Transcription termination and antitermination of bacterial CRISPR arrays
Figures
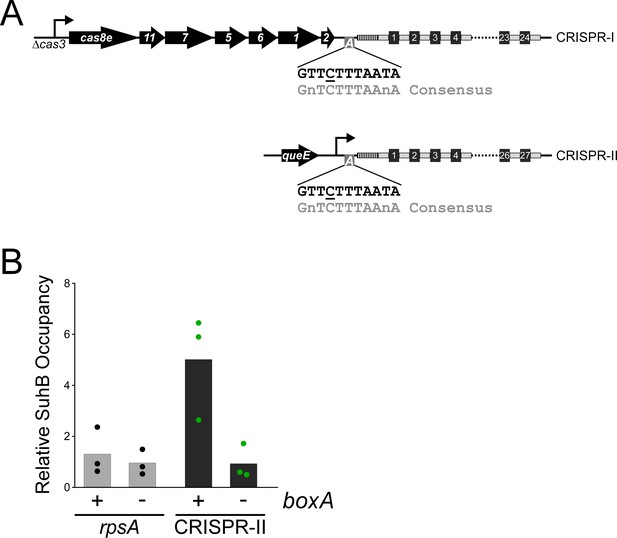
boxA elements upstream of both CRISPR arrays in Salmonella Typhimurium.
(A) Schematic of the two CRISPR arrays in S. Typhimurium. Repeat sequences are represented by gray rectangles and spacer sequences are represented by black squares. Spacers are numbered within the array, with spacer one being closest to the leader sequence (dashed rectangle). The CRISPR-I array is co-transcribed with the upstream cas genes. For the work presented in this study, the cas3 gene was deleted and replaced with a constitutive promoter. Similarly, a constitutive promoter was inserted immediately downstream of queE, upstream of the CRISPR-II array. boxA elements (boxes containing an ‘A’) are located immediately upstream of the leader sequences of both CRISPR arrays. (B) Relative occupancy of SuhB-TAP, determined by ChIP-qPCR, within the highly expressed rpsA gene (gray bars), or at the boxA sequence upstream of CRISPR-II (black bars). Occupancy was measured in a strain with an intact boxA upstream of CRISPR-II (AMD710), or a single base-pair substitution within the boxA (underlined base in (A); AMD711). Values shown with bars are the average of three independent biological replicates, with dots showing each individual datapoint.
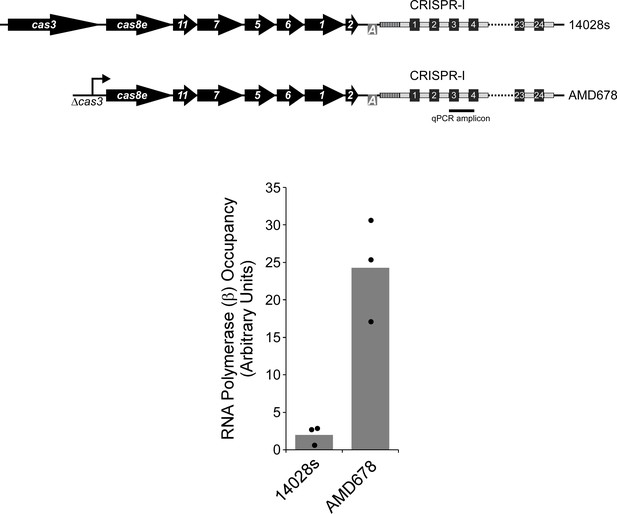
The CRISPR-I array is co-transcribed with the upstream cas gene operon.
Occupancy of RNA polymerase (β subunit) measured by ChIP-qPCR within the CRISPR-I array in wild-type 14028s or AMD678, which has a constitutive promoter in place of cas3, as indicated in the schematic. The location of the qPCR amplicon is indicated on the schematic. Values shown with bars are the average of three independent biological replicates, with dots showing each individual datapoint.
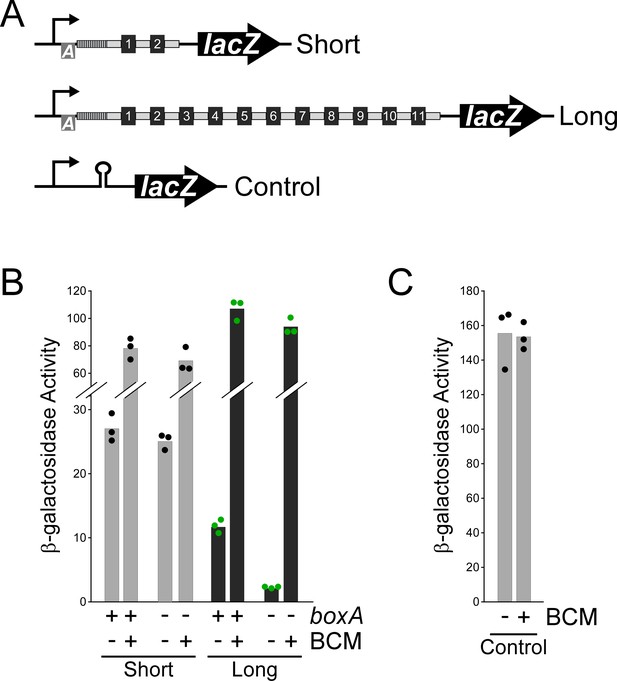
BoxA-mediated antitermination of a CRISPR array.
(A) Schematic of short (pGB231 and pGB237) and long (pGB250 and pGB256) lacZ reporter gene transcriptional fusions to the CRISPR-II array, and a control transcriptional fusion (pJTW060) that includes an intrinsic terminator upstream of lacZ. (B) β-galactosidase activity of the short and long lacZ reporter gene fusions with either an intact (pGB231 and pGB250) or mutated (pGB237 and pGB256) boxA sequence, in cells grown with/without addition of the Rho inhibitor bicyclomycin (BCM). (C) β-galactosidase activity of the control lacZ reporter gene fusion (pJTW060) that includes an intrinsic terminator upstream of lacZ, in cells grown with/without BCM. Values shown with bars are the average of three independent biological replicates, with dots showing each individual datapoint.
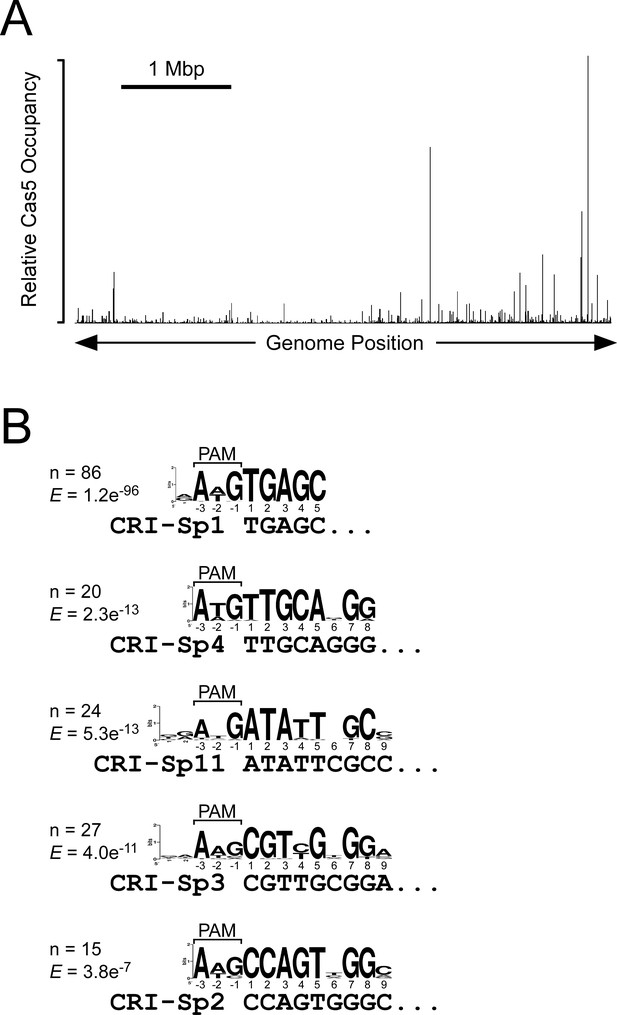
Extensive off-target binding of S. Typhimurium Cascade.
(A) Relative FLAG3-Cas5 occupancy across the S. Typhimurium genome as measured by ChIP-seq in strain AMD678. (B) Enriched sequence motifs in Cascade-bound regions, as identified by MEME (Bailey and Elkan, 1994). Sequence matches to the AWG PAM and to the seed sequence of specific spacers are indicated. The number of Cascade-bound genomic regions containing each sequence motif is indicated, as is the enrichment score ("E") generated by MEME (Bailey and Elkan, 1994).
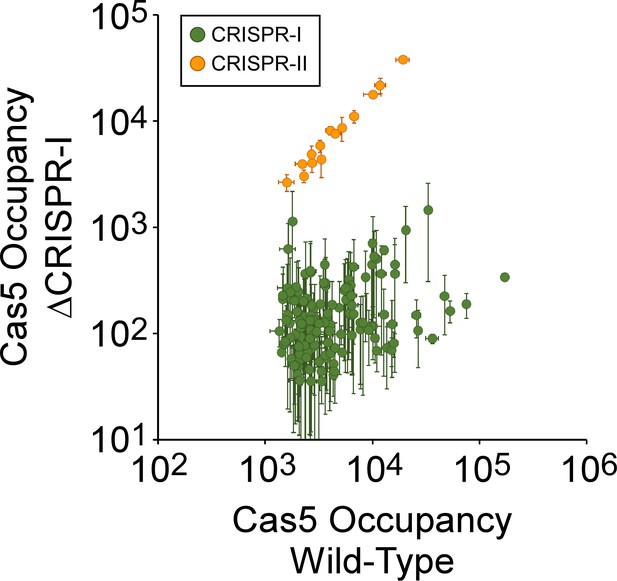
Confirmation of spacer assignments to Cas5- binding sites.
Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-I array (AMD678; x-axis), and ΔCRISPR-I cells (AMD679; y-axis). Cascade binding associated with spacers from CRISPR-I is indicated by green datapoints. Cascade binding associated with spacers from CRISPR-II is indicated by orange datapoints. Values plotted are the average of two independent biological replicates. Error bars represent one standard-deviation from the mean.
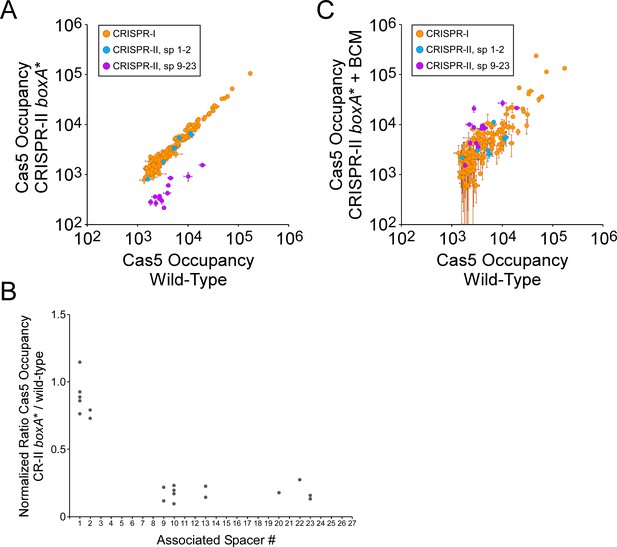
The CRISPR-II boxA facilitates use of all spacers by preventing premature Rho termination.
(A) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-II boxA (AMD678; x-axis), and cells with a single base-pair substitution in the CRISPR-II boxA (‘boxA*”; AMD685; y-axis). Cascade binding associated with spacers from CRISPR-I is indicated by orange datapoints. Cascade binding associated with spacers 1–2 from CRISPR-II is indicated by light blue datapoints. Cascade binding associated with spacers 9–23 from CRISPR-II is indicated by purple datapoints. Values plotted are the average of two independent biological replicates. Error bars represent one standard-deviation from the mean. (B) Normalized ratio of FLAG3-Cas5 occupancy associated with spacers from CRISPR-II. Values are plotted according to the associated spacer, and are normalized to the average value for sites associated with spacers from CRISPR-I. (C) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-II boxA (AMD678; x-axis), and cells with a single base-pair substitution in the CRISPR-II boxA (‘boxA*”; AMD685) that were treated with bicyclomycin (BCM; y-axis).
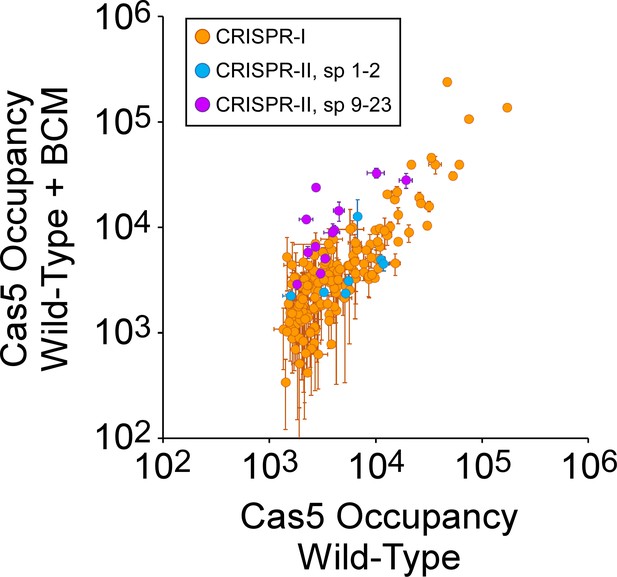
Inhibition of Rho in cells with an intact boxA increases the use of spacers 9–23 of CRISPR-II.
Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-II boxA (AMD678) without (x-axis) and with bicyclomycin (BCM) treatment (y-axis). Cascade binding associated with spacers from CRISPR-I is indicated by orange datapoints. Cascade binding associated with spacers 1–2 from CRISPR-II is indicated by light blue datapoints. Cascade binding associated with spacers 9–23 from CRISPR-II is indicated by purple datapoints. Values plotted are the average of two independent biological replicates. Error bars represent one standard-deviation from the mean.
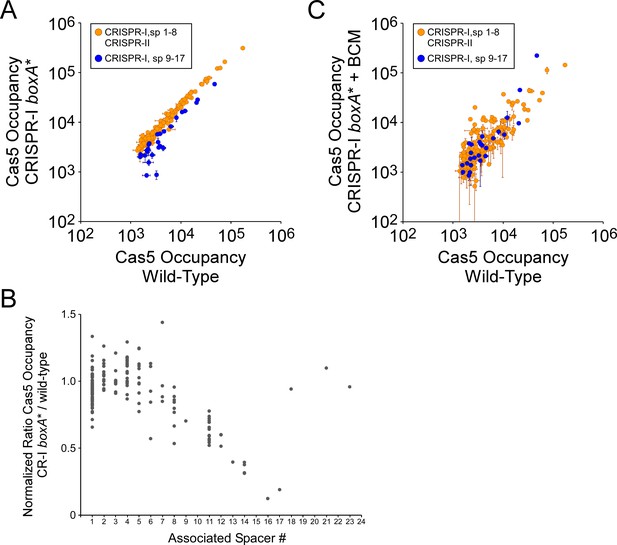
The CRISPR-I boxA facilitates use of all spacers by preventing premature Rho termination.
(A) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-I boxA (AMD678; x-axis), and cells with a single base-pair substitution in the CRISPR-I boxA (‘boxA*”; AMD684; y-axis). Cascade binding associated with spacers from CRISPR-II, and spacers 1–8 from CRISPR-I, is indicated by orange datapoints. Cascade binding associated with spacers 9–17 from CRISPR-I is indicated by dark blue datapoints. Values plotted are the average of two independent biological replicates. Error bars represent one standard-deviation from the mean. (B) Normalized ratio of FLAG3-Cas5 occupancy associated with spacers from CRISPR-I. Values are plotted according to the associated spacer, and are normalized to the average value for sites associated with spacers from CRISPR-II. (C) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in cells with an intact CRISPR-I boxA (AMD678; x-axis), and cells with a single base-pair substitution in the CRISPR-I boxA (‘boxA*”; AMD684) that were treated with bicyclomycin (BCM; y-axis).
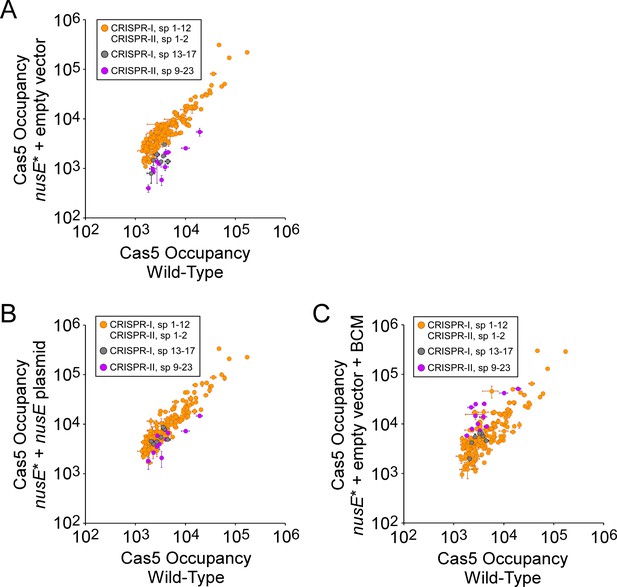
Nus factors facilitate use of all spacers by preventing premature Rho termination.
(A) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in nusE+ (AMD678; x-axis), and nusE mutant cells (‘nusE*”; AMD698) containing an empty vector (pBAD24-amp; y-axis). Cascade binding associated with spacers 1–12 from CRISPR-I, and spacers 1–2 from CRISPR-II, is indicated by orange datapoints. Cascade binding associated with spacers 13–17 from CRISPR-I is indicated by gray datapoints. Cascade binding associated with spacers 9–23 from CRISPR-II is indicated by purple datapoints. Values plotted are the average of two independent biological replicates. Error bars represent one standard-deviation from the mean. (B) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in nusE+ (AMD678; x-axis) or nusE mutant cells (‘nusE*”; AMD698) expressing a plasmid-encoded (pAMD239) copy of wild-type nusE (y-axis). (C) Comparison of FLAG3-Cas5 ChIP-seq occupancy at off-target chromosomal sites in nusE+ cells (AMD678; x-axis), and nusE mutant cells (‘nusE*”; AMD698) containing an empty vector and treated with bicyclomycin (BCM; y-axis).
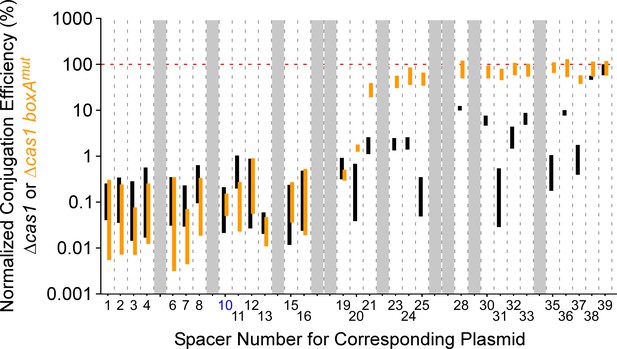
Conjugation efficiencies of protospacer-containing plasmids into Vibrio cholerae.
Normalized conjugation efficiency (%) of protospacer-containing plasmids into Δcas1 (KS2094) or boxAmut Δcas1 (KS2206) V. cholerae A50. Bars indicate the range of conjugation efficiencies across two replicate experiments, with the spacer corresponding to each plasmid indicated on the x-axis. Black and orange bars indicate efficiency values for the Δcas1 strain and boxAmut Δcas1 strain, respectively. Note that spacers 10 and 26 are identical, with data only shown for spacer 10 (blue text). The red horizontal, dashed line indicates 100% conjugation efficiency (i.e. the same efficiency as the control plasmid that lacks a protospacer). Greyed-out regions indicate protospacer-containing plasmids with <50 sequence reads in the control sample for at least one of the two replicates.

Sequence of the CRISPR array and upstream sequence from Vibrio cholerae A50.
The first nucleotide shown is the first nucleotide downstream of cas2. Repeat sequences are highlighted in green. The boxA sequence is highlighted in yellow. Individual spacers and their position in the array are indicated below the sequence of the array. The red highlighted spacers are identical.
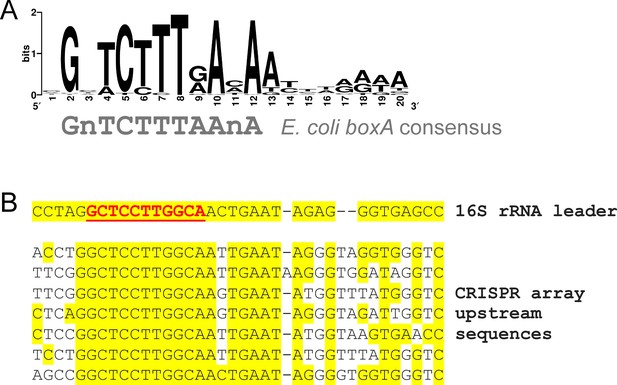
Widespread conservation of boxA sequences upstream of CRISPR arrays in diverse bacterial genera.
(A) Sequence motif found by MEME (Bailey and Elkan, 1994) to be significantly enriched upstream of 52 CRISPR arrays spread across the bacterial kingdom (from 187 tested; MEME E-value = 7.1e−69). (B) A sequence alignment of sequences upstream of CRISPR arrays and ribosomal RNA genes in Aquifex aeolicus. The known boxA sequence upstream of ribosomal RNA genes is underlined and in red text. Yellow shading indicates identical bases.
Conservation of nusB across the bacterial kingdom.
(A) A conservation plot for nusB generated using the Aquerium tool (Adebali and Zhulin, 2017). Each circle represents a taxonomic rank (kingdom, phylum, class, order, family, genus, species), with lower level ranks being further from the center of the plot. The outer, blue circle indicates species that have nusB. Raw data are shown in Supplementary file 3.
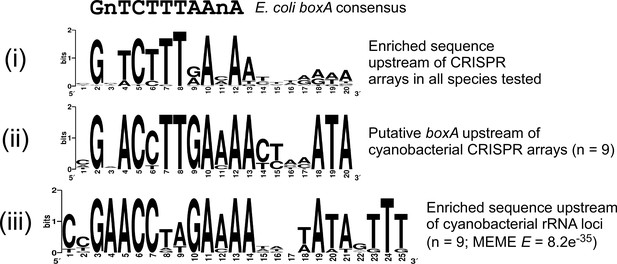
Analysis of putative boxA sequences upstream of cyanobacterial CRISPR arrays and ribosomal RNA loci.
Comparison of sequence motifs for putative boxA sequences from (i) the set of CRISPR array upstream sequences from 187 bacterial species (also shown in Figure 7A), (ii) the subset of (i) from nine cyanobacterial species, and (iii) ribosomal RNA upstream sequences for the nine species analyzed in (ii). The E-value determined by MEME is shown for the motif derived from cyanobacterial ribosomal RNA upstream sequences. The E. coli boxA consensus sequence is shown above the sequence logos.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Salmonella Typhimurium) | 14028s | DOI:10.1128/JB.01233–09 | ||
| Strain (Salmonella Typhimurium) | AMD678 | This study | 14028s PKAB-TG::Δcas3, FLAG3-cas5, PKAB-TG::CRISPR-II | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD679 | This study | AMD678 ΔCRISPR-I::thyA | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD684 | This study | AMD678 CRISPR-I boxA(C4A) | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD685 | This study | AMD678 CRISPR-II boxA(C4A) | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD698 | This study | AMD678 nusE(N3H) | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD710 | This study | 14028s PKAB-TG::Δcas3, FLAG3-cas5, suhB-TAP, PJ23119-thyA::CRISPR-II | Supplementary file 6 |
| Strain (Salmonella Typhimurium) | AMD711 | This study | AMD710 CRISPR-II boxA(C4A) | Supplementary file 6 |
| Strain (Vibrio cholerae) | KS1234 | DOI: 10.1128/JB.00747–15 | ||
| Strain (Vibrio cholerae) | KS2094 | This study | KS1234 Δcas1 | Supplementary file 6 |
| Strain (Vibrio cholerae) | KS2206 | This study | KS1234 Δcas1 boxAmut (C4A, T5G) | Supplementary file 6 |
| Strain (Vibrio cholerae) | BJO185 | DOI: 10.1128/JB.00747–15 | ||
| Strain (Escherichia coli) | S17 | DOI: 10.1016/0378-1119(90)90147-J | ||
| Antibody | Anti-E. coli RNA Polymerase β Antibody (Mouse monoclonal) | BioLegend | Catalog # 663903 | ChIP (1 µL) |
| Antibody | Monoclonal anti-FLAG M2 antibody (Mouse monoclonal) | SIGMA | Catalog # F1804 | ChIP-seq (2 µL) |
| Recombinant DNA reagent | pBAD24-amp (plasmid) | DOI: 10.1128/jb.177.14.4121–4130.1995 | ||
| Recombinant DNA reagent | pAMD239 (plasmid) | This study | pBAD24-nusE | |
| Recombinant DNA reagent | pGEM-T (plasmid) | Promega, Catalog # A1360 | ||
| Recombinant DNA reagent | pVS030 (plasmid) | This study | pGEM-T-TAP-thyA-TAP | |
| Recombinant DNA reagent | pJTW064 (plasmid) | DOI: 10.1128/JB.01007–13 | ||
| Recombinant DNA reagent | pGB231 (plasmid) | This study | pJTW064 with CRISPR-II upstream region and up to spacer 2 | Supplementary file 6 |
| Recombinant DNA reagent | pGB237 (plasmid) | This study | pJTW064 with CRISPR-II upstream region and up to spacer 2, boxA(C4A) | Supplementary file 6 |
| Recombinant DNA reagent | pGB250 (plasmid) | This study | pJTW064 with CRISPR-II upstream region and up to spacer 11 | Supplementary file 6 |
| Recombinant DNA reagent | pGB256 (plasmid) | This study | pJTW064 with CRISPR-II upstream region and up to spacer 11, boxA(C4A) | Supplementary file 6 |
| Recombinant DNA reagent | p958 (plasmid) | DOI: 10.1128/JB.00747–15 | ||
| Recombinant DNA reagent | pAMD244 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 1 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD245 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 2 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD246 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 3 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD247 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 4 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD248 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 5 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD249 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 6 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD250 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 7 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD251 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 8 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD252 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 9 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD253 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 11 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD254 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 12 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD255 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 13 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD256 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 14 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD257 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 15 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD258 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 16 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD259 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 17 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD260 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 19 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD262 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 20 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD263 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 21 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD264 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 22 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD265 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 23 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD266 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 24 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD267 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 25 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD268 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 10/26 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD269 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 27 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD270 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 28 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD271 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 29 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD272 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 30 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD273 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 31 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD274 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 32 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD275 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 33 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD276 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 34 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD277 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 35 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD278 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 36 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD279 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 37 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD280 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 38 | Supplementary file 6 |
| Recombinant DNA reagent | pAMD281 (plasmid) | This study | pKS958 with V. cholerae A50 protospacer 39 | Supplementary file 6 |
| Chemical compound, drug | Bicyclomycin | Santa Cruz Biotechnology, Inc | Catalog # sc-391755 | |
| Chemical compound | Protein A Sepharose CL-4B Medium | Cytiva (Formerly GE Healthcare Life Sciences) | Catalog # 17078001 | |
| Chemical compound | IgG Sepharose 6 Fast Flow Medium | Cytiva (Formerly GE Healthcare Life Sciences) | Catalog # 17096901 |
Additional files
-
Supplementary file 1
List of FLAG3-Cas5 ChIP-seq peaks.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp1-v2.xlsx
-
Supplementary file 2
Assignment of ChIP-seq peak centers to spacers, and relative Cas5 occupancy at each peak center.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp2-v2.xlsx
-
Supplementary file 3
Conservation analysis of nusB across the bacterial kingdom.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp3-v2.xlsx
-
Supplementary file 4
Sequences used for phylogenetic analysis of CRISPR array upstream regions.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp4-v2.xlsx
-
Supplementary file 5
Sequences used for phylogenetic analysis of CRISPR array upstream regions, with oppositely oriented sequences for species with just a type II-C CRISPR-Cas system.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp5-v2.xlsx
-
Supplementary file 6
List of strains and plasmids used in this study.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp6-v2.xlsx
-
Supplementary file 7
List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp7-v2.xlsx
-
Supplementary file 8
Custom python script to count Vibrio cholerae protospacers from a fastq file.
- https://cdn.elifesciences.org/articles/58182/elife-58182-supp8-v2.py.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58182/elife-58182-transrepform-v2.pdf





