Extrinsic activin signaling cooperates with an intrinsic temporal program to increase mushroom body neuronal diversity
Figures
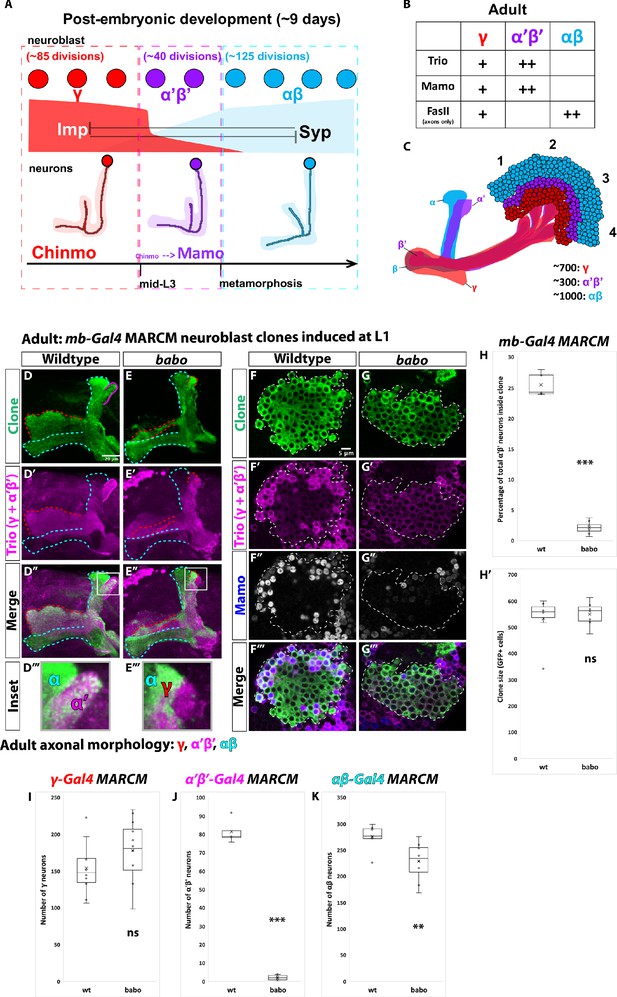
α’β’ neurons are not generated from babo mutant neuroblasts.
(A) Summary of intrinsic temporal patterning mechanism operating during mushroom body development. During early larval stages, mushroom body neuroblasts express high levels of Imp (red) and Chinmo (red) in neurons to specify γ identity for ~85 neuroblast divisions (red-dashed box). From mid-L3 to metamorphosis, when Imp and Syp (cyan) are both at low levels, the same neuroblast divides ~40 times to produce α’β’ neurons (magenta-dashed box). Low Chinmo regulates the expression Mamo, a terminal selector of α’β’ identity. From the beginning of metamorphosis throughout pupal development, high Syp leads to αβ neurons (cyan-dashed outline). (B) Known molecular markers can distinguish between the three mushroom body neuronal types in the adult. (C) Mushroom body projections originating from neurons born from four neuroblasts (numbered 1 to 4) per hemisphere fasciculate into a single bundle (peduncle) before branching into the five mushroom body lobes. The first-born γ neurons (red) remodel during development to project into a single, medial lobe in the adult. This lobe is the most anterior of the medial lobes. Axons from α’β’ neurons (magenta) bifurcate to project into the vertical and medial α’ and β’ lobes. The β’ lobe is posterior to the γ lobe. The last-born αβ neurons (cyan) also bifurcate their axons into the vertical projecting α lobe and medial projecting β lobe. The α lobe is positioned adjacent and medial to the α’ lobe. The β lobe is the most posterior medial lobe. (D-E) Representative max projections showing adult axons of clonally related neurons born from L1 stage in wildtype and babo conditions. UAS-CD8::GFP is driven by mb-Gal4 (OK107-Gal4). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. A white box outlines the Inset panel. Trio (magenta) is used to label all γ and α’β’ axons for comparison to GFP+ axons. (D) In wildtype, GFP+ axons (green, outlined in red, magenta and cyan) are visible in all observable mushroom body lobes. (E) In babo mutant clones, γ neurons (red outline) remain unpruned. GFP+ axons are missing inside the Trio+ α’ lobe, indicating the absence of α’β’ neurons. (F-G) Representative, single z-slices from the adult cell body region of clones induced at L1 in wildtype and babo conditions. UAS-CD8::GFP is driven by mb-Gal4. (F) Wildtype clones show the presence of strongly expressing Trio (magenta) and Mamo (blue, gray in single channel) neurons, indicative of α’β’ identity. (G) In babo mutant clones, cells strongly expressing Trio and Mamo are not present. (H) Quantification of MARCM clones marked by mb-Gal4, which labels all mushroom body neuronal types. The number of α’β’ neurons are quantified in wildtype (n = 7) and babo (n = 8) conditions. Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within clones, consistent with our expectation since each mushroom body is made from four neuroblasts. In babo clones, only 2.2 ± 0.4% of α’β’ neurons are within clones. (H’) There are no significant differences between the average clone sizes (wildtype:533.6 ± 33.3; babo:551.3 ± 17.6). (I) Quantification of γ neurons marked by γ-Gal4 (R71G10-Gal4) in MARCM clones. Plotted is the total number of γ neurons marked by GFP and Trio in wildtype (n = 10) and babo mutant (n = 12) clones. In wildtype, the average number of γ neurons is 154.3 ± 11.4. In babo mutants, the average is 178.4 ± 11.9. (J) Quantification of α’β’ neurons marked by α’β’-Gal4 (R41C07-Gal4) in MARCM clones. Plotted is the total number of α’β’ neurons marked by GFP and strong Trio in wildtype (n = 4) and babo mutant (n = 8) clones. In wildtype, the average number of α’β’ neurons is 81.5 ± 3.4. In babo mutants, the average is 2.1 ± 0.5. (K) Quantification of αβ neurons marked by αβ-Gal4 (R44E04-Gal4) in MARCM clones. Plotted is the total number of GFP+ cells in wildtype (n = 7) and babo mutant (n = 8) clones. In wildtype, the average number is 276 ± 9.1. In babo mutants, the average number is 228.9 ± 13.2. A two-sample, two-tailed t-test was performed. ***p<0.001, **p<0.01, ns: not significant. Scale bars: D, 20 µm; F, 5 µm.
-
Figure 1—source data 1
Neuron number counts for data presented in Figure 1A and Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig1-data1-v2.xlsx
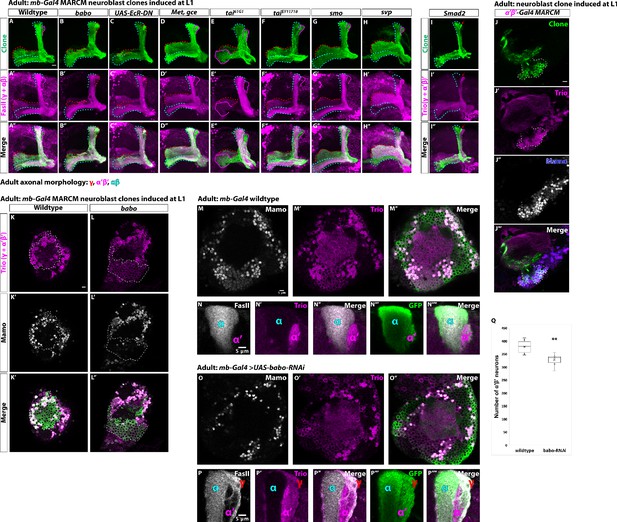
α’β’ neurons are lost from the adult neuropil in activin signaling mutant clones.
(A-H) Representative images of adult mushroom body lobes from MARCM screen in which clones were induced at L1 stage. Clonally related neurons are GFP+ (green). UAS-CD8::GFP is driven by mb-Gal4. All mushroom body axons, both clonal and non-clonal, are marked by FasII (γ and αβ axonal marker, magenta). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. (A) In wildtype clones, all three mushroom body neuronal types are present. α’β’ axons are GFP+ and FasII-. (B) In babo mutants, γ neurons do not remodel (red outline in vertical lobe). α’β’ neurons are also missing, as there are no GFP+, FasII- axons. (C) UAS-EcR-DN expressing clones are missing α’β’ axons and γ neurons do not remodel. (D-G) No loss of neuronal diversity is observed. (H) svp mutant clones (GFP+, green) contain all three mushroom body neuronal types. (I) Smad2 mutant clones (GFP, green) have unpruned γ axons (red arrow) and no α’β’ axons (Trio+, magenta). (J-J’’’) Representative adult wildtype clone labelled with R41C07-Gal4 (α’β’-Gal4). A single z-slice showing that strong Trio and strong Mamo label α’β’ neuronal cell bodies. (K-L) Representative, single z-slice showing strong Trio+ and Mamo+ cells inside and outside a GFP+ clone in wildtype (K) but not in babo (L) clones. Images represent the same sections as in Figure 1F–G. (M-M’’) Representative images showing the presence of α’β’ neurons based on strong Mamo (gray) and Trio (magenta) expression in adult mushroom body neurons labeled by mb-Gal4 driving UAS-CD8::GFP. (N-N’’’) Images highlighting the vertical mushroom body axons. α’ axons are Trio+ and GFP+. FasII (gray) labels α axons. (O-O’’) The majority of α’β’ neurons are still present following expression of UAS-babo-RNAi based on strong Mamo and Trio expression. (P-P’’’) The vertical lobes indicate that γ neurons do not remodel (FasII+/Trio+/GFP+) and that α’β’ neurons are still present (α’ lobe that is FasII-/Trio+/GFP+) following UAS-babo-RNAi expression. (Q) Quantification of the number of α’β’ neurons following expression of UAS-babo-RNAi (329 ± 10.4, n = 6) compared to wildtype controls (379 ± 11, n = 6). A two-sample, two-tailed t-test was performed. **p<0.01. Scale bars: A, 10 µm; J,K,M,N,P, 5 µm.
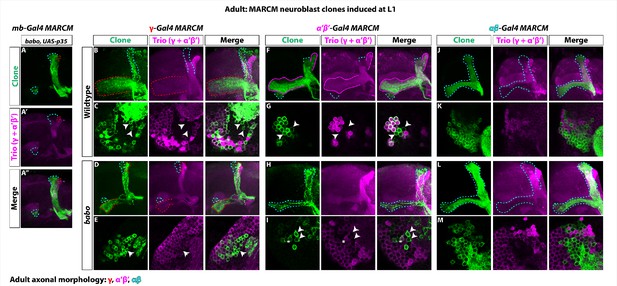
γ neuron numbers likely increase, while αβ numbers decrease, in babo mutant clones.
(A-A’’) The loss of α’β’ neurons in babo mutant clones marked by mb-Gal4 (GFP, green) is not rescued by blocking cell death. γ neurons (red outline) also retain their larval branching. (B-E) Representative wildtype and babo mutant clones made using R71G10-Gal4 (γ-Gal4) driving UAS-CD8::GFP. Note the presence of some GFP+ axons in the αβ lobes (cyan outline). Arrowheads in the cell body region point to αβ neurons based on the absence of Trio expression. Only cells that were both GFP+ and Trio+ were used for quantification. (F-I) Representative wildtype and babo mutant clones made using R41C07-Gal4 (α’β’-Gal4) driving UAS-CD8::GFP. Note the presence of some GFP+ axons in the αβ lobes (cyan outline). Arrowheads in the cell body region point to αβ neurons based on the absence of Trio expression. In babo mutants, the majority of neurons remaining are αβ (GFP+ and Trio-). The ‘*” indicates the presence of an α’β’ neuron cell body based on strong Trio expression. Only cells that were both GFP+ and Trio+ were used for quantification. (J-M) Representative wildtype and babo mutant clones made using R44E04-Gal4 (αβ-Gal4) driving UAS-CD8::GFP.
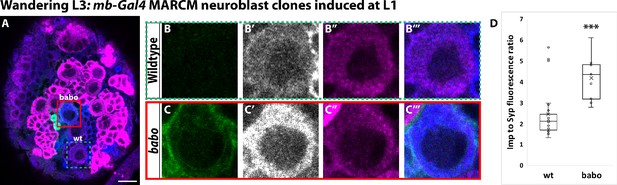
Activin signaling acts in neuroblasts to lower Imp levels.
(A) Representative image of a babo mushroom body neuroblast marked by UAS-CD8::GFP driven by mb-Gal4 (red box) adjacent to a wildtype neuroblast (green-dashed box) in the same focal plane from a wandering L3 stage brain, immunostained for Imp (blue, gray in single channel) and Syp (magenta). (B) Close-up view of wildtype neuroblast (green-dashed box in A). (C) Close up view of babo mutant neuroblast (red box in A). (D) Quantification of the Imp to Syp ratio in babo neuroblasts (4.2 ± 0.4, n = 9 from 4 different brains) compared to wildtype (2.4 ± 0.3, n = 23 from the same 4 brains as babo neuroblasts). A two-sample, two-tailed t-test was performed. ***p<0.001, ns: not significant. Scale bar: 10 µm.
-
Figure 2—source data 1
Imp and Syp fluorescence quantification in babo mutant clones.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig2-data1-v2.xlsx
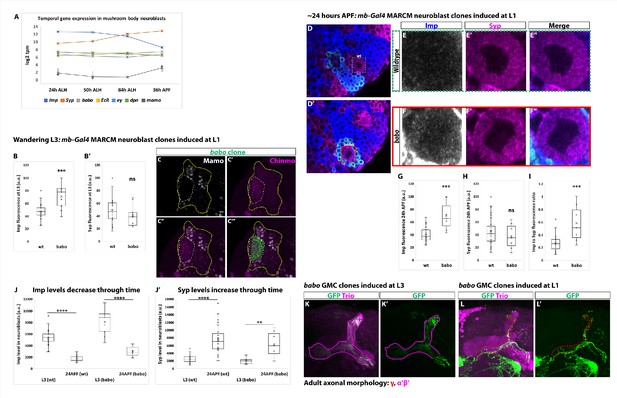
Activin signaling lowers Imp levels but the Imp to Syp transition does not depend on activin signaling.
(A) log2 transcripts per million (tpm) of selected factors expressed in mushroom body neuroblasts through time. babo (gray) is not differentially expressed through time like Imp (blue) and Syp (orange). babo is expressed at similar levels to other important factors in mushroom body neuroblasts (i.e., eyeless (ey) and deadpan (dpn)). mamo served as a control since this gene is known not to be expressed in mushroom body neuroblasts. (B-B’) Arbitrary fluorescent intensity values (scaled to 100) of Imp and Syp in wildtype (n = 23) and babo (n = 9) neuroblasts at wandering L3. The Imp level is significantly higher in babo neuroblasts compared to wildtype neuroblasts. Syp intensity values are not significantly different. (C-C’’’) At wandering L3, babo mutant clones marked by UAS-CD8::GFP (green, white-dashed line) driven by mb-Gal4 do not express Mamo. Mushroom body cells outside the clone (marked by Chinmo (magenta), yellow line) do express Mamo. In addition, Chinmo levels appear higher in babo mutant neurons (n = 3). Scale bar: 5 µm. (D-D’) Representative image of a babo neuroblast ~24 hr After Puparium Formation (APF) marked by UAS-CD8::GFP driven by mb-Gal4 (red box) ventral to a wildtype neuroblast (green-dashed box) immunostained for Imp (blue, gray in single channel) and Syp (magenta). (E-F) Close-up view of wildtype (E) and babo (F) neuroblasts from (D-D’). (G-H) Arbitrary fluorescent intensity values (scaled to 100) of Imp (G) and Syp (H) in wildtype (n = 27 neuroblasts from 6 brains) and babo (n = 7 neuroblasts from the same 6 brains as babo) neuroblasts. Imp but not Syp values are significantly different. (I) Quantification of the Imp to Syp ratio in babo neuroblasts compared to wildtype ~24 hr APF. babo neuroblasts have a significantly higher Imp to Syp ratio. (J) Comparison of Imp fluorescent values in wildtype (wt) and babo mutant (babo) neuroblasts at L3 versus ~24 hr APF. Imp levels decrease through time independent of whether neuroblasts are wildtype or babo mutant. (J’) Comparison of Syp fluorescent values in wildtype (wt) and babo mutant (babo) neuroblasts at L3 versus ~24 hr APF. Syp levels increase through time independent of whether neuroblasts are wildtype or babo mutant. (K-K’) Neurons in the adult neuropil born from babo GMC clones induced at L3 stage and marked by mb-Gal4 driving UAS-CD8::GFP (GFP, green). These neurons project axons into the Trio labeled α’β’ lobes (magenta) (n = 34/34). (L-L’) In contrast, babo GMC clones induced at L1 show that γ neurons do not remodel in the adult neuropil, as expected (n = 8/10). In all cases, a two-sample, two-tailed t-test was performed. ****p<0.0001, ***p<0.001, **p<0.01, ns: not significant.
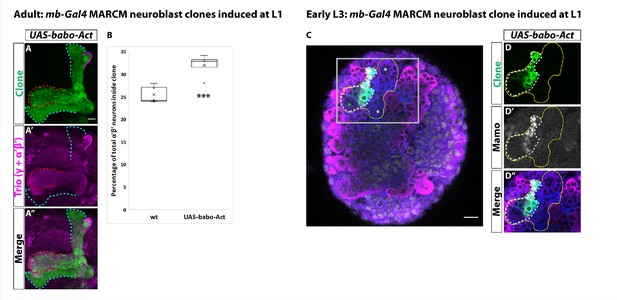
Activin signaling is sufficient to expand production of α’β’ neurons.
(A) Expression of UAS-Babo-Act by mb-Gal4 leads to additional α’β’ neurons but does not convert all mushroom body neurons into this fate. (B) Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. The number of α’β’ neurons is quantified in wildtype (n = 7, replotted from data in Figure 1H) and UAS-babo-Act (n = 4). In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within a clone while precociously activating the activin pathway increased the percentage to 32 ± 1.4%. (C) A representative image of an early L3 brain in which a single mushroom body neuroblast is expressing UAS-babo-Act driven by mb-Gal4 (white-dashed line). Imp (blue) and Syp (magenta), along with GFP (green), are used to identify mushroom body neuroblasts (asterisks) and neurons. (D) Inset (gray box in C) showing that Mamo (gray) is expressed inside GFP+ cells that express UAS-babo-Act but not outside in adjacent wildtype mushroom body neurons (yellow line) (n = 3/3). A two-sample, two-tailed t-test was performed. ***p<0.001.
-
Figure 3—source data 1
Neuron number counts for data presented in Figure 3.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig3-data1-v2.xlsx
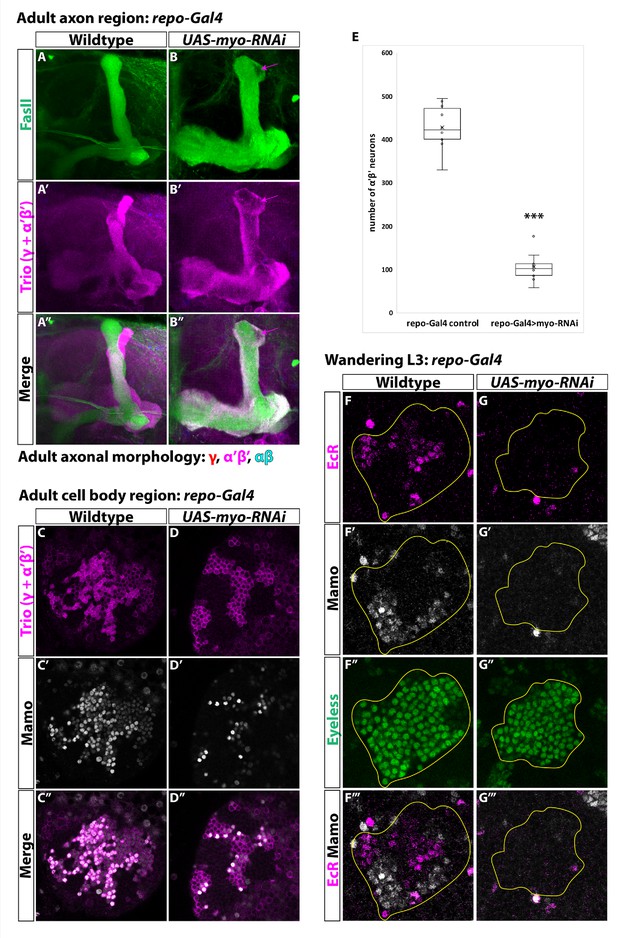
Glia are the source of the activin ligand Myo to specify α’β’ neurons.
(A-B) Representative images of adult mushroom body lobes labeled by FasII (green) and Trio (magenta). (A) In wildtype controls (428.9 ± 16.2, n = 10) (repo-Gal4 only) all three neuronal types are present based on axonal projections. (B) Expressing UAS-myo-RNAi (106.6 ± 11.4, n = 10) causes γ neurons not to remodel and to the loss of the majority of α’β’ neurons, however some still remain (purple arrow, FasII- region). (C-D) Representative images of adult mushroom body cell body region. Trio (magenta) and Mamo (gray) are used to distinguish between the three neuronal types. Expressing UAS-myo-RNAi leads to loss of the majority of strong Mamo+ and Trio+ cells, indicating the loss of α’β’ neurons. (E) Quantification of phenotypes presented in A-D. (F) At L3, EcR (magenta) and Mamo (gray) are expressed in mushroom body neurons labeled by Eyeless (green, yellow outline). Mamo+ cells are newborn α’β’ neurons. G. Expressing UAS-myo-RNAi with repo-Gal4 leads to loss of both Mamo and EcR in mushroom body neurons. A two-sample, two-tailed t-test was performed. ***p<0.001.
-
Figure 4—source data 1
Neuron number counts for data presented in Figure 4.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig4-data1-v2.xlsx
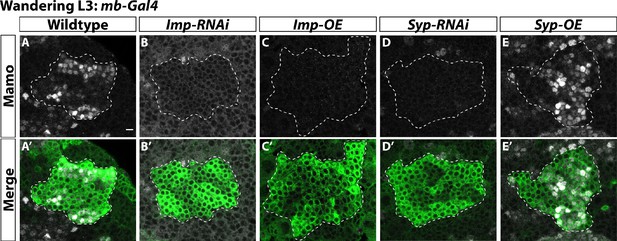
α’β’ neurons are specified by low Imp levels at L3.
(A-A’) Representative image of wildtype mushroom body neurons labeled by mb-Gal4 driving UAS-CD8::GFP (green, white-dashed outline) during the wandering L3 stage. Mamo (gray) is used as a marker for α’β’ neurons. (B-B’) When mb-Gal4 is used to drive UAS-Imp-RNAi, Mamo is not expressed. (C-C’) Similarly, Mamo expression is lost when overexpressing Imp (UAS-Imp-overexpression (OE)). (D-D’). Expressing UAS-Syp-RNAi also leads to the loss of Mamo. (E) Expressing UAS-Syp-overexpression (OE) does not affect Mamo. Scale bar: 5 µm.
-
Figure 5—source data 1
Neuron number counts for data presented in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig5-data1-v2.xlsx
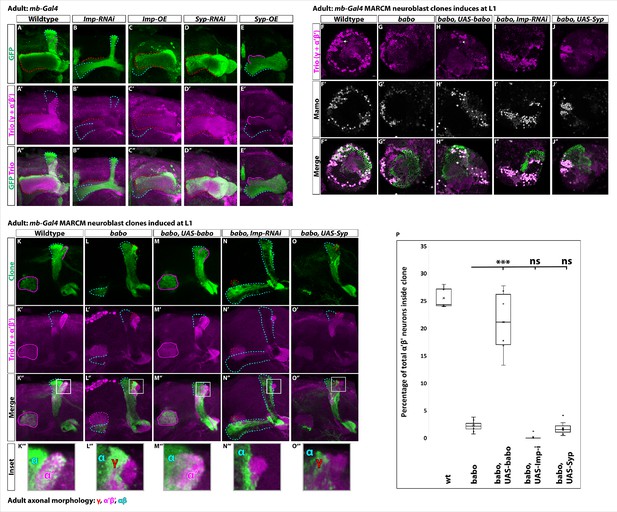
Low Imp levels are required for α’β’ specification.
(A-E) Representative images of adult mushroom body lobes labeled by mb-Gal4 driving UAS-CD8::GFP (green) in wildtype (A), UAS-Imp-RNAi (B), UAS-Imp-OE (C), UAS-Syp-RNAi (D) or UAS-Syp-OE (E). When visible, γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. Trio (magenta) labels γ and α’β’ axons. (A) In wildtype, all three axonal types are present. (B) With Imp-RNAi, the majority of γ axons (red outline) are lost and α’β’ axons are completely missing. (C) Imp-OE leads to mushroom body axons projecting almost entirely into the γ lobe (red outline). Some αβ axons (cyan outline) are present but are missing the vertical α projection. α’β’ axons are completely lost. (D) Syp-RNAi is similar but more severe than Imp-OE as mushroom body axons project entirely into the γ lobe (red outline). (E) Syp-OE does not abolish α’β’ axons (magenta outline). αβ axons (cyan outline) are also present. However, the vertical α’ and α lobes are both missing. (F-J) Representative images of single z slices. Strong Trio (magenta) labels α’β’ neurons while weak Trio labels γ neurons. Strong Mamo (gray) also labels α’β’ neurons while weak Mamo labels γ neurons. Arrows points to α’β’ neurons inside the clone. F. In wildtype clones driven by mb-Gal4, GFP+ cells (green) contain strong Trio+ and Mamo+ cells. (G-H) These cells are lost in babo clones (G) but rescued in clones expressing UAS-babo (H). (I-J) Expressing UAS-Imp-RNAi (I) or UAS-Syp (J) does not rescue the loss of α’β’ neurons. (K-O). Representative maximum-projection images of adult mushroom body lobes from mb-Gal4 MARCM clones induced at L1, focused on the α’ and β’ lobes. Clonally related neurons are GFP+ (green). All mushroom body axons, both clonal and non-clonal, are marked by Trio (magenta). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. A gray box outlines the Inset panel. (K) In wildtype, GFP+ axons are observed in the α’β’ lobes (magenta outline). (L) In babo mutant clones, γ neurons do not remodel (red outline) and α’β’ neurons are missing. (M) These phenotypes are rescued by expressing UAS-babo inside mutant clones, as GFP+ axons colocalize within the Trio labeled α’β’ lobes (magenta outline). (N-O) Neither reducing Imp with UAS-Imp-RNAi or decreasing the Imp to Syp ratio by expressing UAS-Syp rescues the loss of α’β’ neurons. (P) Quantification of MARCM clones represented in K-O. Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. The number of α’β’ neurons is quantified in wildtype (n = 7, replotted from data in Figure 1H), babo (n = 8, replotted from data in Figure 1H), babo, UAS-babo (n = 6), babo, UAS-imp-RNAi (n = 7), and babo, UAS-Syp (n = 7), In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within a clone while they only represent 2.2 ± 0.4% in babo clones. Expressing UAS-babo rescues to 21.1 ± 2.4%. In contrast, expression of UAS-Imp-RNAi (0.17 ± 0.17%) or UAS-Syp (1.8 ± 0.5%) is not statistically different from babo. Significance values were determined using a Tukey test. ***p<0.001, ns: not significant.
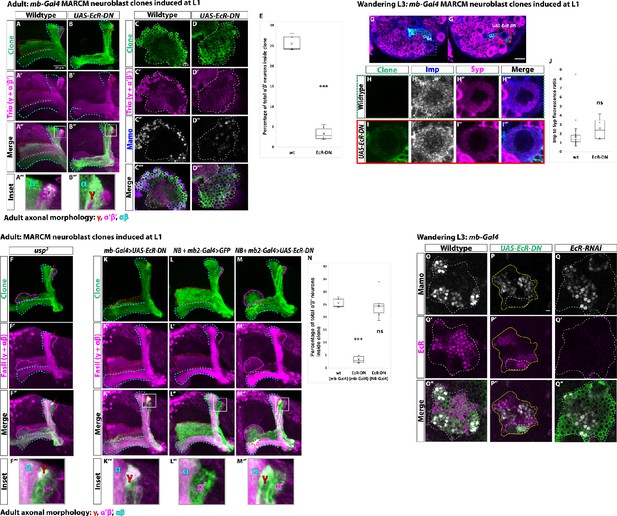
Ecdysone signaling is not necessary for α’β’ specification.
(A-B) Representative max projections showing adult axons of clonally related neurons born from L1 stage in wildtype and UAS-EcR-DN conditions. UAS-CD8::GFP is driven by mb-Gal4 (OK107-Gal4). Outlines mark GFP+ axons, where γ axons are outlined in red, α’β’ axons are outlined in magenta, and αβ axons are outlined in cyan. A white box outlines the Inset panel. Trio (magenta) is used to label all γ and α’β’ axons for comparison to GFP+ axons. (A) In wildtype, GFP+ axons are visible in all mushroom body lobes. (B) α’β’ axons are lost, and γ neurons do not remodel, in UAS-EcR-DN expressing clones. (C-D) Representative, single z-slices from the adult cell body region of clones induced at L1 in wildtype and UAS-EcR-DN conditions. UAS-CD8::GFP is driven by mb-Gal4. (C) Wildtype clones show the presence of strongly expressing Trio (magenta) and Mamo (blue, gray in single channel) neurons, indicative of α’β’ identity. (D) In UAS-EcR-DN clones, strong Trio and Mamo cells are not present. (E) Quantification of MARCM clones marked by mb-Gal4, which labels all mushroom body neuronal types. The number of α’β’ neurons are quantified in wildtype (n = 7, replotted from data in Figure 1H) and UAS-EcR-DN (n = 6) conditions. Plotted is the percentage of strong Mamo+ and GFP+ cells (clonal cells) versus all Mamo+ cells (clonal and non-clonal cells) within a single mushroom body. In wildtype, 25.5 ± 0.7% of the total strong Mamo expressing cells (α’β’ neurons) are within clones. In UAS-EcR-DN clones, only 3.4 ± 0.6% of α’β’ neurons are within clones. (F) usp mutant clones contain α’β’ neurons. FasII (magenta) is used to label γ and αβ lobes. Red arrow indicates unpruned γ neurons. (G) Representative image of an UAS-EcR-DN expressing neuroblast marked by UAS-CD8::GFP driven by mb-Gal4 (red box) ventral to a wildtype neuroblast (green-dashed box) from the same wandering L3 stage brain, immunostained for Imp (blue, gray in single channel) and Syp (magenta). (H) Close-up view of wildtype neuroblast (green-dashed box in G). (I) Close-up view of UAS-EcR-DN neuroblast (red box in G). (J) Quantification of the Imp to Syp ratio in UAS-EcR-DN neuroblasts (n = 4 from four different brains) compared to wildtype (n = 27 from the same four brains as UAS-EcR-DN neuroblasts). (K) A representative adult mushroom body clone (green) induced at L1 expressing UAS-EcR-DN driven by mb-Gal4. α’β’ neurons (GFP+ (green), FasII- (magenta)) are not observed and γ neurons do not remodel (GFP+, FasII+, red outline). (L) A representative adult wildtype clone induced at L1 driven by NB + mb2-Gal4. All three neuron types are present, including α’β’ neurons (GFP+, FasII-, magenta outline). (M) α’β’ neurons are also present when UAS-EcR-DN is driven by NB + mb2-Gal4 although γ neurons do not remodel. (N) Quantification of MARCM clones in which UAS-EcR-DN is driven by mb-Gal4 (n = 6, replotted from data in E) or NB + mb2-Gal4 (n = 6) compared to wildtype (n = 7, replotted from data in Figure 1H). In UAS-EcR-DN clones driven by NB + mb2-Gal4, 24.6 ± 2.1% of α’β’ neurons are within a clone, similar to wildtype. O. At L3, Mamo (gray) is expressed in young mushroom body neurons (α’β’) while EcR (magenta) can only be detected in more mature neurons (mainly γ at this stage). Note that there is no overlap between Mamo and EcR. (P) Expressing UAS-EcR-DN with mb-Gal4 (green, white outline) leads to the loss of Mamo expression (gray) inside the clone but not in surrounding wildtype mushroom body neurons. (Q) In contrast, expressing UAS-EcR-RNAi drivenE by mb-Gal4 abolishes EcR expression but does not affect Mamo. For E and J a two-sample, two-tailed t-test was performed. For N, a Tukey test was performed. ***p<0.001, ns: not significant. Scale bars: A, 20 µm; G, 10 µm; P, 5 µm.
-
Figure 6—source data 1
Neuron number counts for data presented in Figure 6 and Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Imp and Syp fluorescence quantification when expressing UAS-EcR-DN.
- https://cdn.elifesciences.org/articles/58880/elife-58880-fig6-data2-v2.xlsx
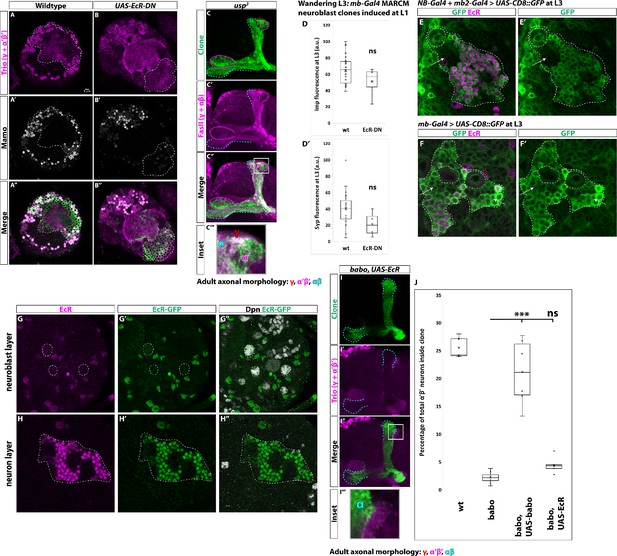
Ecdysone signaling is not necessary for α’β’ specification.
(A) Representative, single z-slice showing strong Trio+ and Mamo+ cells both inside and outside a GFP+ clone. (B) Expressing UAS-EcR-DN leads to the loss of α’β’ neurons. Images in A and B represent the same sections as in Figure 6C–D. (C) usp3 mutant clones contain α’β’ neurons, similar to usp2 mutant clones. (D-D’) Arbitrary fluorescent intensity values (scaled to 100) of Imp and Syp in wildtype (n = 27) and UAS-EcR-DN (n = 4) neuroblasts. Neither Imp nor Syp intensity values are different compared to wildtype. (E-E’) Insc-Gal4 + R13F02-Gal4 (NB-Gal4 + mb2-Gal4) driving expression of UAS-CD8::GFP at L3 stage. Strong GFP (green) is detected in mushroom body neuroblasts (white-dashed circle) but not in newborn neurons (arrows) positioned adjacent to mushroom body neuroblasts. GFP is strongly expressed in more distally positioned, mature neurons marked by EcR (magenta). (F-F’) OK107-Gal4 (mb-Gal4) driving expression of UAS-CD8::GFP at L3. GFP (green) is detected in mushroom body neuroblasts (white-dashed circles), newborn neurons (arrows), and mature neurons marked by EcR expression (magenta). (G-G’’) Mushroom body neuroblasts (white-dashed circles) marked by Dpn (gray) do not express EcR based on antibody staining (magenta) or an EcR-GFP (green) at the wandering L3 stage. (H-H’’) Mushroom body neurons (white-dashed outline) positioned ventrally to the mushroom body neuroblasts in G do express EcR but not in young neurons (black regions inside white-dashed line). (I) α’β’ neurons are not rescued in babo clones by expressing UAS-EcR. (J) Quantification of phenotype presented in (I) The number of α’β’ neurons is quantified in wildtype (replotted from data in Figure 1H), babo (replotted from data in Figure 1H), babo, UAS-babo (replotted from Figure 5—figure supplement 1), and babo, UAS-EcR (n = 8, 4.4 ± 0.4%). Significance values were determined using a Tukey test. ***p<0.001, ns: not significant.
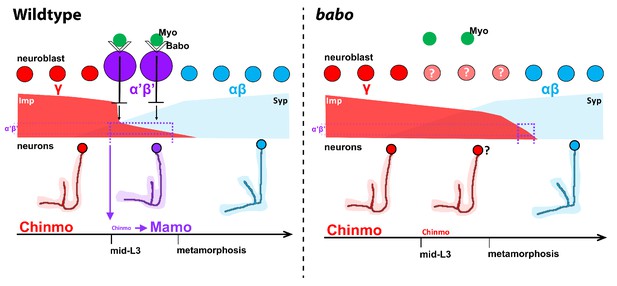
Model of how activin signaling defines the α’β’ temporal identity window.
In wildtype, as development proceeds, mushroom body neuroblasts incorporate an activin signal (Myo) from glia through Babo to lower the level of the intrinsic temporal factor Imp (magenta dashed line). The lower Imp levels inherited by newborn neurons leads to lower Chinmo levels to control the expression of the α’β’ effector Mamo, defining the mid-temporal window (magenta dashed lines). In babo mutants, Imp remains higher for longer, leading to the loss of Mamo (and likely many other targets) during mid-late L3 in neurons. In this model, γ neuron numbers increase, α’β’ neurons are lost, and fewer αβ neurons are produced. Nonetheless, the Imp to Syp transition still occurs, allowing for young (γ) and old (αβ) fates to be produced.

Representative images showing the presence of α’β’ neurons based on strong Mamo (gray) and Trio (magenta) expression in adult mushroom body neurons labeled by mb-Gal4 driving UAS-CD8::GFP.
N-N’’’. Images highlighting the vertical mushroom body axons. α’ axons are Trio+ and GFP+. FasII (gray) labels α axons. O-O’’. The majority of α’β’ neurons are still present following expression of UAS-babo-RNAi based on strong Mamo and Trio expression. P-P’’’. The vertical lobes indicate that γ neurons do not remodel (FasII+/Trio+/GFP+) and that α’β’neurons are still present (α’ lobe that is FasII-/Trio+/GFP+) following UAS-babo-RNAi expression. Q. Quantification of the number of α’β’ neurons following expression of UAS-babo-RNAi (n=6) compared to wildtype controls (n=6). A two-sample, two-tailed t-test was performed.





