SUMO is a pervasive regulator of meiosis
Figures
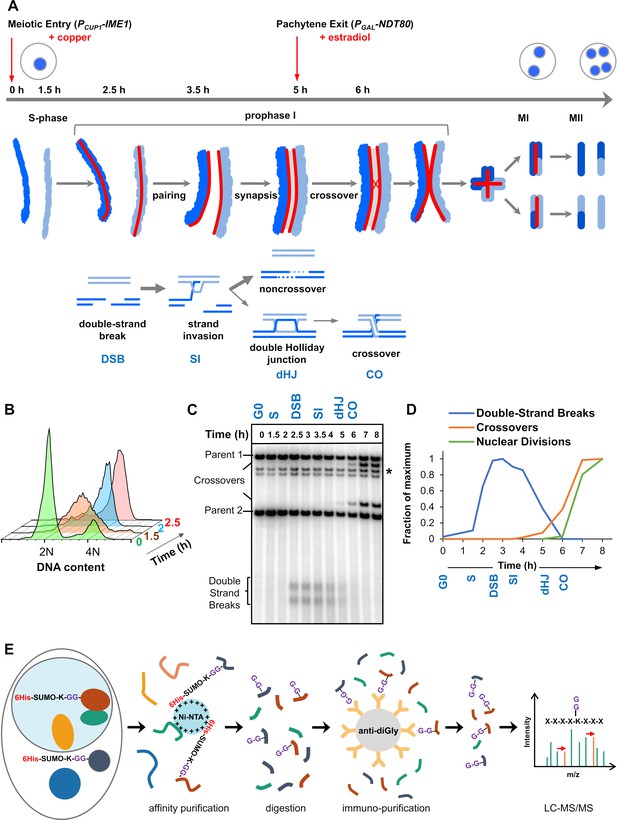
Experimental approach for profiling SUMOylation during meiosis.
(A) Cell synchronization regimen showing the timing of nuclear (top), chromosomal (middle) and recombination (bottom) events. Samples were collected at the indicated timepoints (h, hours). Red arrows denote induction points of IME1 and NDT80 expression. (B) Flow cytometry data illustrating the progression of meiotic S-phase following PCUP1-IME1 induction. (C) Southern blot image showing the progression of recombination at the HIS4::LEU2 hotspot. Asterisk indicates cross-hydridizing bands. (D) Timing and synchrony of meiotic cultures. Levels of DSBs and crossovers at HIS4::LEU2, and nuclear divisions (MI±MII) were normalized to 1. The timing of SEIs and dHJs was determined using 2D gels (Figure 1—figure supplement 1). (E) Regimen for purification of peptides harboring K-ε-GG SUMO remnants (G-G-). (Ni-NTA, nickel-nitrilotriacetic acid resin; anti-diGly, anti-K-ε-GG antibody beads).
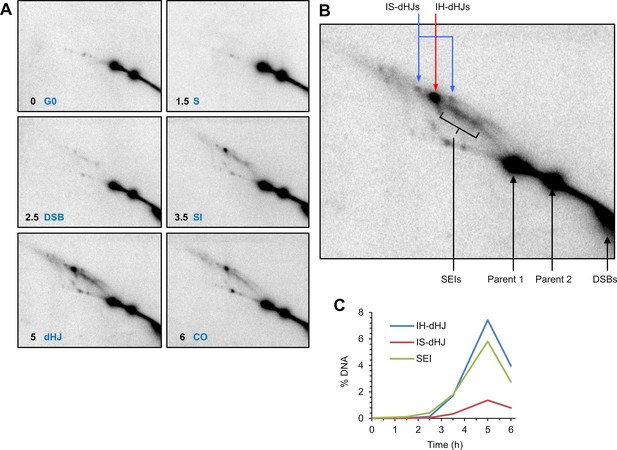
Two-dimensional Southern blot analysis of joint-molecule recombination intermediates.
(A) 2D gels Southern images representative of each timepoint collected for proteomics analysis. G0, G0 phase before cells enter meiosis; S, S-phase; DSB, initiation of recombination via DNA double-stranded break formation; SI, strand invasion and ongoing homolog synapsis; dHJ, double-Holliday junction formation and full synapsis; CO, crossover formation and desynapsis. (B) Single panel highlighting specific recombination intermediates. IH-dHJ, inter-homolog double-Holliday junction; IS-dHJ, inter-sister double-Holliday junction; SEI, single-end invasion; DSB, double-strand break. (C) Quantification of SEIs and dHJs over the entire meiotic time course.
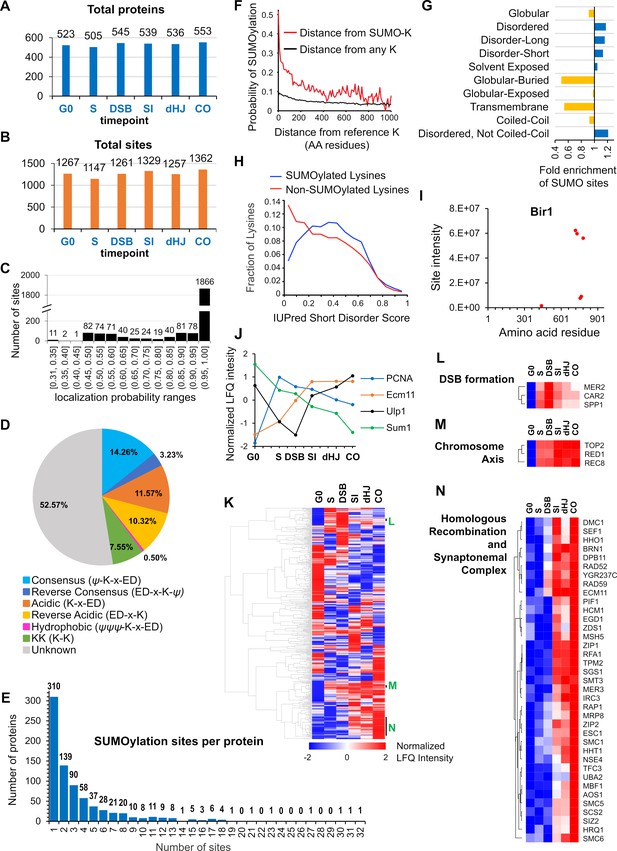
Characteristics of SUMOylation sites and temporal LFQ profiles of SUMOylated proteins.
(A-B) Total numbers of proteins (A) and SUMO sites (B) identified from each time point in the triplicate experiments used for quantitative analysis. (C) Localization probabilities for identified SUMO sites as calculated by MaxQuant. (D) Proportions of identified SUMOylation sites that conform to indicated consensus sequences. Ψ, hydrophobic amino acid; x: any amino acid. (E) Distribution of SUMOylation sites per protein. (F) Probability of a lysine being SUMOylated as a function of its distance from either a SUMOylated lysine (red line) or any lysine (black line). (G) Enrichment or depletion of SUMOylation sites relative to protein secondary structure. (H) Distributions of all lysines (red line) and SUMOylated lysines (blue line) relative to IUPred short disorder score. (I) Site intensity profile of chromosomal passenger complex component Bir1 illustrating site clustering. (J) Normalized temporal LFQ profiles for PCNA, Ecm11, Ulp1 and Sum1 highlighting the diverse dynamics of SUMOylation during meiosis. (K) Hierarchical clustering of normalized LFQ profiles. (L) (M) (N) Clustering of SUMO targets involved in (L) DSB formation, (M) chromosome axes, and (N) recombination and synapsis.
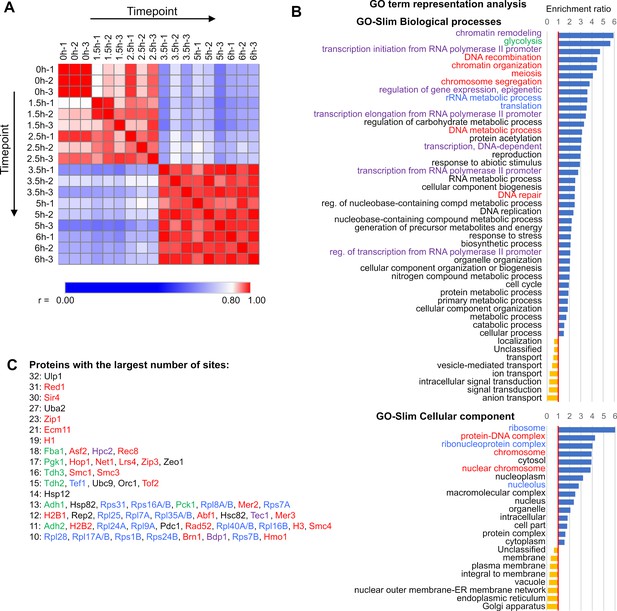
Characteristics of the the meiotic SUMO-modified proteome.
(A) Similarity matrix of samples used for label-free quantification (LFQ). Heatmap representing Pearson correlation coefficient (r) values for protein LFQ (scale at the bottom). Labels indicate timepoint (hours) and replicate serial number. (B) Over-representation analysis of GO-Slim Biological processes and GO-Slim Cellular component showing categories that are significantly over- (blue bars) or under-represented (gold bars). Related processes are highlighted with colored text. (C) Proteins in descending order of SUMOylation site number, color-coded to match the GO categories that they represent.
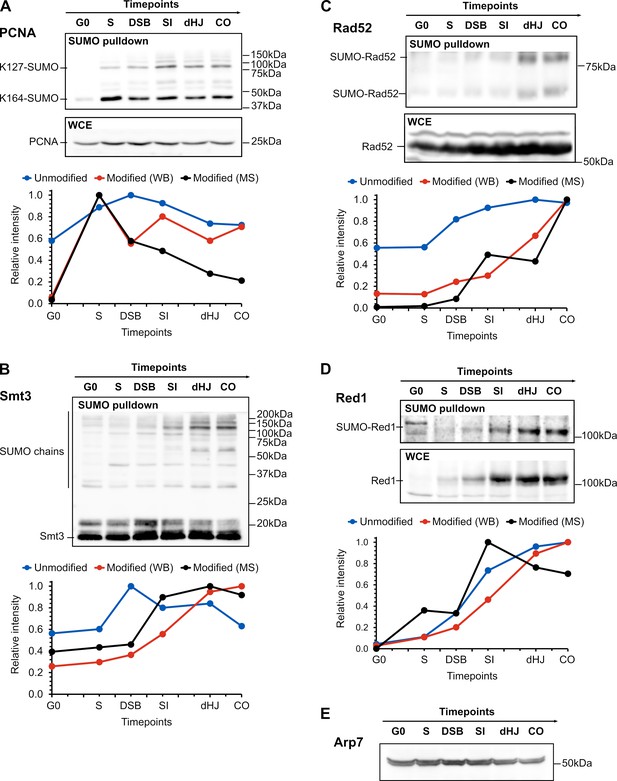
Western blot validation of SUMO targets.
Western blot analysis of SUMO-modified and unmodified forms of: (A) PCNA, (B) Smt3, (C) Rad52, and (D) Red1 in SUMO enriched (6His-Smt3 Ni-NTA purified ‘SUMO pulldown’) and whole-cell extract (‘WCE’) samples, respectively. Timepoints correspond to those used for proteomics analysis. Graphs compare the relative intensity profiles of unmodified and SUMO-modified forms, estimated from quantifying western blots (WB), with the averaged relative LFQ intensity profiles for the SUMOylated protein (Modified [MS]). For quantification of western blots, intensities were adjusted to the loading control (Arp7) shown in panel E.
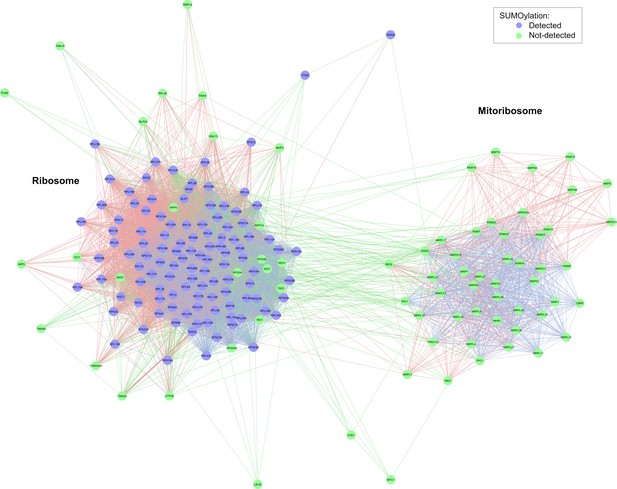
Ribosomal protein network.
Network diagram highighting SUMOylated components of the ribosome. In stark contrast, the mito-ribosome is not SUMOylated.
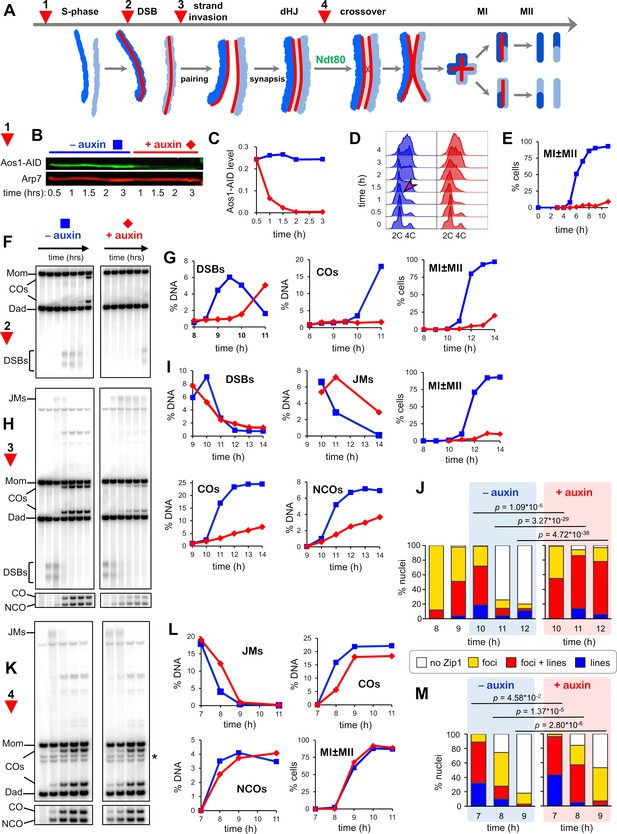
SUMOylation functions throughout meiosis.
(A) Cartoon showing the four points (experiments 1–4) at which de novo SUMOylation was acutely inactivated. (B) Immunoblot of Aos1-AID with and without the addition of auxin at 30 mins (experiment 1). Arp7 was used as loading control. (C) Quantification of the blot shown in (B). (D) Flow cytometry analysis of S-phase progression for experiment 1. The magenta arrow indicates the onset of S-phase in control cells. (E) Meiotic nuclear divisions (MI ± MII) for experiment 1. (F) 1D gel Southern blot image for analysis of DSBs and crossovers in experiment 2. (G) Quantification of DSBs, crossovers and meiotic divisions for experiment 2. (H) 1D gel Southern blot images for experiment 3. The top gel was used to quantify DSBs and crossovers (COs). The bottom gel was used to quantify non-crossover products (NCOs). (I) Quantification of DSBs, joint molecules (JMs), COs, NCOs, and meiotic divisions for experiment 3. JMs were quantified from 2D gel Southern analysis (Figure 3—figure supplement 4). (J) Quantification of synapsis (Zip1-staining classes) for experiment 3 (Figure 3—figure supplement 1F). (K) 1D gel Southern blot images for experiment 4. The top gel was used to quantify DSBs and crossovers (COs). The bottom gel was used to quantify non-crossovers (NCOs). (L) Quantification of DSBs, joint molecules (JMs), COs, NCOs and meiotic divisions for experiment 4. JMs were quantified from 2D gel Southern analysis (Figure 3—figure supplement 4). (M) Quantification of synapsis (Zip1-staining classes) for experiment 4.
Analysis of meiotic recombination and synapsis.
(A–E) Physical assay system for monitoring recombination at the HIS4::LEU2 locus. (A) Map of the locus showing diagnostic restriction sites, hybridization probe, detectable DNA species and their molecular weights. X, XhoI site; B, BamHI site; N, NgoMIV site; DSB, double-strand break; IH-dHJ, interhomolog double-Holliday junction; IS-JM, inter-sister joint molecule (likely IS-dHJ); mc-JM, multi-chromatid joint molecule; SEI, single-end invasion. (B) Southern blot image of 1D gel analysis of DSBs and crossovers. (C) Inferred structures of SEI and dHJ intermediates. (D) Southern blot image of native/native 2D gel analysis of recombination intermediates. (E) 1D Southern blot image of 1D gel crossover/non-crossover analysis. (F) Representative immunofluorescence images of Zip1 immunostained nuclear spreads illustrating the different classes of Zip1 staining scored in Figure 3 and Figure 3—figure supplement 2.
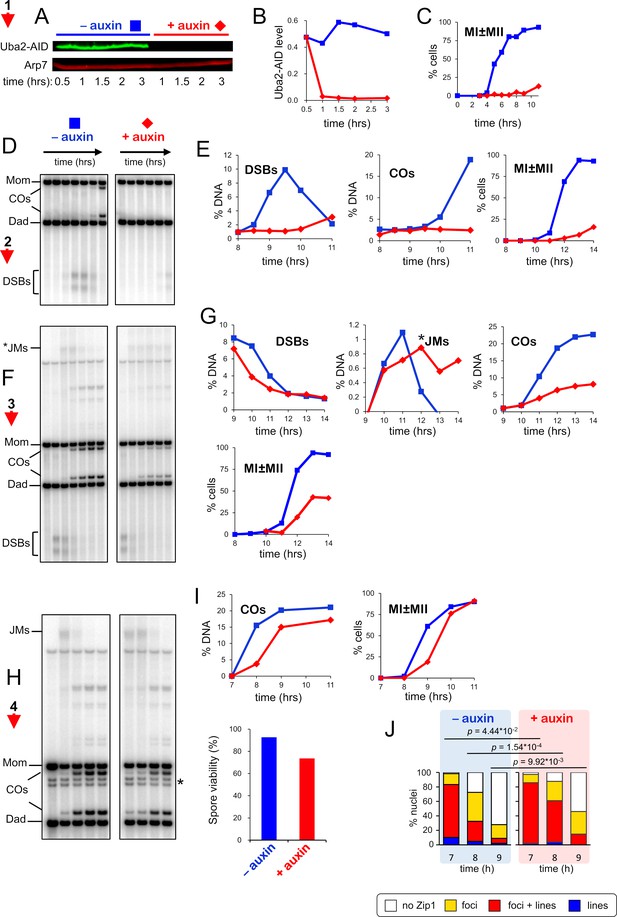
SUMO execution-point analysis using the Uba2-AID degron allele.
(A) Immunoblot showing the degradation of Uba2-AID after addition of auxin. (B) Quantification of the blot shown in (A). (C) Meiotic nuclear divisions (MI ± MII) for experiment 1 (see Figure 3A for details of experiments 1–4) (D) 1D gel Southern blot image for analysis of DSBs and crossovers in experiment 2. (E). Quantification of DSBs, crossovers, and meiotic divisions for experiment 2. (F) 1D gel Southern blot images for experiment three used to quantify DSBs, crossovers (COs), and joint-molecules (JM) recombination intermediates. (G) Quantification of DSBs, JMs, COs, and meiotic divisions for experiment 3. *JMs were quantified from the 1D Southern analysis. (H) 1D gel Southern blot image for experiment 4. Asterisk indicates cross-hybridizing bands. (I) Quantification of COs and meiotic divisions for experiment 4. Spore viability was also analyzed (lower bar graph) and shown to be statistically lower following Uba2-AID degradation at the time of IN-NDT80 expression (91.5% vs 76.4%, p<0.04, t-test). (J) Quantification of synapsis (Zip1-staining classes) for experiment 4 indicates that SC disassembly is delayed (p values are shown for G-test analysis).
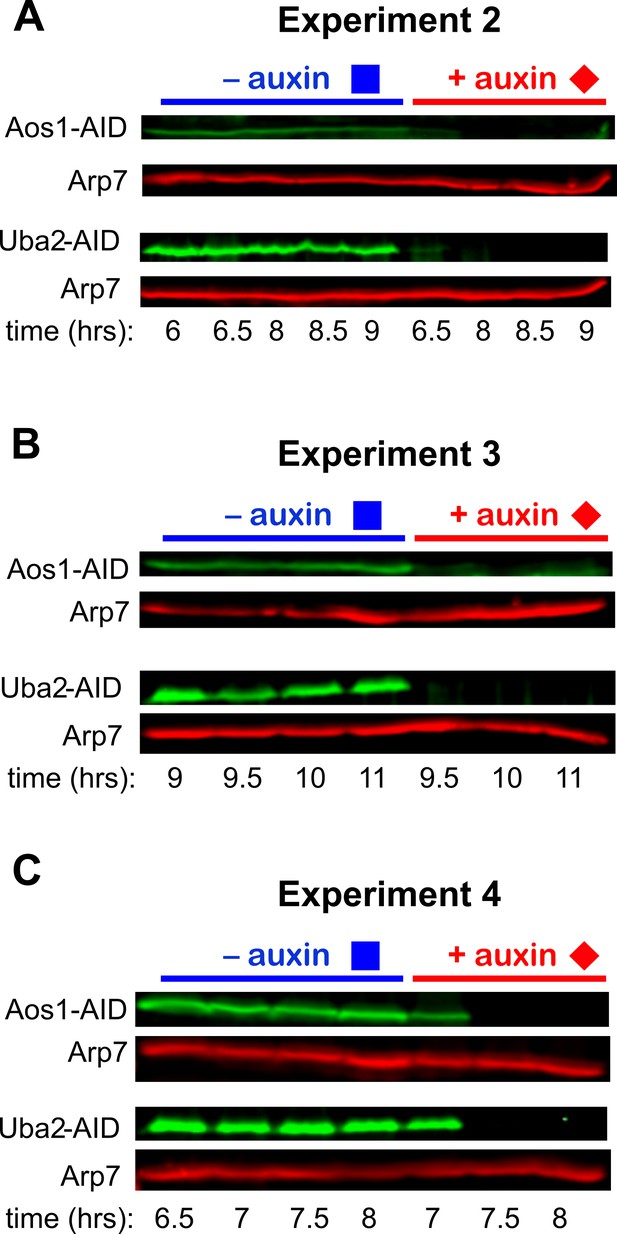
Degron-mediated depletion of Aos1 and Uba2.
Immunoblots showing degradation of Aos1-AID and Uba2-AID after addition of auxin in (A) experiment 2, at the time of DSB formation; (B) experiment 3 at the time of strand invasion and synapsis; and (C) experiment 4 as cells resolve dHJs and exit pachytene. See Figure 3A for details.
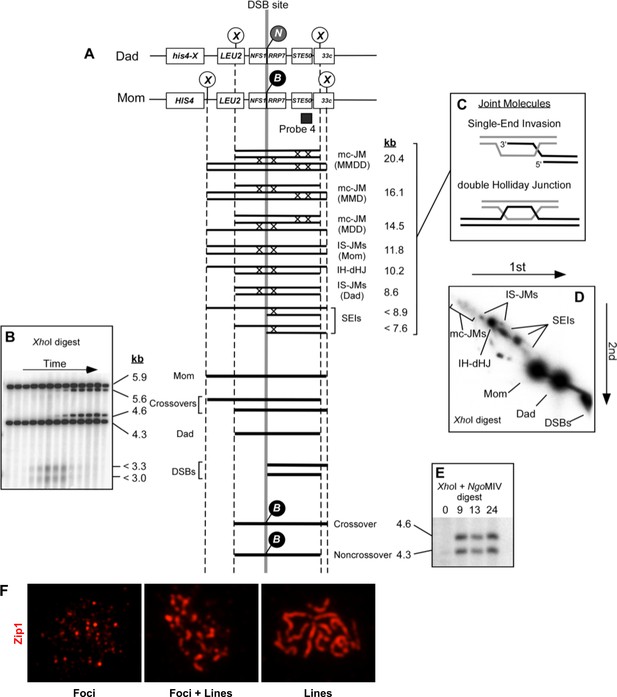
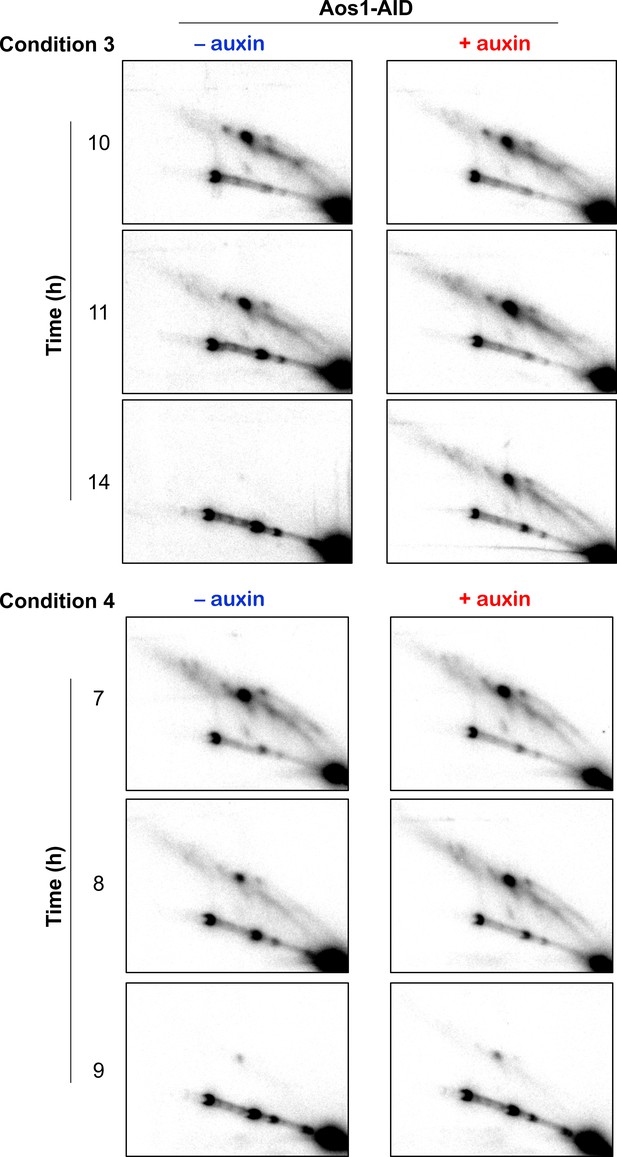
2D Southern blot analysis of joint molecule recombination intermediates.
2D gel Southern blot images of joint-molecule analysis for experiments 3 and 4 in Figure 3H and K, with or without the addition of auxin to degrade Aos1-AID. Quantification is shown in Figure 3I and L.
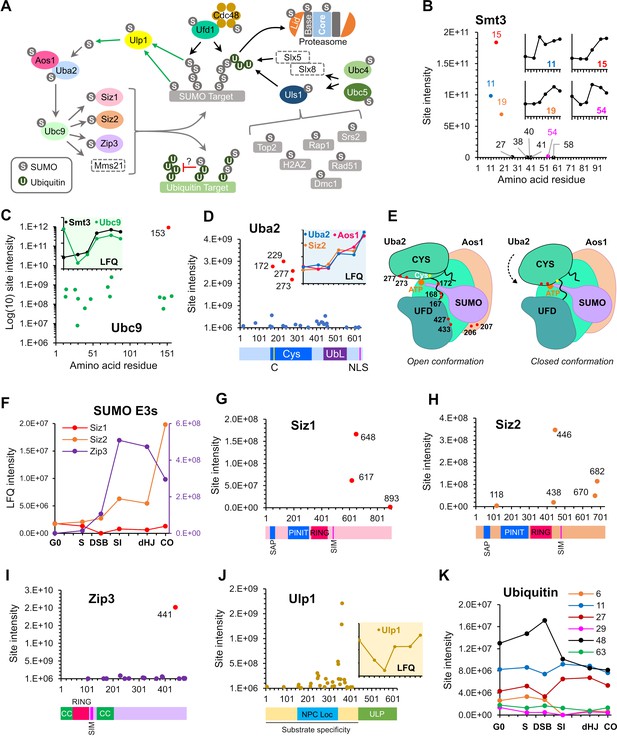
SUMOylation of the SUMO and Ubiquitin-proteasome machinery.
(A) Summary of targets in the SUMO and ubiquitin-proteasome systems and the relationships between these factors. Dashed rectangles indicate SUMOylation was not detected. Note that although SUMOylation of Mms21 was not detected, the associated Smc5/6 complex was modified. (B) Intensity plot of SUMOylated sites on Smt3. Insets show temporal profiles of the four most prominent SUMO-chain linkages. (C) Intensity plot of SUMOylated sites on E2 conjugase Ubc9. Inset shows normalized LFQ profiles of Ubc9 and Smt3. (D) SUMO-site intensities on Uba2. The cartoon below shows key domains along the peptide backbone. C: active-site cysteine, NLS: nuclear localization sequence. Inset shows normalized LFQ profiles of Uba2, Aos1, and Siz2 (E) Cartoon of the Aos1-Uba2 structure highlighting the locations of SUMOylation sites. (F) LFQ profiles of the SUMO E3 ligases. The Zip3 profile is plotted on the purple Y-axis on the right-hand side. (G) (H) (I) Site intensities of Siz1 (G), Siz2 (H) and Zip3 (I) plotted over their respective domain structures. SAP, SAF-A/B, Acinus, and PIAS domain; PINIT, PINIT motif-containing domain; RING: SpRING domain; SIM, SUMO interacting motif; CC, coiled coil. (J) SUMO-site intensities of Ulp1 plotted over its domain structure. NPC Loc, nuclear pore complex localization domain; ULP, Ubiquitin-like protease domain. Inset shows the normalized LFQ profile. (K) Intensity profiles of SUMOylation sites on Ubiquitin.
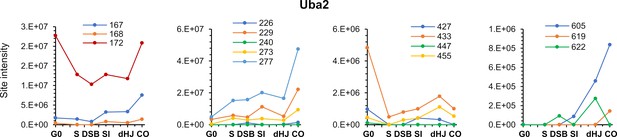
Diverse temporal profiles of SUMOylation sites on the SUMO E1 subunit Uba2.
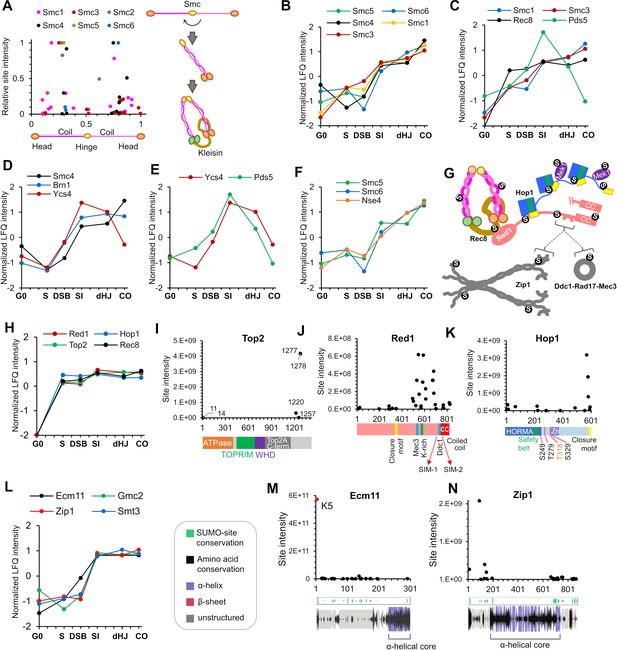
SUMOylation of homolog axes and synaptonemal complex.
(A) Relative positions of SUMO sites on the SMC proteins. Adjacent cartoon illustrates how SMCs are assembled into ring structures. (B–F) Normalized LFQ profiles of (B) the SMCs (note that Smc2 did not have measurable LFQ intensities); (C) components of Rec8-cohesin; (D) condensin; (E) HEAT-repeat proteins Ycs4 and Pds5; and (F) components of the Smc5/6 complex. (G) Summary of SUMOylated axis proteins and pertinent interaction partners. Arcs in Red1 and Zip1 indicate SIMs. (H) Normalized LFQ profiles of axis components Red1, Hop1, Top2, and Rec8. (I) Site intensities of Top2 plotted along its domain structure. TOPRIM, Topoisomerase-primase domain; WHD, winged-helix domain. (J) Site intensities of Red1 plotted along its domain structure. (K) Site intensities of Hop1 plotted along its domain structure. S249, T279 T318, and S329 are key phosphorylation sites important for Hop1 function. (L) Normalized LFQ profiles of SC central region components Ecm11, Zip1, and Gmc2 relative to SUMO (Smt3). (M) (N) Site intensity plots for (M) Ecm11 and (N) Zip1 plotted along predicted secondary structures.
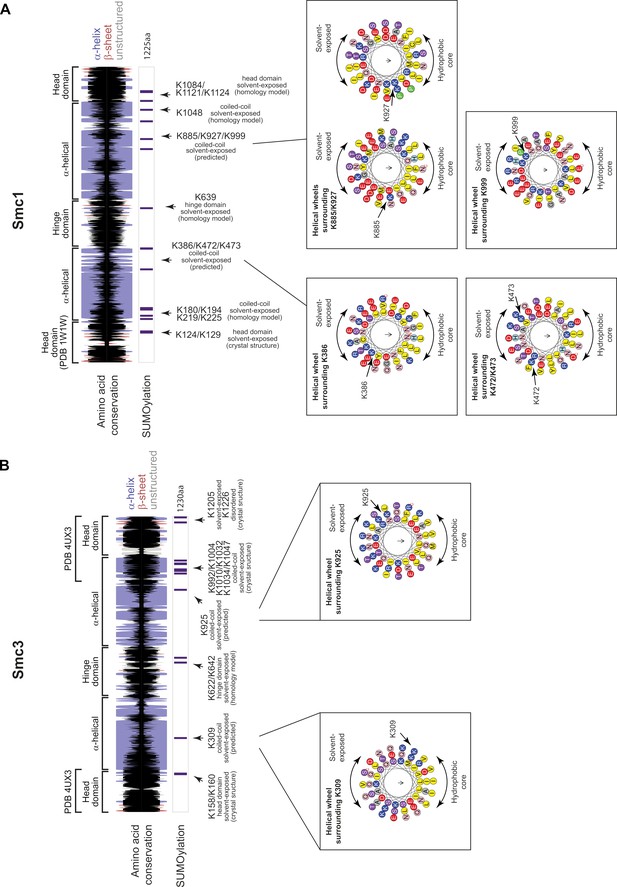
Secondary structure and helical projections of Cohesin SMC proteins.
Cartoons showing secondary structure predictions (α-helix: purple bar, β-sheet: red bar, unstructured: gray bar) and amino acid conservation (black bar) for SMC proteins: (A) Smc1, (B) Smc3. Height of the black bars represents the level of amino-acid conservation. SUMO sites are shown below together with information regarding location in structure and solvent exposure. Insets show helical projections in cases where SUMO sites lie on an α-helix.
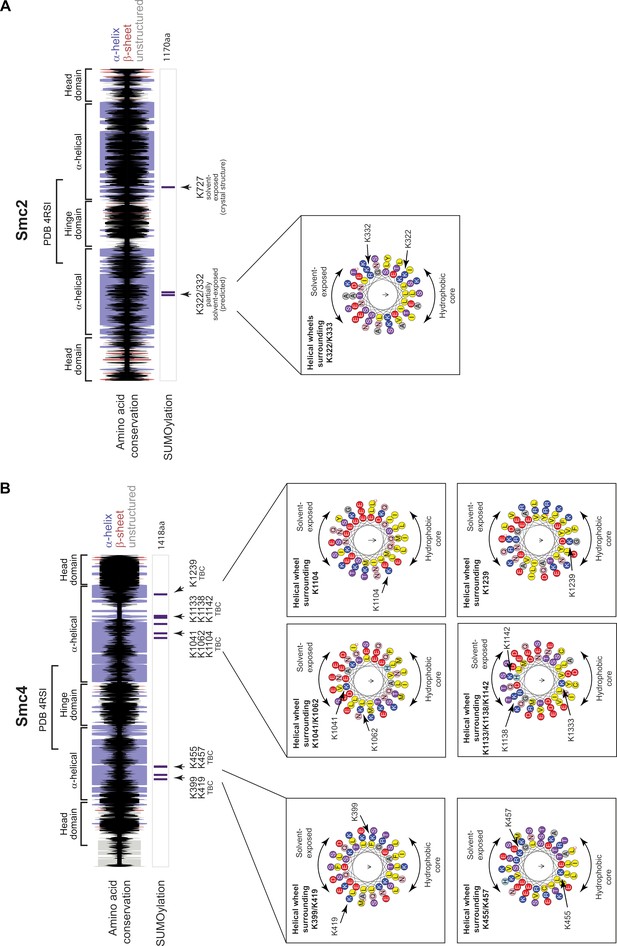
Secondary structure and helical projections of Condensin SMC proteins.
Cartoons showing secondary structure predictions (α-helix: purple bar, β-sheet: red bar, unstructured: gray bar) and amino acid conservation (black bar) for SMC proteins: (A) Smc2, (B) Smc4. Height of the black bars represents the level of amino-acid conservation. SUMO sites are shown below together with information regarding location in structure and solvent exposure. Insets show helical projections in cases where SUMO sites lie on an α-helix.
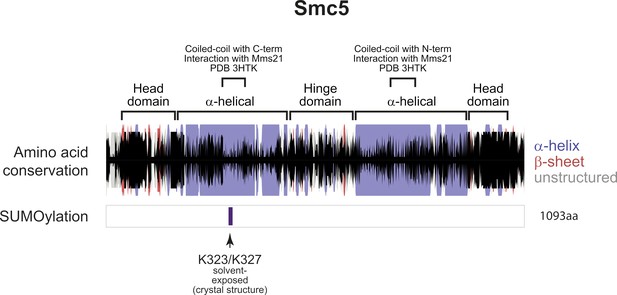
Secondary structure and helical projections of Smc5.
Cartoons showing secondary structure predictions (α-helix: purple bar, β-sheet: red bar, unstructured: gray bar) and amino acid conservation (black bar). Height of the black bars represents the level of amino-acid conservation. SUMO sites are shown below together with information regarding location in structure and solvent exposure.
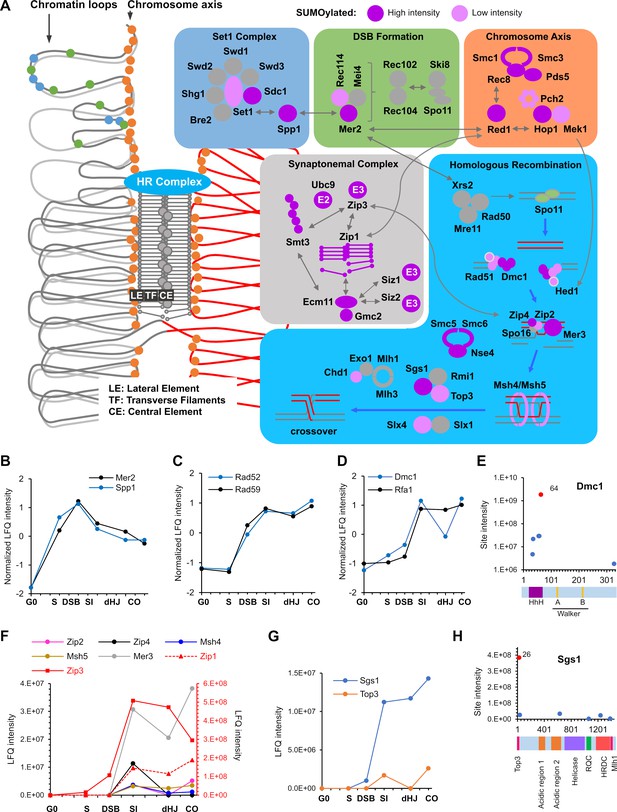
SUMOylation of the recombination machinery.
(A) Summary of SUMOylated meiotic HR factors and their functional integration with other pertinent SUMO targets in the chromosome axes and SCs. The schematic on the left-hand side illustrates the linear chromatin loop array of prophase-I chromosomes and relative localizations of SUMOylated factors illustrated in the color-coded boxes. (B–D) Normalized LFQ profiles of: (B) DSB factors Mer2 and Spp1; (C) Rad51-mediator and DNA strand-annealing proteins Rad52 and Rad59; and (D) the strand exchange factors Dmc1 and Rfa1. (E) Site intensities of Dmc1 plotted along its domain structure. HhH: Helix-hairpin-helix. (F) LFQ profiles of the ZMM proteins (SUMOylation of Spo16 was not detected). Zip1 and Zip3 are plotted on the right-hand side Y-axis in red. (G) Normalized LFQ profiles of DNA helicase Sgs1 and topoisomerase Top3. (H) Sgs1 site intensities mapped along its domain structure. Top3, Top3 interacting domain; RQC, RecQ C-terminal domain; HRDC, helicase and RNaseD C-terminal domain; Mlh1, Mlh1 interacting domain.
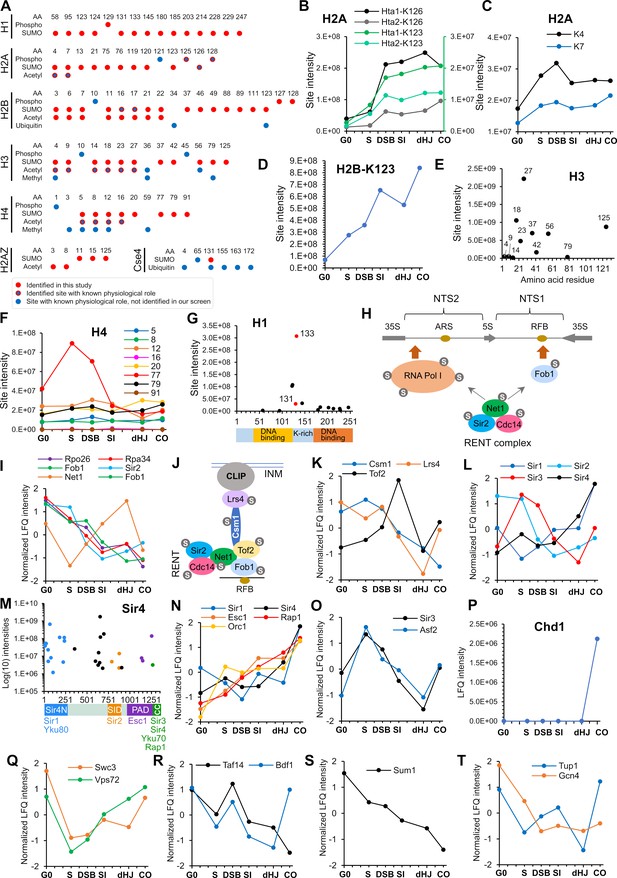
SUMOylation of chromatin and associated factors.
(A) Chart summarizing histone SUMOylation, phosphorylation and acetylation sites identified in this study, together with previously identified modifications. AA, amino acid position. (B) Temporal profiles of SUMOylation on K123 and K126 of histones H2A1 and H2A2. The K123 profiles are plotted on the right-hand side Y-axis in green. (C) Temporal profiles of K4 and K7 SUMOylation on histone H2A. (D) Temporal profile of histone H2B-K123 SUMOylation. (E) SUMO site intensities on histone H3. (F) Temporal profiles of histone H4 SUMOylation sites. (G) Site intensities of histone H1 SUMOylation sites plotted along its domain structure. Two sites that are co-modified with adjacent phosphorylation are indicated in red. (H) Summary of SUMOylated proteins involved in rDNA silencing. NTS1/2, non-transcribed sequences; RFB, replication fork barrier. (I) Normalized LFQ profiles of RENT complex components. (J) Cartoon illustrating the recruitment of rDNA loci to the inner nuclear membrane (INM) via RENT, cohibin (Csm1 and Lrs4), and CLIP (chromosome linkage inner-nuclear membrane proteins) complexes. INM, inner-nuclear membrane; RFB, replication fork barrier. (K) Normalized LFQ profiles of Cohibin (Csm1 and Lrs4) and Tof2. (L) Normalized LFQ profiles of the SIR proteins. (M) Site intensity plot of Sir4 plotted along its domain structure. (N) Normalized LFQ profiles of Sir4 and its binding partners. (O) Normalized LFQ profiles of Sir3 and its paralog Asf2. (P) LFQ profile of chromatin remodeler Chd1. (Q) Normalized LFQ profiles of SWR1-chromatin remodeler components Swc3 and Vps72 (R) Normalized LFQ profiles of chromatin remodeler components Bdf1 (SWR subunit that binds acetylated H4) and Taf14 (subunit of TFIID, TFIIF, INO80, Swi/Snf, and NuA3m that binds acetylated H3). (S) Normalized LFQ profile of Sum1, a component of the Sum1-Rfm1-Hst1 deacetylase that represses middle-meiotic genes. (T) Normalized LFQ profiles of Tup1, a constituent of the Tup1-Cyc8 transcriptional repressor, and the transcriptional activator Gcn4.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Other | Histrap FF 1 ml | GE Healthcare | 17-5319-01 | AKTA FPLC cartridge |
| Peptide, recombinant protein | Lysyl Endopeptidase (Lys-C) | Wako Chemicals | 125–02541 | |
| Peptide, recombinant protein | Glu-C, sequencing grade | Promega | V1651 | |
| Other | Sep-Pak tC18 1cc vac cartridge 50 mg | Waters | WAT054960 | Desalting cartridge |
| Commercial assay, kit | PTMScan Ubiquitin Remnant Motif (K-ε-GG) | Cell Signaling technology | 5562S | Antibody beads and buffer kit |
| Chemical compound, drug | PP1 | Tocris | 1397 | Synthetic ATP analog |
| Chemical compound, drug | 3-Indoleacetic acid (Auxin) | Sigma-Aldrich | I3750-25G-A | Protein alkylation agent |
| Chemical compound, drug | β-Estradiol | Sigma-Aldrich | E2758 | |
| Antibody | Anti-c-Myc ‘mouse monoclonal’ antibody | Roche | 11667149001, RRID:AB_390912 | ‘1:5000’ |
| Antibody | Anti-Arp7 ‘goat polyclonal’ antibody | Santa Cruz Biotechnology | y-C20, RRID:AB_671730 | ‘1:2000’ |
| Antibody | ‘Donkey polyclonal’ anti-goat, IRDye 680 | LI-COR Biosciences | 926–68074, RRID:AB_10956736 | ‘1:5000’ |
| Antibody | ‘Donkey polyclonal’ anti-mouse, IRDye 800 | LI-COR Biosciences | 926–32212, RRID:AB_621847 | ‘1:5000’ |
| Antibody | ‘Goat polyclonal’ anti-guinea pig, Alexa Fluor 555 | Thermo Fisher | A-21435, RRID:AB_2535856 | ‘1:200’ |
| Antibody | Anti-Zip1 ‘goat polyclonal’ | Santa Cruz Biotechnology | y-N16, RRID:AB_794259 | ‘1:50’ |
| Antibody | Anti-Zip1 ‘guinea pig polyclonal’ | Dr. Scott Keeney | gp3 | ‘1:500’ |
| Antibody | ‘Donkey polyclonal’ anti-goat, Alexa Fluor 555 | Thermo Fisher | A-21432, RRID:AB_141788 | ‘1:1000’ |
| Software, algorithm | MaxQuant 1.6.1 | Max Planck Institute | RRID:SCR_014485 | MS raw data search |
Additional files
-
Supplementary file 1
Comparison of SUMOylated proteins identified in this study with data from previous studies.
- https://cdn.elifesciences.org/articles/57720/elife-57720-supp1-v2.docx
-
Supplementary file 2
Key MaxQuant Tables.
Compilation of useful tables generated by MaxQuant is a single combined search of all samples: ProteinGroups: All proteins including unmodified proteins, contaminants and false positives GlyGly sites: SUMO sites Phospho (STY) sites: phosphorylated serine, threonine and tyrosine residues ModificationSpecific Peptides: All identified peptides and their relevant modifications Oxidation (M) sites: Oxidized methionine residues.
- https://cdn.elifesciences.org/articles/57720/elife-57720-supp2-v2.xlsx
-
Supplementary file 3
Comparison of SUMO sites identified in this study with sites predicted by SUMOsp.
- https://cdn.elifesciences.org/articles/57720/elife-57720-supp3-v2.docx
-
Supplementary file 4
Annotated list of SUMO targets.
- https://cdn.elifesciences.org/articles/57720/elife-57720-supp4-v2.xlsx
-
Supplementary file 5
Protein diagrams mapping SUMO sites and SIMs on protein secondary structure.
Upper Track: Red line – SUMOylated lysine, Black line – lysine, Predicted SUMO Interaction Motifs (gray boxes): Dark gray – high threshold, Medium gray – medium threshold, Light gray – low threshold; Middle Track: Blue box - PFAM domains; Lower Track - Protein Secondary Structure: Purple – Globular Domains, Orange-red – Disorder score, Red – coiled-coil, Black – Transmembrane; Horizontal Axis: amino acid residue number
- https://cdn.elifesciences.org/articles/57720/elife-57720-supp5-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57720/elife-57720-transrepform-v2.docx





