adjusted p-value<0.05) are highlighted in red and green, respectively. Experiments were performed in at least two biological replicates.

Identification of polymorphic ETn and MuLV retrotransposon insertions in Chr4-cl KO and WT mice.
Heatmaps show normalized capture-seq read counts in RPM (Read Per Million) for identified polymorphic ETn (A) and MuLV (B) loci in different mouse strains. Only loci with strong support for germ line ETn or MuLV insertions (at least 100 or 3000 ETn or MuLV RPM, respectively) in at least two animals are shown. Non-polymorphic insertion loci with high read counts in all screened mice were excluded for better visibility. The sample information (sample name and cell type/tissue) is annotated at the bottom, with the strain information indicated by color at the top. The color gradient indicates log10(RPM+1).
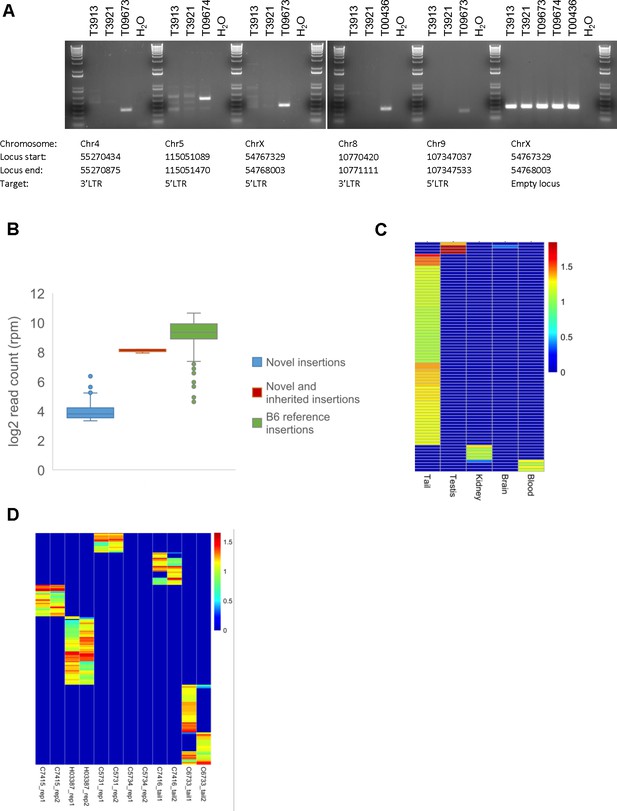
Confirmation of novel ETn insertions identified by capture-seq.
(A) PCR validation of novel ETn insertions in genomic DNA of three littermates (IDs: T09673, T09674 and T00436) and their parents (T3913 and T3921). Primer sequences are shown in Supplementary file 3. (B) ETn capture-seq read counts (RPM) at putative novel somatic (loci identified exclusively in one single animal), novel germ line (loci identified in several littermates) insertions, and at B6 reference ETn elements. (C) Heatmap shows capture-seq read counts (RPM) of a Chr4-cl KO mouse (ID: C6733) as determined in different tissues. Each row represents a novel ETn locus that was identified in at least one tissue. The color gradient indicates log10(RPM+1). (D) Heatmap shows the capture-seq RPM in technical replicates using the same Chr4-cl KO DNA sample (rep1/rep2) or replicates with DNA samples prepared from different sections of the tail from the same mouse at different ages (tail1/tail2). Each row represents a novel ETn locus that was identified in at least one of the displayed samples. The color gradient indicates log10(RPM+1).
Tables
KRAB-ZFP genes clusters in the mouse genome that were investigated in this study.
* Number of protein-coding KRAB-ZFP genes identified in a previously published screen (Imbeault et al., 2017) and the ChIP-seq data column indicates the number of KRAB-ZFPs for which ChIP-seq was performed in this study.
| Cluster | Location | Size (Mb) | # of KRAB-ZFPs* | ChIP-seq data |
|---|---|---|---|---|
| Chr2 | Chr2 qH4 | 3.1 | 40 | 17 |
| Chr4 | Chr4 qE1 | 2.3 | 21 | 19 |
| Chr10 | Chr10 qC1 | 0.6 | 6 | 1 |
| Chr13.1 | Chr13 qB3 | 1.2 | 6 | 2 |
| Chr13.2 | Chr13 qB3 | 0.8 | 26 | 12 |
| Chr8 | Chr8 qB3.3 | 0.1 | 4 | 4 |
| Chr9 | Chr9 qA3 | 0.1 | 4 | 2 |
| Other | - | - | 248 | 4 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | 129 × 1/SvJ | The Jackson Laboratory | 000691 | Mice used to generate mixed strain Chr4-cl KO mice |
| Cell line (Homo-sapiens) | HeLa | ATCC | ATCC CCL-2 | |
| Cell line (Mus musculus) | JM8A3.N1 C57BL/6N-Atm1Brd | KOMP Repository | PL236745 | B6 ES cells used to generate KO cell lines and mice |
| Cell line (Mus musculus) | B6;129‐ Gt(ROSA)26Sortm1(cre/ERT)Nat/J | The Jackson Laboratory | 004847 | ES cells used to generate KO cell lines and mice |
| Cell line (Mus musculus) | R1 ES cells | Andras Nagy lab | R1 | 129 ES cells used to generate KO cell lines and mice |
| Cell line (Mus musculus) | F9 Embryonic carcinoma cells | ATCC | ATCC CRL-1720 | |
| Antibody | Mouse monoclonal ANTI-FLAG M2 antibody | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 | ChIP (1 µg/107 cells) |
| Antibody | Rabbit polyclonal anti-HA | Abcam | Cat# ab9110, RRID:AB_307019 | ChIP (1 µg/107 cells) |
| Antibody | Mouse monoclonal anti-HA | Covance | Cat# MMS-101P-200, RRID:AB_10064068 | |
| Antibody | Rabbit polyclonal anti-H3K9me3 | Active Motif | Cat# 39161, RRID:AB_2532132 | ChIP (3 µl/107 cells) |
| Antibody | Rabbit polyclonal anti-GFP | Thermo Fisher Scientific | Cat# A-11122, RRID:AB_221569 | ChIP (1 µg/107 cells) |
| Antibody | Rabbit polyclonal anti- H3K4me3 | Abcam | Cat# ab8580, RRID:AB_306649 | ChIP (1 µg/107 cells) |
| Antibody | Rabbit polyclonal anti- H3K4me1 | Abcam | Cat# ab8895, RRID:AB_306847 | ChIP (1 µg/107 cells) |
| Antibody | Rabbit polyclonal anti- H3K27ac | Abcam | Cat# ab4729, RRID:AB_2118291 | ChIP (1 µg/107 cells) |
| Recombinant DNA reagent | pCW57.1 | Addgene | RRID:Addgene_41393 | Inducible lentiviral expression vector |
| Recombinant DNA reagent | pX330-U6-Chimeric_BB-CBh-hSpCas9 | Addgene | RRID:Addgene_42230 | CRISPR/Cas9 expression construct |
| Sequence-based reagent | Chr2-cl KO gRNA.1 | This paper | Cas9 gRNA | GCCGTTGCTCAGTCCAAATG |
| Sequenced-based reagent | Chr2-cl KO gRNA.2 | This paper | Cas9 gRNA | GATACCAGAGGTGGCCGCAAG |
| Sequenced-based reagent | Chr4-cl KO gRNA.1 | This paper | Cas9 gRNA | GCAAAGGGGCTCCTCGATGGA |
| Sequence-based reagent | Chr4-cl KO gRNA.2 | This paper | Cas9 gRNA | GTTTATGGCCGTGCTAAGGTC |
| Sequenced-based reagent | Chr10-cl KO gRNA.1 | This paper | Cas9 gRNA | GTTGCCTTCATCCCACCGTG |
| Sequenced-based reagent | Chr10-cl KO gRNA.2 | This paper | Cas9 gRNA | GAAGTTCGACTTGGACGGGCT |
| Sequenced-based reagent | Chr13.1-cl KO gRNA.1 | This paper | Cas9 gRNA | GTAACCCATCATGGGCCCTAC |
| Sequenced-based reagent | Chr13.1-cl KO gRNA.2 | This paper | Cas9 gRNA | GGACAGGTTATAGGTTTGAT |
| Sequenced-based reagent | Chr13.2-cl KO gRNA.1 | This paper | Cas9 gRNA | GGGTTTCTGAGAAACGTGTA |
| Sequenced-based reagent | Chr13.2-cl KO gRNA.2 | This paper | Cas9 gRNA | GTGTAATGAGTTCTTATATC |
| Commercial assay or kit | SureSelectQXT Target Enrichment kit | Agilent | G9681-90000 | |
| Software, algorithm | Bowtie | http://bowtie-bio.sourceforge.net | RRID:SCR_005476 | |
| Software, algorithm | MACS14 | https://bio.tools/macs | RRID:SCR_013291 | |
| Software, algorithm | Tophat | https://ccb.jhu.edu | RRID:SCR_013035 |
Additional files
-
Source code 1
Custom Perl script used to get methylation pattern for each CpG dyads from Bismark methylation calling results.
- https://cdn.elifesciences.org/articles/56337/elife-56337-code1-v2.pl
-
Supplementary file 1
Experimental parameters, gene-centered informa347tion and summary of KRAB-ZFP ChIP-seq analysis.
- https://cdn.elifesciences.org/articles/56337/elife-56337-supp1-v2.xlsx
-
Supplementary file 2
Differential CpG methylation status of TEs in WT and Chr4-cl KO ES cells in serum and 2i culture conditions.
- https://cdn.elifesciences.org/articles/56337/elife-56337-supp2-v2.xlsx
-
Supplementary file 3
Sequence information of used PCR primers, gRNAs and cloned oligos for luciferase repression assays.
- https://cdn.elifesciences.org/articles/56337/elife-56337-supp3-v2.xlsx
-
Supplementary file 4
Overview of generated NGS data.
- https://cdn.elifesciences.org/articles/56337/elife-56337-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56337/elife-56337-transrepform-v2.docx





