A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies
Figures
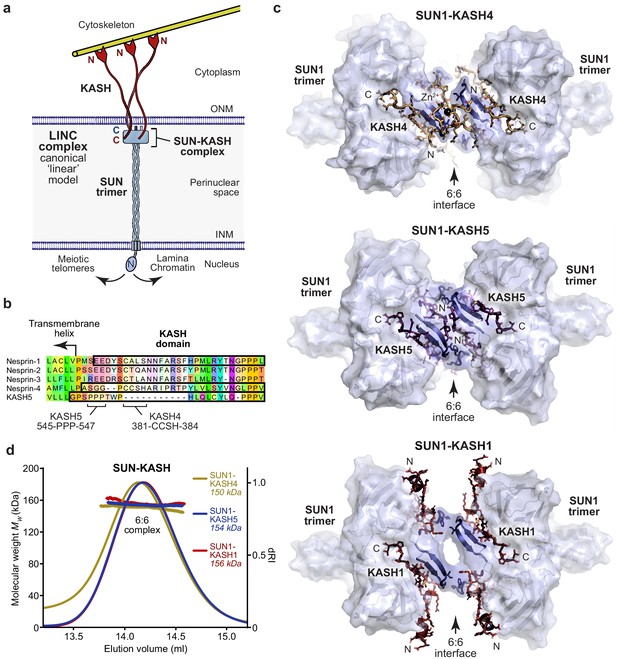
SUN-KASH complexes are 6:6 head-to-head assemblies.
(a) The Linker of Nucleoskeleton and Cytoskeleton (LINC) complex traverses the nuclear envelope to transmit forces between the cytoskeleton and nuclear components. The canonical model of the LINC complex is a linear structure formed of SUN and Nesprin proteins, which interact via a 3:3 complex between their SUN and KASH domains within the peri-nuclear space, and cross the inner and outer nuclear membranes (INM and ONM), respectively. (b) Sequence alignment of the KASH domains of human Nesprins 1–4 and KASH5. In this study, KASH1, KASH4, and KASH5 refer to the C-terminal KASH domains of Nesprin-1, Nesprin-4, and KASH5, respectively, which are highlighted (black outline), and key amino acids within KASH4 and KASH5 are indicated. (c) Crystal structures of human SUN1-KASH4 (top), SUN1-KASH5 (middle), and SUN1-KASH1 (bottom). The SUN1 molecular surface is displayed with SUN1 KASH-lids highlighted in blue as cartoons, and KASH sequences are represented as sticks (yellow, purple, and red, respectively). All structures are 6:6 complexes in which KASH proteins lie at the midline head-to-head interface between SUN1 trimers. (d) SEC-MALS analysis showing differential refractive index (dRI) profiles with fitted molecular weights (Mw) plotted as diamonds across elution peaks. SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 form 6:6 complexes in solution, with experimental molecular weights of 150, 154, and 156 kDa, respectively (theoretical 6:6 – 155, 155, and 157 kDa). Representative of more than three replicates using different protein preparations. Full elution profiles are shown in Figure 1—figure supplement 2.
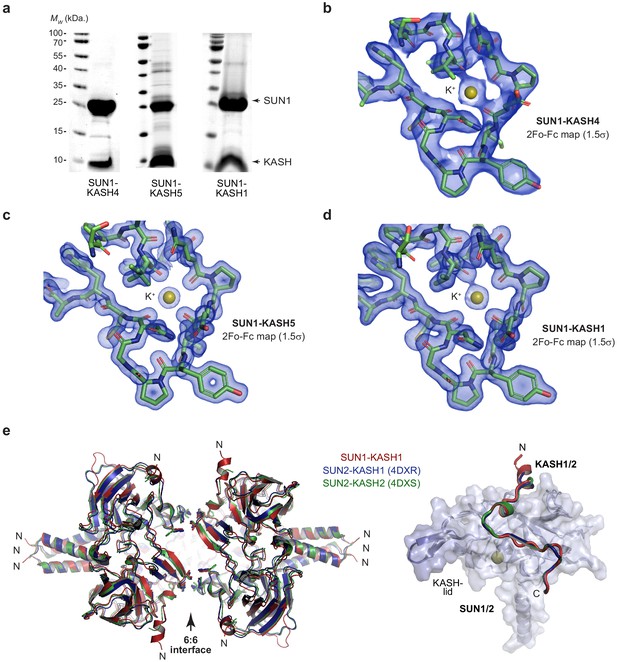
Crystal structures of SUN-KASH complexes.
(a) SDS-PAGE of purified SUN1-KASH4 (left), SUN1-KASH5 (middle) and SUN1-KASH1 (right) complexes used for crystallographic and biophysical experiments. (b–d) 2Fo-Fc electron density maps contoured at 1.5 σ of the same orientation of the SUN1 K+-binding site for (b) SUN1-KASH4, (c) SUN1-KASH5, and (d) SUN1-KASH1. (e) Superposition of SUN1-KASH1 (red) with SUN2-KASH1 (blue; PDB accession 4DXR) and SUN2-KASH2 (green; PDB accession 4DXS) (Sosa et al., 2012), showing the common 6:6 assembly that is present within their crystal lattices (left) and their constituent 1:1 protomers (right).
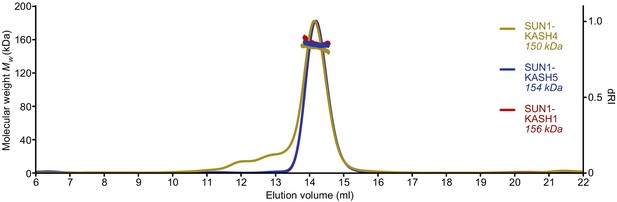
SUN-KASH complexes are 6:6 assemblies.
Full SEC-MALS elution profiles corresponding to the peaks shown in Figure 1d. SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 form 6:6 complexes in solution, with experimental molecular weights of 150, 154, and 156 kDa, respectively (theoretical 6:6 – 155, 155, and 157 kDa).
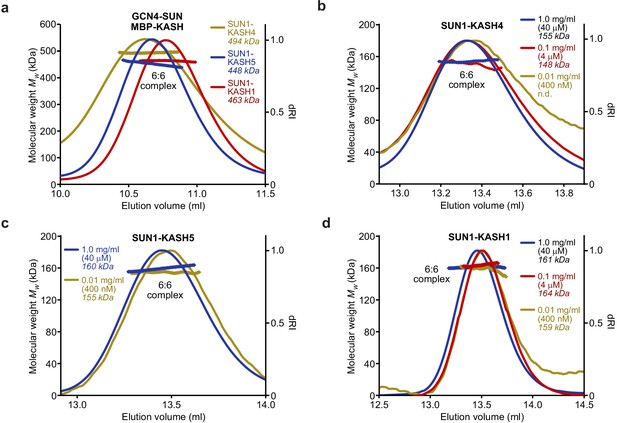
SUN-KASH 6:6 complexes are stable in solution.
(a–d) SEC-MALS analysis performed in 20 mM Tris pH 8.0, 150 mM KCl, 2 mM DTT. (a) GCN4-SUN1 and MBP-KASH form 6:6 complexes of 494 kDa (KASH4, yellow), 448 kDa (KASH5, blue), and 463 kDa (KASH1, red) (theoretical 6:6 – 464, 464, and 466 kDa). (b–d) Dilution series of SUN-KASH complexes analysed at 1.0 mg/ml (blue), 0.1 mg/ml (red), and 0.01 mg/ml (yellow) for (b) SUN1-KASH4 (theoretical 6:6 – 155 kDa), (c) SUN1-KASH5 (theoretical 6:6 – 155 kDa), and (d) SUN1-KASH1 (theoretical 6:6 – 157 kDa).
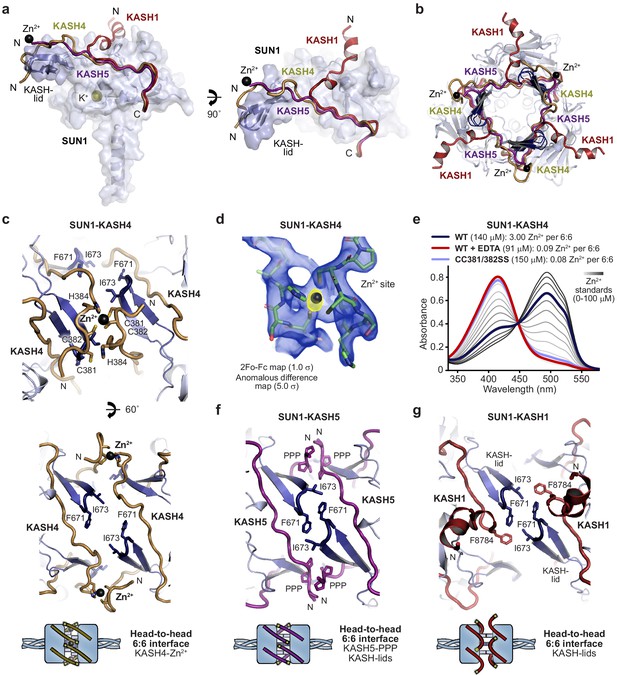
Specialised KASH sequences provide distinct SUN-KASH 6:6 assembly mechanisms.
(a) SUN-KASH 1:1 protomers from SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 crystal structures, superposed, and displayed as the SUN1 molecular surface with KASH-lids highlighted in blue as cartoons, and KASH sequences represented as cartoons (yellow, purple, and red, respectively). (b) Cross-section through the head-to-head interface of superposed SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 6:6 assemblies such that their constituent 3:3 complexes are visible. (c) Structural details of the SUN1-KASH4 6:6 interface, showing a zinc-binding site in which opposing KASH4 chains provide asymmetric ligands C381 and C382, and C382 and H384 (top), and the lack of interface-spanning interactions between opposing SUN1 KASH-lids (bottom). (d) 2Fo-Fc (blue) and anomalous difference (yellow) electron density maps contoured at 1.0 σ and 5.0 σ, respectively, at a zinc-binding site of SUN1-KASH4. (e) Spectrophotometric determination of zinc content for SUN1-KASH4 wild-type (dark blue; 3.00 Zn2+ per 6:6), wild-type with EDTA treatment prior to gel filtration (red; 0.09 Zn2+ per 6:6), and CC381/382SS (light blue; 0.08 Zn2+ per 6:6), using metallochromic indicator PAR, with zinc standards shown in a gradient from light to dark grey (0–100 µM). Representative of three replicates. (f) Structural details of the SUN1-KASH5 6:6 interface, demonstrating interface-spanning interactions between PPP-motifs (amino acids 545-PPP-547) of opposing KASH5 chains, and between amino acids F671 and I673 of opposing SUN1 KASH-lids. (g) Structural details of the SUN1-KASH1 6:6 interface showing interactions between amino acids F671 and I673 of opposing SUN1 KASH-lids that are supported by KASH1 amino acid F8784, but with no interface-spanning interactions between opposing KASH1 chains.
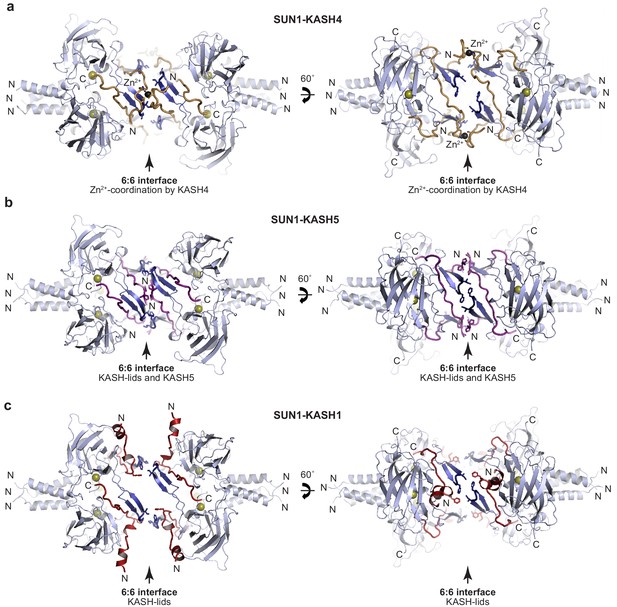
SUN-KASH crystal structures.
(a–c) Crystal structures of (a) SUN1-KASH4, (b) SUN1-KASH5, and (c) SUN1-KASH1 shown in cartoon representation with SUN1 KASH-lids highlighted in blue and KASH4, KASH5, and KASH1 shown in yellow, purple, and red, respectively.
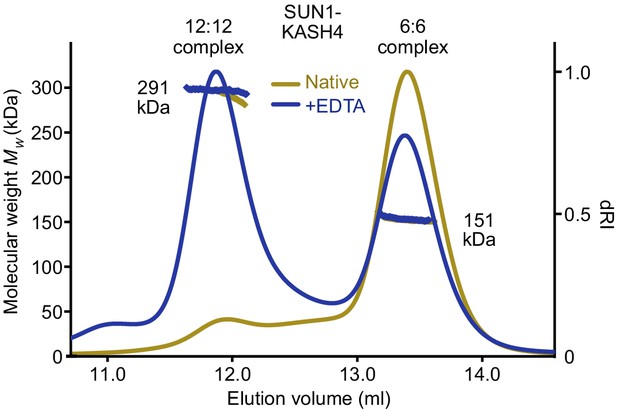
SUN1-KASH4 forms 6:6 and 12:12 complexes upon sequestration of bound zinc SEC-MALS analysis of SUN1-KASH4 (yellow) and following the removal of bound zinc (demonstrated in Figure 3e) by treatment with EDTA (blue).
Following EDTA treatment, SUN1-KASH4 forms two distinct species corresponding to the 6:6 complex of the native protein (151 kDa; theoretical – 150 kDa) and an additional 12:12 complex (291 kDa; theoretical – 300 kDa), in a ratio of 2:3 (by mass).
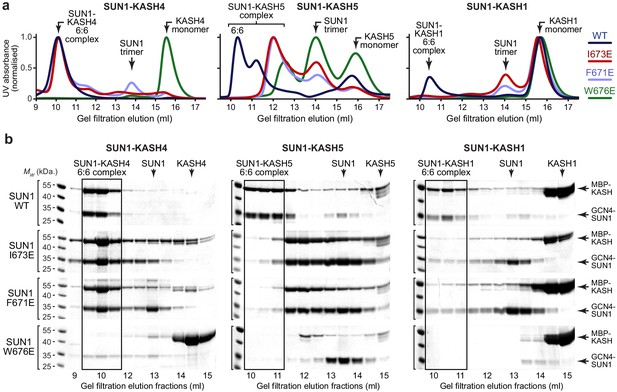
SUN1 KASH-lid residues involved in 6:6 assembly are essential for KASH1-binding.
(a,b) Gel filtration analysis. GCN4-SUN1 and MBP-KASH proteins were co-expressed and purified by amylose affinity (utilising non-specific binding by SUN1 for non-interacting mutants) and ion exchange (Figure 4—figure supplement 1d), and all fractions containing SUN-KASH complexes and dissociated proteins were concentrated and loaded onto an analytical gel filtration column. The elution profiles were validating by SEC-MALS in which wild-type fusion complexes and dissociated GCN4-SUN1 and MBP-KASH1 proteins were found to be 6:6 complexes, trimers and monomers, respectively (Figure 4—figure supplement 1f). (a) Gel filtration chromatograms (UV absorbance at 280 nm) across elution profiles for SUN1 wild-type (WT; dark blue), I673E (red), F671E (light blue), and W676E (green), with KASH4 (left), KASH5 (middle), and KASH1 (right), and (b) SDS-PAGE of their corresponding elution fractions. Representative of three replicates using different protein preparations. Source data are provided in Figure 4—source data 1.
-
Figure 4—source data 1
Uncropped gel images relating to Figure 4b.
- https://cdn.elifesciences.org/articles/60175/elife-60175-fig4-data1-v2.xlsx
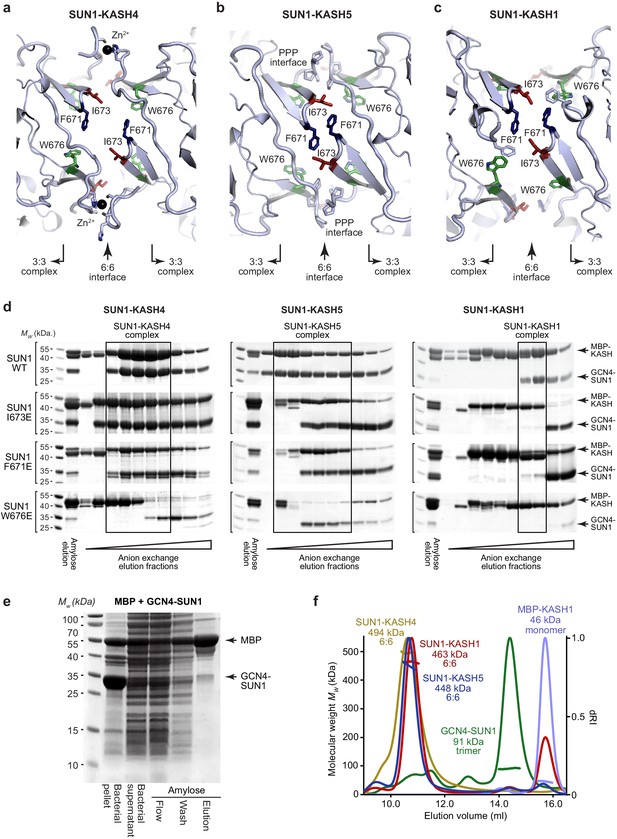
SUN-KASH complex formation upon SUN1 KASH-lid mutagenesis.
(a–c) Structural details of the 6:6 interface of (a) SUN1-KASH4, (b) SUN1-KASH5, and (c) SUN1-KASH1, highlighting SUN1 KASH-lid residues I673 (red), F671 (blue), and W676 (green). (d) SDS-PAGE of anion exchange elution profiles of GCN4-SUN1 and MBP-KASH proteins that were co-expressed and purified by amylose affinity (utilising non-specific binding by SUN1 for non-interacting mutants), comprising SUN1 wild-type, I673E, F671E, and W676E, with KASH4 (left), KASH5 (middle), and KASH1 (right). All fractions containing SUN-KASH complexes or dissociated proteins were pooled, concentrated and loaded onto an analytical gel filtration column for the analyses shown in Figure 4a,b. (e) Amylose pulldown following co-expression of MBP and GCN4-SUN1, showing that GCN4-SUN1 binds non-specifically to amylose resin. (f) SEC-MALS analysis demonstrating that SUN1-KASH1/4/5 fusion complexes, GCN4-SUN1 and MBP-KASH1 are 6:6 complexes, trimers and monomers, respectively (theoretical masses – 466, 464, 464, 88, and 48 kDa). This analysis provides validation for the gel filtration elution profiles shown in Figure 4a,b.
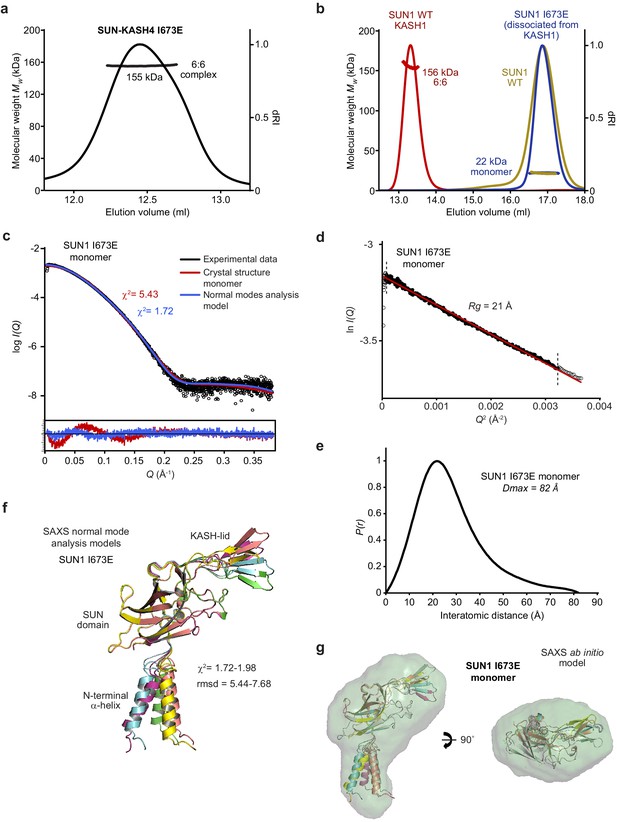
Biophysical analysis of the SUN1 I673E mutant.
(a) SEC-MALS analysis demonstrating that SUN1 I673E and KASH4 form a 155 kDa 6:6 complex (theoretical 6:6 – 155 kDa). (b) SEC-MALS analysis demonstrating that isolated SUN1 wild-type (WT; yellow), and SUN1 I673E that has dissociated following co-expression with KASH1 (blue), are 22 kDa monomers (theoretical – 23 kDa). The wild-type SUN1-KASH1 6:6 complex is shown in red for comparison. (c–g) SAXS analysis of the SUN1 I673E monomer. (c) SAXS scattering curve overlaid with theoretical scattering curves of a SUN1 protomer from the SUN1-KASH1 crystal structure (red; χ2 = 5.43) and its normal mode analysis model (blue; χ2 = 1.72). (d) SAXS Guinier analysis revealing a radius of gyration (Rg) of 21 Å; the linear fit is shown in black and demarcated by dashed vertical lines (Q.Rg values were <1.3). (e) SAXS P(r) distribution showing a maximum dimension of 82 Å. (f) SAXS normal mode analysis in which the five highest scoring models are displayed, based on their fit to the experimental SAXS data (χ2 = 1.72–1.98), and demonstrate hinge-like motion of the N-terminal α-helix relative to the SUN domain. (g) SAXS ab initio model (NSD = 0.645; reference model χ2 = 1.85) into which the normal mode analysis models are docked.
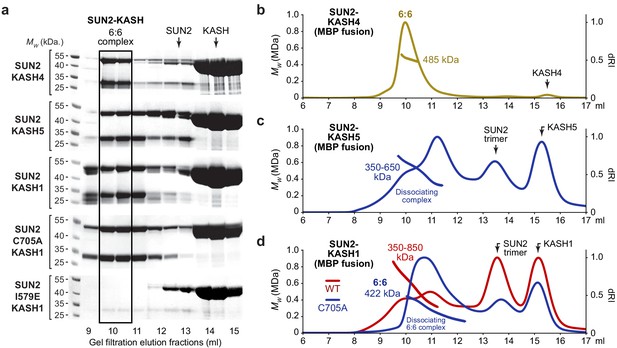
SUN2 forms 6:6 and higher molecular weight SUN-KASH complexes through head-to-head assembly.
(a) Gel filtration analysis shown as SDS-PAGE of elution fractions. GCN4-SUN2 (wild-type, C705A and I579E) and MBP-KASH proteins were co-expressed and purified by amylose affinity (utilising non-specific binding by SUN2 for non-interacting mutants) and ion exchange, and all fractions containing SUN-KASH complexes and dissociated proteins were concentrated and loaded onto an analytical gel filtration column. Source data are provided in Figure 5—source data 1. (b–d) SEC-MALS analysis of SUN2-KASH (MBP fusion) complexes following gel filtration elution (a). (b) SUN2-KASH4 is a 6:6 complex of 485 kDa (theoretical – 463 kDa). (c) SUN2-KASH5 forms a range of molecular species of at least 350–650 kDa, suggesting dissociation across the elution profile of 6:6 and larger complexes (theoretical 3:3 and 6:6–232 kDa and 463 kDa). (d) SUN2-KASH1 wild-type (red) forms a range of molecular species of at least 350–850 kDa, whilst the SUN2 C705A mutation (blue) stabilises a 6:6 complex of 422 kDa, suggesting dissociation across the elution profile of 6:6 and larger complexes (theoretical 3:3 and 6:6–232 kDa and 465 kDa).
-
Figure 5—source data 1
Uncropped gel images relating to Figure 5a.
- https://cdn.elifesciences.org/articles/60175/elife-60175-fig5-data1-v2.xlsx
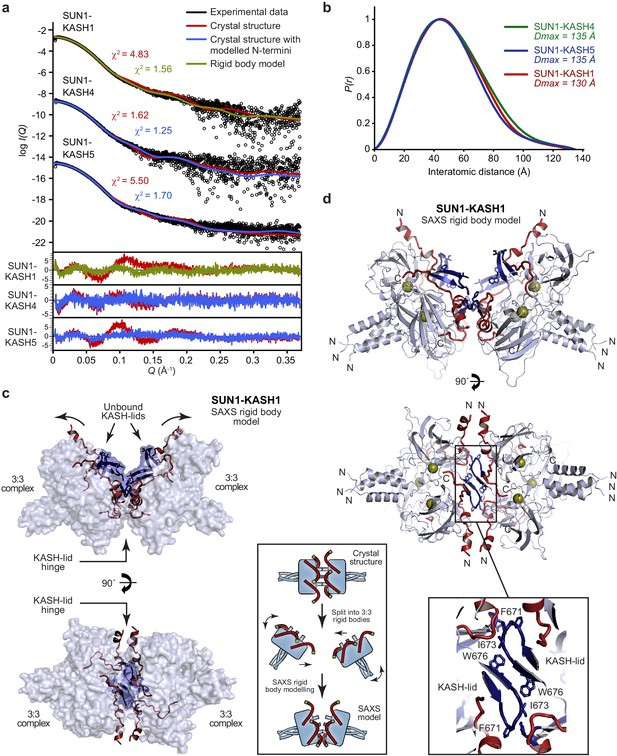
SEC-SAXS analysis of SUN-KASH 6:6 complexes.
(a) SAXS scattering curves of SUN1-KASH4, SUN1-KASH5, and SUN1-KASH1 overlaid with theoretical scattering curves of their crystal structures (red), crystal structures with KASH flexible N-termini modelled by CORAL (blue) and rigid body model of two 3:3 complexes (green). Residuals for each fit are shown (inset). Representative of more than three replicates using different protein preparations. (b) SAXS P(r) distributions showing maximum dimensions of 135 Å, 135 Å, and 130 Å, respectively. (c–d) SAXS rigid body model of SUN1-KASH1 shown as (c) surface and (d) cartoon representation, in which two constituent 3:3 complexes from its crystal structure were assigned as rigid bodies, with the 6:6 assembly generated by fitting to experimental SAXS data of solution SUN1-KASH1 (χ2 = 1.56). The inlet schematic illustrates the SAXS rigid body modelling procedure in which the crystal structure was split into its constituent 3:3 complexes, which were rotated as rigid bodies in three dimensions and allowed to interact, whilst fitted against experimental SAXS data. (d) The cartoon representation highlights structural details of the predicted KASH-lid interface, including the presence of unbound KASH-lids, and the close approximation of opposing KASH-lids, which achieve an asymmetric positioning of the N-termini of KASH domains in locations and orientations compatible with their upstream sequences crossing the outer nuclear membrane.
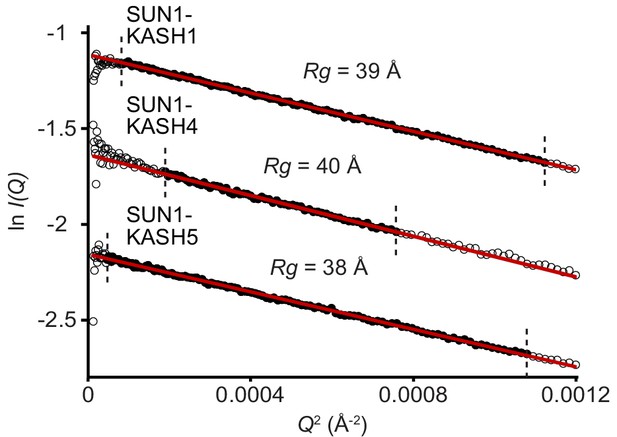
SAXS analysis of SUN-KASH complexes.
SAXS Guinier analysis revealing radius of gyration (Rg) values of 38, 40, and 39 Å, respectively; the linear fits are shown in black and demarcated by dashed vertical lines (Q.Rg values were <1.3).
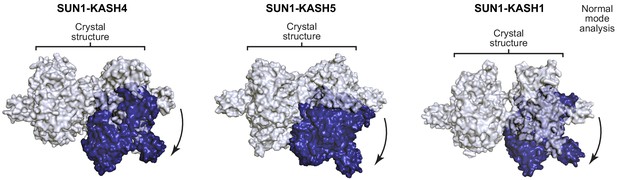
Hinge-link conformational flexibility within SUN-KASH 6:6 assemblies.
Normal mode analysis of SUN-KASH complexes in which non-linear normal modes calculated by the NOLB algorithm are shown as the largest amplitude of motion of one constituent 3:3 complex (blue) relative to its original position and its stationary opposing 3:3 complex within the crystal structure (grey) for SUN1-KASH4 (left), SUN1-KASH5 (middle), and SUN1-KASH1 (right).

The Linker of Nucleoskeleton and Cytoskeleton (LINC) complex as a branched network of SUN-KASH assemblies.
Model of the LINC complex as a branched network in which SUN-KASH 6:6 complexes act as nodes for force integration and distribution between two SUN trimers and three KASH dimers, which can bind to spatially separated and distinct nuclear and cytoskeletal components, respectively. This model enables cooperation between adjacent molecules within a LINC complex network to facilitate the transduction of large and coordinated forces across the nuclear envelope.
Tables
Data collection, phasing, and refinement statistics.
| Sun1-kash4 | Sun1-kash5 | Sun1-kash1 | |
|---|---|---|---|
| PDB accession | 6R16 | 6R2I | 6R15 |
| Data collection | |||
| Space group | P212121 | P6322 | P6322 |
| Cell dimensions | |||
| a, b, c (Å) | 104.37, 117.21, 138.42 | 80.16, 80.16, 177.62 | 80.45, 80.45, 182.55 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Wavelength (Å) | 0.9795 | 0.9282 | 0.9282 |
| Resolution (Å) | 48.83–2.75 (2.85–2.75)* | 88.81–1.54 (1.57–1.54)* | 65.09–1.82 (1.87–1.82)* |
| Rmeas | 0.111 (1.355) | 0.070 (1.551) | 0.085 (2.192) |
| Rpim | 0.056 (0.741) | 0.015 (0.329) | 0.019 (0.465) |
| Completeness (%) | 99.7 (97.5) | 97.5 (100.0) | 100.0 (100.0) |
| I/σ(I) | 15.4 (1.4) | 23.5 (2.2) | 21.5 (1.7) |
| CC1/2 | 0.999 (0.488) | 1.000 (0.801) | 1.000 (0.776) |
| Multiplicity | 7.1 (5.6) | 21.3 (22.1) | 20.6 (21.9) |
| Refinement | |||
| Resolution (Å) | 47.67–2.75 | 23.99–1.54 | 65.09–1.82 |
| No. reflections | 44658 | 49372 | 32230 |
| Rwork / Rfree | 0.2190/0.2549 | 0.1495/0.1683 | 0.1587/0.1817 |
| Cruickshank DPI (Å) | 0.25 | 0.06 | 0.06 |
| No. atoms | 10562 | 2127 | 2107 |
| Protein | 10451 | 1817 | 1845 |
| Ligand/ion | 21 | 1 | 26 |
| Water | 90 | 309 | 236 |
| B factors | 80.64 | 36.45 | 48.87 |
| Protein | 80.87 | 35.09 | 47.73 |
| Ligand/ion | 68.03 | 18.87 | 119.12 |
| Water | 56.37 | 44.50 | 50.06 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.002 | 0.011 | 0.013 |
| Bond angles (°) | 0.444 | 1.076 | 0.995 |
-
* Values in parentheses are for highest-resolution shell.
Summary of SEC-SAXS data.
| SUN1 I673E (monomer) | Sun1-kash4 (6:6) | Sun1-kash5 (6:6) | Sun1-kash1 (6:6) | |
|---|---|---|---|---|
| SASDBD accession | SASDJF5 | SASDJC5 | SASDJD5 | SASDJE5 |
| Guinier analysis | ||||
| I(0) (cm−1) | 0.042 | 0.045 | 0.100 | 0.130 |
| Rg (Å) | 21 | 40 | 38 | 39 |
| qmin (Å−1) | 0.0080 | 0.0014 | 0.0070 | 0.0090 |
| P(r) analysis | ||||
| I(0) (cm−1) | 0.042 | 0.045 | 0.102 | 0.132 |
| Rg (Å) | 22 | 40 | 39 | 39 |
| Dmax (Å) | 82 | 135 | 135 | 130 |
| Porod volume (Å3) | 39,367 | 292,301 | 274,824 | 303,602 |
| MW from Porod volume (kDa) | 23 | 172 | 162 | 179 |
| VC (Å2) | 238 | 825 | 784 | 853 |
| MW from VC (kDa) | 22 | 139 | 131 | 152 |
| DAMMIF ab initio modelling (30 models) | ||||
| Symmetry | P1 | N/A | N/A | N/A |
| NSD mean | 0.645 | N/A | N/A | N/A |
| χ2 (reference model) | 1.85 | N/A | N/A | N/A |
| Structural modelling | ||||
| CRYSOL - crystal structure (χ2) | 5.43 | 1.62 | 5.50 | 4.83 |
| CORAL - modelling of N-termini (χ2) | N/A | 1.25 | 1.70 | 4.55 |
| CORAL - rigid body modelling (χ2) | N/A | N/A | N/A | 1.56 |
| SREFLEX - normal mode analysis (χ2) | 1.72–1.98 | N/A | N/A | N/A |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | SUN1 | GeneArt | O94901 | |
| Gene (Homo sapiens) | SUN2 | GeneArt | Q9UH99 | |
| Gene (Homo sapiens) | Nesprin-1 | GeneArt | Q8NF91 | |
| Gene (Homo sapiens) | Nesprin-4 | GeneArt | Q8N205 | |
| Gene (Homo sapiens) | KASH4 | GeneArt | Q8N6L0 | |
| Recombinant DNA reagent | pRSF-Duet1-SUN1 (plasmid) | This paper | SUN1 (616–812) cloned into a pRSF-Duet1 vector | |
| Recombinant DNA reagent | pRSF-Duet1-SUN1 I673E (plasmid) | This paper | SUN1 (616–812) I673E cloned into a pRSF-Duet1 vector | |
| Recombinant DNA reagent | pRSF-Duet1-SUN1 F671E (plasmid) | This paper | SUN1 (616–812) F671E cloned into a pRSF-Duet1 vector | |
| Recombinant DNA reagent | pRSF-Duet1-SUN1 W676E (plasmid) | This paper | SUN1 (616–812) W676E cloned into a pRSF-Duet1 vector | |
| Recombinant DNA reagent | pRSF-Duet1-SUN2 (plasmid) | This paper | SUN2 (522–717) cloned into a pRSF-Duet1 vector | |
| Recombinant DNA reagent | pMAT11-KASH1 (plasmid) | This paper | Nesprin-1 (8769–8797) cloned into a pMAT11 vector | |
| Recombinant DNA reagent | pMAT11-KASH4 (plasmid) | This paper | Nesprin-4 (376–404) cloned into a pMAT11 vector | |
| Recombinant DNA reagent | pMAT11-KASH5 (plasmid) | This paper | KASH5 (542–562) cloned into a pMAT11 vector | |
| Strain, strain background (Escherichia coli) | Rosetta2 (DE3) | Thermo Fisher | EC0114 | Chemically competent cells |
| Software, algorithm | XDS | http://xds.mpimf-heidelberg.mpg.de/ | ||
| Software, algorithm | XSCALE | http://xds.mpimf-heidelberg.mpg.de/html_doc/xscale_program.html | ||
| Software, algorithm | Phaser | PHENIX | ||
| Software, algorithm | PHENIX Autobuild | PHENIX | ||
| Software, algorithm | PHENIX refine | PHENIX | ||
| Software, algorithm | AutoPROC | Global phasing | ||
| Software, algorithm | ASTRA 6 | Wyatt Technology | ||
| Software, algorithm | ScÅtter 3.0 | http://www.bioisis.net | ||
| Software, algorithm | PRIMUS | Atsas | ||
| Software, algorithm | DAMMIF | Atsas | ||
| Software, algorithm | CRYSOL | Atsas | ||
| Software, algorithm | SREFLEX | Atsas | ||
| Software, algorithm | CORAL | Atsas | ||
| Software, algorithm | SAMSON element | https://www.samson-connect.net |





