Peroxiredoxin promotes longevity and H2O2-resistance in yeast through redox-modulation of protein kinase A
Figures
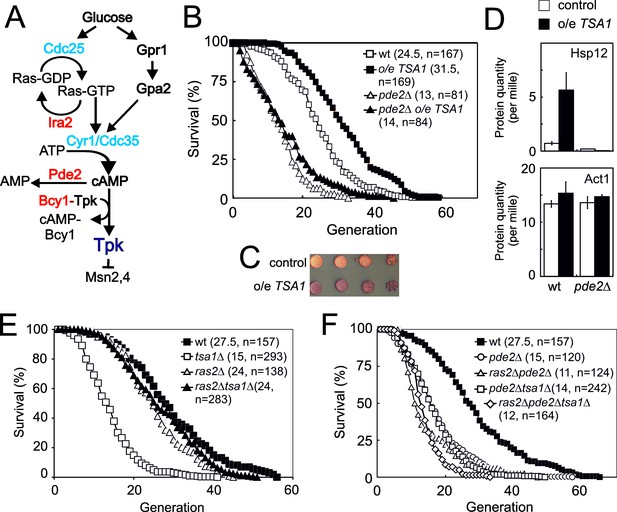
The 2-cys peroxiredoxin Tsa1 slows down aging via inhibiting protein kinase A signaling.
(A) Overview of the Ras-cAMP-PKA signaling pathway. In blue stimulatory components and in red inhibitory. (B) Lifespans of cells expressing an extra copy of the TSA1 gene or not (vector control) in combination with the deletion of PDE2 to induce high PKA signaling (pde2Δ). (C) Accumulation of glycogen in vector control cells or cells expressing an extra copy of the TSA1 gene as assayed by iodine vapor. (D) Expression of Hsp12 in the indicated mutant strains (n = 3). (E–F) Lifespan of cells lacking Tsa1, Ras2, Pde2 or combinations thereof.
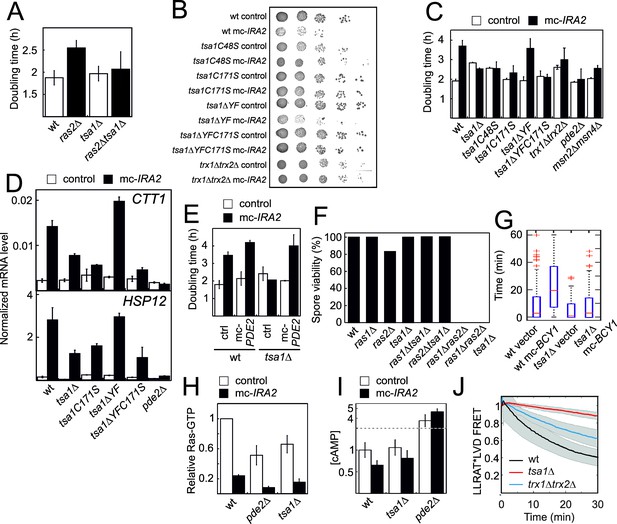
The Tsa1 catalytic cysteines affect protein kinase A dependent proliferation downstream of cAMP but not downstream of the catalytic subunits.
(A) Growth of cells lacking Ras2, Tsa1 or both (n = 3, error bars indicate SD). (B–C) Growth of cells overexpressing IRA2 in the indicated mutants of the Tsa1 catalytic cycle or the PKA signaling pathway on solid (B) or in liquid medium (C), n = 3–15). (D) Expression of the PKA repressed CTT1 or HSP12 genes in the indicated mutants in the Tsa1 catalytic cycle overexpressing IRA2 (mc-IRA2) or not (instead expressing the vector, control, n = 3 ± SD) sampled during mid-exponential growth. (E) Growth of Tsa1-proficient or deficient (tsa1Δ) cells overexpressing IRA2 (mc-IRA2) or PDE2 (mc-PDE2), both or the corresponding vector control plasmids (control) in liquid medium (n = 3 ± SD). (F) Spore germination in cells deficient in Ras1, Ras2, Tsa1 or combinations thereof. Spore germination was estimated in 32 tetrads where genotypes could be assigned to all spores (128 in total, 8–23 spores per genotype). (G) Total time of nuclear Msn2 localization in the indicated mutant strains for 60 min following the addition of 0.3 mM H2O2 (n = 46–82). (H–I) Ras-GTP (H) or cAMP (I) levels in the wild-type or the indicated mutant strains overexpressing IRA2 (mc-IRA2) or not (expressing the vector control, control, n = 3). (J) Phosphorylation of the ectopic AKAR4 PKA site upon H2O2 addition (0.4 mM) in wt, tsa1Δ and trx1Δtrx2Δ cells. (n = 85, 71 and 32, respectively). Error bars indicate SD.
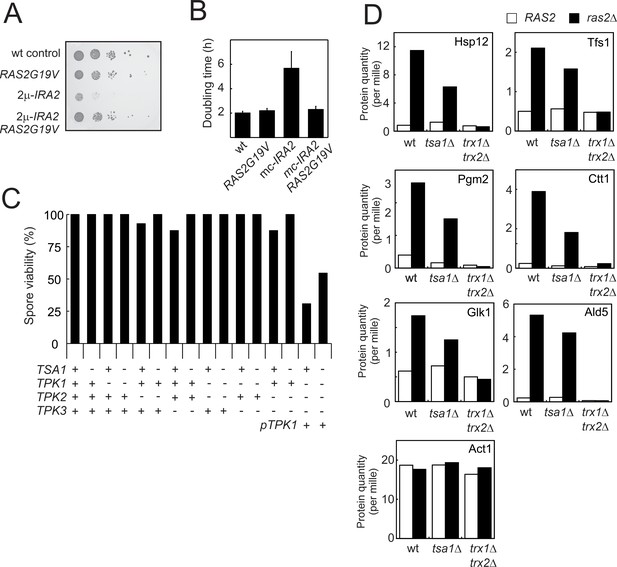
Tsa1 and the cytosolic thioredoxins Trx1 and Trx2 impact on PKA related growth signaling but lack of Tsa1 cannot overcome the requirement for a PKA catalytic subunit for spore viability.
(A-B) Growth of cells expressing the oncogenic RAS2G19V allele, overexpressing IRA2 (mc-IRA2) or both. (C) Spore viability in mutants segregating in a tsa1Δ x tpk1Δtpk2Δtpk3Δ mutant cross. The tpk1Δtpk2DΔtpk3Δ mutant was kept alive by a Tpk1-expressing plasmid (pRS313-TPK1). Spore viability was estimated in 43 tetrads where genotypes could be assigned to all spores (172 spores in total and in 8-15 spores per genotype). (D) Expression of PKA repressed Msn2/4 targets (Hasan et al., 2002; Molin et al., 2011) in wild-type, tsa1Δ or trx1Δtrx2Δ cells deficient in RAS2 (ras2Δ) or not (RAS2).
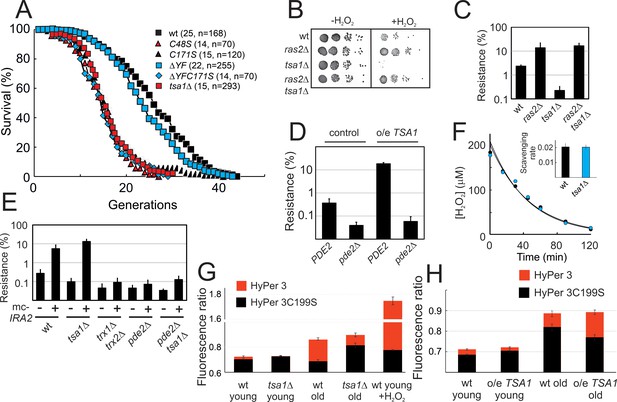
Tsa1 catalytic cysteines slow down aging and increase H2O2-resistance via inhibiting protein kinase A.
(A) Life spans of wild-type or the indicated genomic tsa1 mutant strains. In brackets median life-spans and n. (B) Spot-test assay of growth in the presence and absence of 1.5 mM H2O2 in YPD plates. (C) Quantification of H2O2 resistance in (B) (n = 3). (D) H2O2 resistance (1.5 mM H2O2, YPD medium) in the indicated mutants (n = 3). (E) H2O2 resistance in cells overexpressing IRA2 (mc-IRA2 +) or vector control (-) 0.4 mM H2O2, SD medium (n = 3). (F) Culture medium H2O2 removal assay of wt (black) and tsa1Δ cells (blue) to which 200 μM was added. Inset shows average scavenging rates for cultures upon the addition of 400 μM (n = 3). Error bars indicate SD. (G) Average HyPer3 (red) or HyPer3 C199S (black) fluorescence ratio (500 nm/420 nm) in young or aged wild-type or tsa1Δ cells +/- 400 μM H2O2 for 10 min. Cells of about 10–12 generations of replicative age (aged) or young control cells (young) were assayed. Error bars indicate SEM (n = 231, 170, 319, 236 and 202, respectively). (H) Average HyPer3 (red) or HyPer3 C199S (black) fluorescence ratio (500 nm/420 nm) in young or aged wild-type (YMM130) and o/e TSA1 cells as in (G) Error bars indicate SEM (n = 404, 579, 190 and 204, respectively).
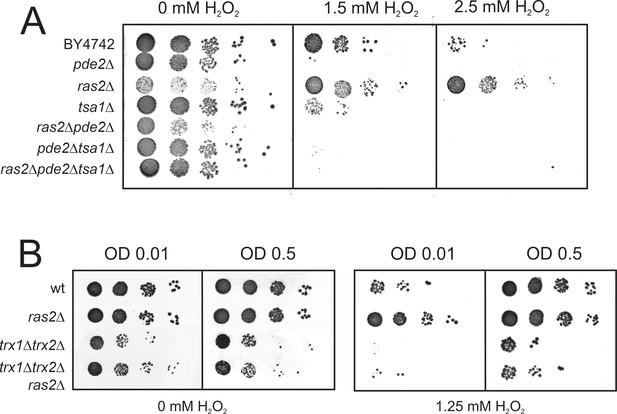
Reduced Ras activity can overcome H2O2 sensitivity of cells lacking Tsa1 but not that of cell lacking the cytosolic thioredoxins Trx1 and Trx2.
(A) H2O2 resistance in the indicated mutant strains strains grown to mid exponential phase (OD 0.3) and spotted onto plates with or without the indicated amounts of H2O2. (B) H2O2 resistance in the indicated mutant strains strains grown to early (OD0.01) and mid exponential phase (OD0.5) and spotted onto plates with or without the indicated amounts of H2O2.
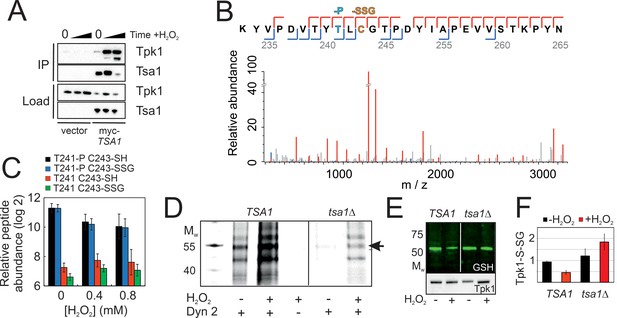
Tsa1 interacts with the PKA catalytic subunit Tpk1 and stimulates Tpk1 cysteine sulfenylation by H2O2.
Tpk1 is glutathionylated at a conserved cysteine. (A) Tpk1 interacts with myc-Tsa1 in a coimmunoprecipitation assay and in a manner strongly stimulated by H2O2. (B) MS-MS spectrum showing the matching b-ion (blue) and y-ion (red) series following fragmentation of the Thr241 phosphorylated and C243 glutathionylated peptide encompassing amino acid residues Y239-K261 in Tpk1. T-P = phospho threonine, C-SSG = glutathionylated cysteine. (C) PRM-based quantification of the indicated Thr241 and Cys243 containing Y239-K261 peptides in Tpk1, in the absence or presence of the indicated amount of H2O2, respectively (n = 3). Error bars indicate SD. (D) DYn-2 assay showing Tpk1 cysteine sulfenylation in the presence and absence of TSA1 and +/- 0.5 mM H2O2 for 5 min. Tpk1-HB was immunoprecipitated from tpk2Δtpk3Δ (TSA1) and tpk2Δtpk3Δtsa1Δ (tsa1Δ) cells and analyzed in gel for cyanine5 fluorescence. (E–F) Glutathionylation of Tpk1-HB in strains in (D) as assayed by anti-glutathione immunoblot of immunoprecipitated Tpk1-HB in the absence of or 10 min following the addition of 0.4 mM H2O2. Extracts were separated under non-reducing conditions (n = 3).
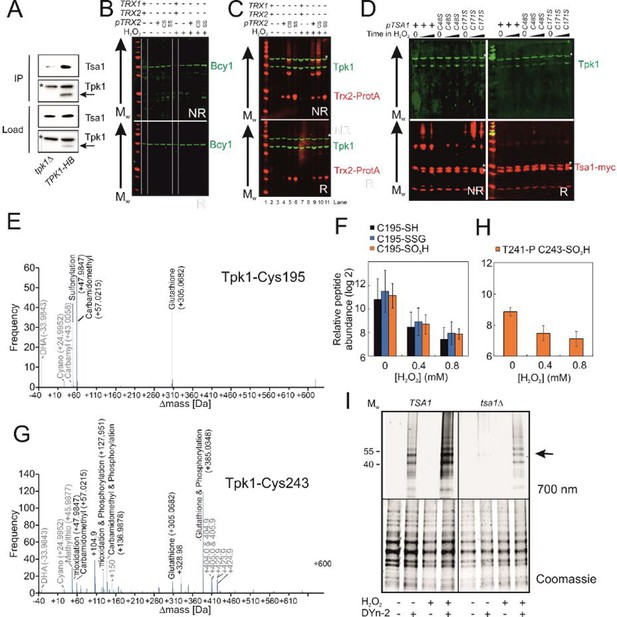
Tsa1 interacts with the PKA catalytic subunits Tpk1, controls Tpk1 cysteine sulfenylation independent on disulphide formation and a significant proportion of Tpk1 cysteines are glutathionylated under basal conditions.
(A) Tsa1 interacts with Tpk1 in a Ni2+-sepharose coimmunoprecipitation assay (Tpk1-HB tpk2Δtpk3Δ strain or tpk1Δtpk3Δ strain used as a negative control). An arrow indicates the Tpk1 specific band, whereas * indicates an unspecific band. (B-C) Bcy1 (B) or Tpk1 (C) redox immunoblots of protein extracts isolated from the indicated thioredoxin mutant strains in the absence of stress (H2O2 -) or following the addition of 0.4 mM H2O2 for 20 min (H2O2 +). NR = non-reducing R = reducing CS = trx2C34S SS = trx2C31SC34S. (D) Tpk1 redox immunoblots of protein extracts isolated from the indicated myc-tsa1 mutant strains in the absence of stress (Time in H2O2 = 0) or 10 or 120 min following the addition of 0.4 mM H2O2. (E, G) Mass-shifts in peptides covering the indicated Tpk1 cysteines detected using unbiased open search approaches. Tpk1-Cys195 denotes the F189-K204 peptide whereas Tpk1-Cys243 the Y239-K261 peptide. (F) PRM-based quantification of the indicated C195 containing Tpk1 peptides (n=3). Error bars indicate SD. (H) PRM-based quantification of the Thr241 phosphorylated and Cys243 sulfinic acid containing Y239-K261 peptide in Tpk1 (n=3). Error bars indicate SD. (I) DYn-2 sulfenylation assay depicting oxidation of Tpk1 following the addition of 0.5 mM of H2O2 for 5 min or not in the presence and absence of TSA1. Tpk1-HB was immunoprecipitated from tpk2Δtpk3Δ (TSA1) and tpk2Δtpk3Δ tpk2Δtsa1Δ (tsa1Δ) cells and analyzed in gel for cyanine5 fluorescence. Arrows indicate Tpk1. Coomassie staining was used to assess total protein used in the assay.
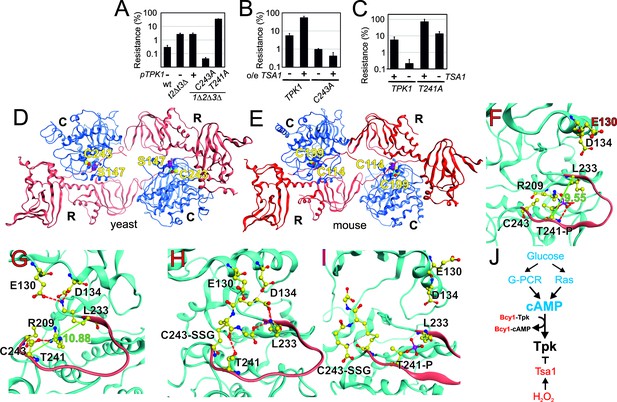
Tpk1 Cys243 redox-modification and Tsa1 inhibits PKA activity by dephosphorylating and destabilizing the activation loop.
(A–B) H2O2 resistance of the wild-type vector control (A, pRS313 or B, pRS403) or the indicated tsa1- or tpk-mutant strains in SD medium, 0.6 mM H2O2. Strains in (B) carry pRS316-TPK1 or pRS316-tpk1C243A as the only PKA catalytic subunit peroxiredoxin Tsa1 slows down (genomic tpk1Δtpk2Δtpk3Δ deletions, n = 3). (C) H2O2 resistance of tpk1Δtpk2Δtpk3Δ and tpk1Δtpk2Δtpk3Δtsa1Δ cells transformed with pRS313-TPK1 or pRS313-tpk1T241A as indicated in SD medium 0.6 mM H2O2 (n = 3). (D–E) Structural homology model of yeast Tpk1 (D) based on the structure of mouse type II PKA holoenzyme (E) [PDB ID 3TNP, (Zhang et al., 2012). (F–I) Amino acids in the activation loop (in red) of Tpk1 in the Thr241 phosphorylated Cys243 non-modified (F), Thr241 non-phosphorylated Cys243 non-modified (G), Thr241 non-modified Cys243 glutathionylated (H) and Thr241 phosphorylated Cys243 glutathionylated (I) states in the Tpk1 structural homology model. The backbones are colored in light blue, carbon atoms in yellow, nitrogen atoms in blue, oxygen atoms in red and phosphor atoms in scarlet. The distance between Lys233 and phosphorylated Thr241 is 9.55 Å (F) whereas Lys233 and non-phosphorylated Thr241 reside 10.88 Å apart (G). (J) Overview of mechanisms by which glucose and H2O2 control PKA activity. In blue activators and in red inhibitors. See also Figure 5—figure supplement 1.
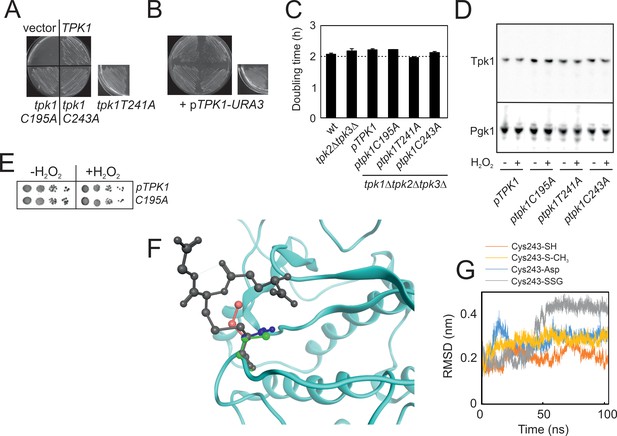
Substitution of Cys195, Thr241 and Cys243 by alanine in the yeast.
PKA catalytic subunit Tpk1 neither affects viability nor growth, whereas in silico simulation suggest that glutathionylation, but not sulfenylation, of Tpk1Cys243 significantly impacts on Tpk1 structure.
(A) Growth of tpk1Δtpk2Δtpk3Δ cells transformed with the vector (vector) or the indicated pRS313-TPK1 plasmids and pRS316-TPK1 (pTPK1-URA3) on solid synthetic defined (-HIS, 5-FOA) medium to counterselect pTPK1-URA3. (B) Growth of the strains in (A) on solid synthetic defined selective (-HIS, URA) medium. Cells in A) and B) were left to grow for 3 days before photographed. (C) Doubling time of the indicated tpk-mutant strains in synthetic defined -HIS medium. (D) Tpk1 levels are not significantly altered in Tpk1 substitution mutants neither with nor without H2O2 (0.4 mM. 10 min). Pgk1 levels were used to indicate protein loading. (E) H2O2 resistance of TPK1 tpk2Δ tpk3Δ and tpk1C195A tpk2Δ tpk3Δ-mutants as indicated. (F) Alignment of cysteine (green), aspartate (blue), methylthiolated (pink) or glutathionylated cysteine (grey) in position 243 in the Tpk1 homology model. (G) Root-mean-square deviation of the C-alpha distances in C243-SH (orange), Cys243Asp (blue), Cys243 methylthiolated (yellow) and C243 glutathionylated enzyme (Cys243-SSG, grey) upon molecular dynamic simulation.
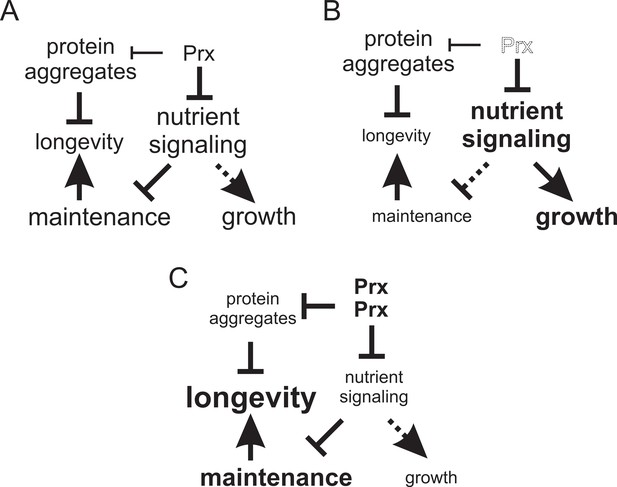
Model of the mechanisms by which altered peroxiredoxin levels impacts on aging.
In the first mechanism peroxiredoxin-dependent redox-signaling impacts in an unconventional manner on the PKA nutrient signaling kinase (this study) and in the other on proteostasis (Hanzén et al., 2016). (A) In wild-type cells Tsa1 catalytic cycling maintains longevity by decreasing PKA-dependent nutrient signaling leading to the stimulation of maintenance but at the expense of growth. (B) In cells lacking Tsa1, nutrient signaling is aberrantly increased leading to reduced maintenance and increased growth. (C) Enforced expression of the peroxiredoxin Tsa1 slows down aging both by repressing nutrient signaling (this study) and by stimulating protein quality control mechanisms to reduce the levels of damaged and aggregated protein (Hanzén et al., 2016).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | E. coli BL21 strain expressing pGEX2T-1-GST-RBD | This paper, 10.1038/s41467-017-01019-z | To purify GST-RBD for Ras-GTP assays | |
| Strain, strain background (Saccharomyces cerevisiae) | wt control | 10.1016/j.cell.2016.05.006 | YMM130 | MAT alpha his3Δ1::pRS403, leu2Δ0 lys2Δ0 ura3Δ0 |
| Strain, strain background (Saccharomyces cerevisiae) | o/e TSA1 | 10.1016/j.cell.2016.05.006 | o/e TSA1 | MAT alpha his3Δ1::pRS403-Myc-TSA1, leu2Δ0 lys2Δ0 ura3Δ0 |
| Strain, strain background (Saccharomyces cerevisiae) | pde2Δ control | This paper | YMM175 | MAT alpha his3Δ1::pRS403, leu2Δ0 lys2Δ0 ura3Δ0 pde2Δ::kanMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | pde2Δ o/e TSA1 | This paper | YMM176 | MAT alpha his3Δ1::pRS403-Myc-TSA1, leu2Δ0 lys2Δ0 ura3Δ0 pde2Δ::kanMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | wt | 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2–2. | BY4742 | MAT alpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1Δ | 10.1016/j.molcel.2011.07.027 | YMM114 | BY4742 tsa1Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | ras2Δ | 10.1016/j.molcel.2011.07.027 | YMM113 | BY4742 ras2Δ::kanMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | ras2Δtsa1Δ | This paper | YMM170 | BY4742 ras2Δ::kanMX4 tsa1Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | pde2Δ | Research Genetics, 10.1038/nature00935. | pde2Δ | BY4742 pde2Δ::kanMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | ras2Δpde2Δ | This paper | YMM171 | BY4742 ras2Δ::kanMX4 pde2Δ::hphMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | pde2Δtsa1Δ | This paper | YMM172 | BY4742 pde2Δ::kanMX4 tsa1Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | ras2Δpde2Δtsa1Δ | This paper | YMM173 | BY4742 ras2Δ::kanMX4 pde2Δ::hphMX4 tsa1Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1C48S | 10.1038/ncomms14791 | YMM145 | BY4742 tsa1C48S |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1C171S | 10.1038/ncomms14791 | YMM146 | BY4742 tsa1C171S |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1ΔYF | 10.1038/ncomms14791 | YMM147 | BY4742 tsa1(1-184) |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1C171SΔYF | 10.1038/ncomms14791 | YMM148 | BY4742 tsa1(1-184)C171S |
| Strain, strain background (Saccharomyces cerevisiae) | trx1Δtrx2Δ | 10.1038/ncomms14791 | YMM143 | BY4742 trx1Δ::hphMX4 trx2Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | msn2Δmsn4Δ | This paper | YMM174 | BY4742 msn2Δ::hphMX4 msn4Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | ras1Δ::hphMX4 | This paper | YMM177 | MAT a, his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ras1Δ::hphMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | This paper | YMM178 | BY-2n met15Δ0/MET15 lys2Δ0/LYS2 tpk1Δ::kanMX4/TPK1 tpk2Δ::natMX4/TPK2 tpk3Δ::hphMX4/TPK3 | |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk3Δ | This paper | YMM179 | BY4742 tpk1Δ::kanMX4 tpk3Δ::hphMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk2Δtpk3Δ | This paper | YMM180 | BY4742 tpk2Δ::natMX4 tpk3Δ::hphMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA | This paper | YMM181 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS316-TPK1 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA vector control | This paper | YMM182 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313 pTPK1-URA3 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA pTPK1 | This paper | YMM183 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-TPK1 pTPK1-URA3 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA3 ptpk1C243A | This paper | YMM184 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1C243A pTPK1-URA3 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA3 ptpk1C243D | This paper | YMM185 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1C243D pTPK1-URA3 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA3 ptpk1T241A | This paper | YMM186 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1T241A pTPK1-URA3 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1 | This paper | YMM187 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-TPK1 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ ptpk1C243A | This paper | YMM188 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1C243A |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ ptpk1C243D | This paper | YMM189 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1C243D |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ ptpk1T241A | This paper | YMM190 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS313-tpk1T241A |
| Strain, strain background (Saccharomyces cerevisiae) | ras2Δtrx1Δtrx2Δ | This paper | YMM191 | BY4742 ras2Δ::kanMX4 trx1Δ::hphMX4 trx2Δ::natMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tsa1Δ::bleMX4 | This paper | YMM192 | BY4741 tsa1Δ::bleMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk2Δtpk3Δtsa1Δ | This paper | YMM193 | BY4741 tpk2Δ::natMX4 tpk3Δ::hphMX4 tsa1Δ::bleMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | TPK1-HBH tpk2Δtpk3Δ | This paper | WR1832 | BY4742 TPK1-HBH::TRP1 tpk2Δ::natMX4 tpk3Δ::hphMX4 trp1Δ::kanMX4 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA vector control | This paper | yCP101 | MAT a his3Δ1::pRS403, leu2Δ0 lys2Δ0 ura3Δ0 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS316-TPK1 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ ptpk1C243A-URA vector control | This paper | yCP102 | MAT alpha his3Δ1::pRS403, leu2Δ0 lys2Δ0 ura3Δ0 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS316-tpk1C243A |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ pTPK1-URA o/e TSA1 | This paper | yCP103 | MAT alpha his3Δ1::pRS403-myc-TSA1, leu2Δ0 lys2Δ0 ura3Δ0 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS316-TPK1 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δ ptpk1C243A-URA o/e TSA1 | This paper | yCP104 | MAT alpha his3Δ1::pRS403-myc-TSA1, leu2Δ0 lys2Δ0 ura3Δ0 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 pRS316-tpk1C243A |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δtsa1Δ pTPK1 | This paper | yCP105 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 tsa1Δ::bleMX4 pRS313-TPK1 |
| Strain, strain background (Saccharomyces cerevisiae) | tpk1Δtpk2Δtpk3Δtsa1Δ ptpk1T241A | This paper | yCP106 | BY4742 tpk1Δ::kanMX4 tpk2Δ::natMX4 tpk3Δ::hphMX4 tsa1Δ::bleMX4 pRS313-tpk1T241A |
| Strain, strain background (Saccharomyces cerevisiae) | TPK1-HBH tpk2Δtpk3Δtsa1Δ | This paper | yCP107 | BY4742 TPK1-HBH::TRP1 tpk2Δ::natMX4 tpk3Δ::hphMX4 tsa1Δ::bleMX4 trp1Δ::kanMX4 tsa1Δ::bleMX4 |
| Antibody | (mouse monoclonal) anti-Tpk1 | Santa Cruz Biotechnology | Sc-374592, RRID:AB_10990730 | (1:1000) |
| Antibody | (goat polyclonal) anti-Bcy1 | Santa Cruz Biotechnology | Sc-6734, RRID:AB_671758 | (1:2000) |
| Antibody | (rabbit) IgG; anti-Protein A | Sigma Aldrich | I5006, RRID:AB_1163659 | 1 μg/ml |
| Antibody | (goat polyclonal) anti-Ras2 | Santa Cruz Biotechnology | Sc-6759, RRID:AB_672465 | (1:2000) |
| Antibody | (mouse monoclonal) anti-Glutathione (D8) | Abcam | ab19534, RRID:AB_880243 | (1:1000) |
| Antibody | (mouse monoclonal) anti-Pgk1 (22C5D8) | Thermo Fisher | 459250, RRID:AB_2532235 | (1:500) |
| Antibody | (mouse monoclonal) anti-2 Cys Prx (6E5); (anti-Tsa1) | Abcam | ab16765, RRID:AB_443456 | (1:1000) |
| Recombinant DNA reagent | yEP24 | 10.1016/0378-1119(79)90004-0 | yeast 2μ, URA3 vector plasmid | |
| Recombinant DNA reagent | pKF56 | 10.1128/mcb.10.8.4303. | IRA2 in yEP24 | |
| Recombinant DNA reagent | pRS425 | 10.1016/0378-1119(92)90454w. | yeast 2μ, LEU2 vector plasmid | |
| Recombinant DNA reagent | yEP13-PDE2 | 10.1093/emboj/cdg314. | PDE2 in yeast 2μ, LEU2 plasmid | |
| Recombinant DNA reagent | yEPlac195 | 10.1016/0378-1119(88)90185-0. | yeast 2μ, URA3 vector plasmid | |
| Recombinant DNA reagent | pXP1 | 10.1128/mcb.19.7.4874. | BCY1 in yEPlac195 | |
| Recombinant DNA reagent | pRS315 | PMID:2659436 | yeast CEN/ARS, LEU2 empty vector plasmid | |
| Recombinant DNA reagent | B561 (pRS315-RAS2G19V) | 10.1128/mcb.19.10.6775. | RAS2G19V in pRS315 | |
| Recombinant DNA reagent | pHyPer3C199S (pRS416-GPD-HyPer3C199S) | This paper, 10.1021/cb300625g | HyPer3C199S | |
| Recombinant DNA reagent | pRS416-GPD-AKAR4 | Molin et al., 2020 | AKAR4 in pRS416-GPD [CEN/ARS, pGPD promotor, URA3] | |
| Recombinant DNA reagent | pRS316 | PMID:2659436 | yeast CEN/ARS, URA3 empty vector plasmid | |
| Recombinant DNA reagent | pRS316- myc-TSA1 | 10.1038/nature02075. | Myc-TSA1 in pRS316 | |
| Recombinant DNA reagent | pRS316- myc-tsa1C48S | 10.1016/j.molcel.2011.07.027 | Myc-tsa1C48S in pRS316 | |
| Recombinant DNA reagent | pRS316- myc-tsa1C171S | 10.1016/j.molcel.2011.07.027 | Myc-tsa1C171S in pRS316 | |
| Recombinant DNA reagent | pRS315-ProtA | This paper | ProteinA in pRS315 | |
| Recombinant DNA reagent | pRS315-TRX2-ProteinA | 10.1038/ncomms14791 | TRX2-ProtA in pRS315 | |
| Recombinant DNA reagent | pRS315-trx2C34S-ProteinA | This paper | trx2C34S-ProtA in pRS315 | |
| Recombinant DNA reagent | pRS315-trx2C31SC34S-ProteinA | This paper | trx2C31SC34S-ProtA in pRS315 | trx2C31SC34S-ProtA in pRS315 |
| Recombinant DNA reagent | pRS313 | PMID:2659436 | yeast CEN/ARS, HIS3 empty vector | yeast CEN/ARS, HIS3 empty vector |
| Recombinant DNA reagent | pRS313-TPK1 | 10.1074/jbc.M110.200071. | TPK1 in pRS313 | TPK1 in pRS313 |
| Recombinant DNA reagent | pRS313-tpk1C243A | This paper | tpk1C243A in pRS313 | tpk1C243A in pRS313 |
| Recombinant DNA reagent | pRS313-tpk1C243D | This paper | tpk1C243D in pRS313 | tpk1C243D in pRS313 |
| Recombinant DNA reagent | pRS313-tpk1T241A | This paper | tpk1T241A in pRS313 | tpk1T241A in pRS313 |
| Recombinant DNA reagent | pTPK1-URA3 (pRS316-TPK1) | Karin Voordeckers | TPK1 in pRS316 | TPK1 in pRS316 |
| Recombinant DNA reagent | ptpk1C243A-URA3 | This paper | tpk1C243A in pRS316 | tpk1C243A in pRS316 |
| Sequence-based reagent | ACT1F | 10.1016/j.molcel.2011.03.021 | For Q-PCR of ACT1 | CTGCCGGTATTGACCAAACT |
| Sequence-based reagent | ACT1R | 10.1016/j.molcel.2011.03.021 | For Q-PCR of ACT1 | CGGTGAATTTCCTTTTGCATT |
| Sequence-based reagent | CTT1F | This paper | For Q-PCR of CTT1 | GCTTCTCAATACTCAAGACCAG |
| Sequence-based reagent | CTT1R | This paper | For Q-PCR of CTT1 | GCGGCGTATGTAATATCACTC |
| Sequence-based reagent | HSP12F | 10.1016/j.molcel.2011.03.021 | For Q-PCR of HSP12 | AGGTCGCTGGTAAGGTTC |
| Sequence-based reagent | HSP12R | 10.1016/j.molcel.2011.03.021 | For Q-PCR of HSP12 | ATCGTTCAACTTGGACTTGG |
| Peptide, recombinant protein | Glutathione-S-Transferase-Raf1-Binding-Domain (GST-RBD) | This paper, 10.1038/s41467-017-01019-z | For Ras-GTP assay | Purified from E. coli strain BL21 expressing pGEX2T-1-GST-RBD |
| Commercial assay or kit | PureLink RNA Mini kit | Thermo-Fisher | Cat #: 12183025 | |
| Commercial assay or kit | QuantiTect Reverse Transcription Kit | Qiagen | Cat #: 205313 | |
| Commercial assay or kit | iQ SYBR Green Supermix | BioRad | Cat #: 170–8882 | |
| Commercial assay or kit | LANCE cAMP 384 kit | Perkin Elmer | Cat #: AD0262 | |
| Chemical compound, drug | G418 | Acros Organics | Cat #: 329400050 | |
| Chemical compound, drug | ClonNAT | Werner Bioagents | Cat #: 5.005.000 | |
| Chemical compound, drug | Hygromycin B | Formedium | Cat #: HYG5000 | |
| Chemical compound, drug | Phleomycin | Sigma Aldrich | P9564 | |
| Chemical compound, drug | 5-fluoroorotic acid | Sigma Aldrich | F5013 | |
| Chemical compound, drug | EZ-Link Sulfo-NHS-LC Biotin | Thermo Fisher | Cat #: 21335 | |
| Chemical compound, drug | Trichloroacetic acid | Sigma Aldrich | Cat #: T6399 | |
| Chemical compound, drug | KSCN | Sigma Aldrich | Cat #: P2713 | |
| Chemical compound, drug | (NH4)2Fe(SO4)2 • 6 H2O | Sigma Aldrich | Cat #: 215406 | |
| Chemical compound, drug | TRIzol Reagent | Thermo Fisher | Cat #: 15596026 | |
| Chemical compound, drug | DNase, RNase-free set | Qiagen | Cat #: 79254 | |
| Chemical compound, drug | cOmplete Mini EDTA-free protease inhibitor | Roche Applied Science | Cat #: 11873580001 | |
| Chemical compound, drug | Glutathione Sepharose beads | GE Healthcare | Cat #: 17-0756-01 | |
| Chemical compound, drug | 12% Bis-Tris NUPAGE gels | Thermo FisherArch Biochem Biophys | Cat #: NP0349BOX | |
| Chemical compound, drug | MOPS running buffer | Thermo Fisher | Cat #: NP0001 | |
| Chemical compound, drug | Immobilon-FL PVDF membrane | Millipore | Cat #: IPFL00010 | |
| Chemical compound, drug | Ni2+-Sepharose beads | GE Healthcare | Cat #: 17-5318-06 | |
| Chemical compound, drug | Anti-c-myc, agarose conjugated | Sigma-Aldrich | Cat #: A7470 | |
| Chemical compound, drug | Trypsin Gold, mass spectrometry grade | Promega | Cat #: V5280 | |
| Chemical compound, drug | N-ethylmaleimide | Sigma-Aldrich | Cat #: E3876 | |
| Chemical compound, drug | DYn-2 | Cayman Chemical | Cat #: 11220 | |
| Chemical compound, drug | 10% Criterion TGX Precast Midi Protein Gel | Bio-Rad | Cat #: 5671034 | |
| Chemical compound, drug | Peptide Retention Time Calibration Mixture | Pierce, Thermo Fisher | Cat #: 88320 | |
| Software, algorithm | MATLAB | Mathworks | version 2016b | |
| Software, algorithm | CellX | 10.1002/0471142727.mb1422s101 | ||
| Software, algorithm | Scrödinger Suite | Schrödinger LLC | ||
| Software, algorithm | GROMACS | 10.1016/j.softx.2015.06.001 | ||
| Software, algorithm | Amber tools | 10.1002/wcms.1121 |
Additional files
-
Supplementary file 1
Mass spectrometry data.
- https://cdn.elifesciences.org/articles/60346/elife-60346-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60346/elife-60346-transrepform-v3.pdf





