FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices
Figures
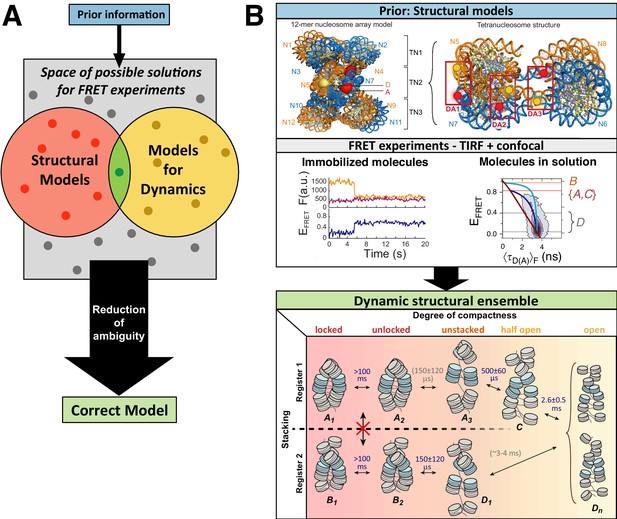
Workflow of modeling dynamic structures from FRET measurements.
(A) Integrative modeling requires structural and dynamic information. Prior information from conventional approaches (X-ray, NMR, cryoEM) together with computational tools defines the space of possible solutions for FRET-assisted structural modeling. The combination of structural (inter-dye distances) and dynamic information (kinetic connectivity and exchange rates) enables identification of a consistent model. (B) Study of structure and dynamics of chromatin fibers. A combined TIRF and confocal FRET study of structure and dynamics of chromatin fibers using three FRET labeling positions (DA1-3) for two pairs of dyes with distinct Förster distances. Förster distances ( is defined in section Inter-dye distances, Equation 6). Prior structural information provided by cryo-electron microscopy (top, left) (Song et al., 2014) and X-ray crystallography (top, right PDB ID: 1ZBB Schalch et al., 2005) is combined with the structural and dynamic information obtained by FRET experiments on immobilized molecules measured by total internal reflection (TIRF) microscopy and on freely diffusing molecules by confocal microscopy (Kilic et al., 2018). From the combined information, a consistent model is derived for chromatin fiber conformations with shifted registers, which are connected by slow (>100 ms) and fast de-compaction processes (150 µs) that do not proceed directly, but rather through an open fiber conformation. Figure 1B was reproduced from Figures 1, 3, and 6 in Kilic et al., 2018, Nature Communications with permission, published under the Creative Commons Attribution 4.0 International Public License (CC BY 4.0; https://creativecommons.org/licenses/by/4.0/).
© 2018, Kilic et al. Panel B was reproduced from Figures 1, 3 and 6 in Kilic et al., 2018 , with permission, published under the Creative Commons Attribution 4.0 International Public License.
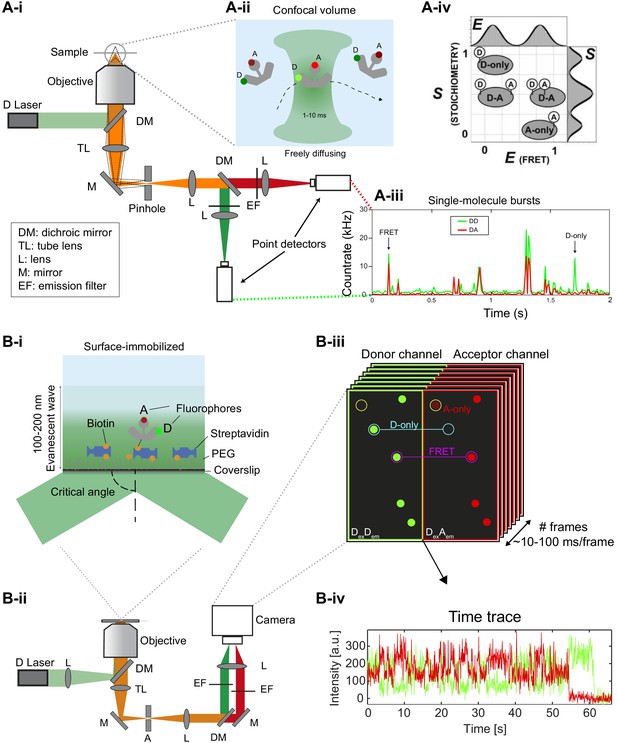
Different smFRET modalities.
(A) Confocal smFRET measurements on freely-diffusing molecules. (i) A schematic of a single-color excitation confocal microscope with point detectors used for two-color detection. The excitation light is guided to the microscope body and reflected by a dichroic mirror (DM) toward a high numerical aperture (NA) objective lens that focuses the light in solution. The fluorescence emission is collected through the same objective lens, passes through the DM and pinhole and is spectrally split into donor and acceptor detection channels by a second DM in the detection path. After passing through emission filters (EF), single photons are detected on point detectors with high quantum efficiency, typically avalanche photodiodes (APD). (ii) Illustration of a double-labeled molecule freely diffusing through the confocal excitation spot. (iii) Exemplary confocal smFRET measurement showing photon bursts arising from single-molecules diffusing through the confocal volume. Green: Donor emission. Red: Acceptor emission. Exemplary bursts belonging to a single- or a double-labeled molecule are indicated with arrows. (iv) In ALEX or PIE experiments, the two-dimensional histogram of the molecule-wise FRET efficiency and stoichiometry S allows one to separate single- and double-labeled populations (2005 Elsevier Ltd. All rights reserved. The figure was originally published as Figure 2A in Lee et al., 2005. Biophysical Journal, 88(4): 2939–2953. Further reproduction of this panel would need permission from the copyright holder). (B) TIRF-based smFRET experiments on surface-immobilized molecules. (i) Illustration of a surface-immobilized sample labeled with donor and acceptor fluorophores. (ii) Scheme of a single-color objective-type TIRF excitation two-color wide-field detection microscope. A: Aperture, TL: Tube lens, L: Lens, M: Mirror, DM: Dichroic mirror, EF: Emission filter. (iii) Illustration of an image of single molecules, in which the donor and acceptor (FRET) signals are split onto two halves of the camera. Mapping between the two channels is typically done using fluorescent beads (Joo and Ha, 2012; Roy et al., 2008; Zhuang et al., 2000) or zero-mode waveguides (Salem et al., 2019). (iv) Single-molecule fluorescence trajectory of the donor and acceptor (FRET) dyes, illustrating an anti-correlation indicative of FRET dynamics.
© 2005, Elsevier. All rights reserved. Panel Aiv was originally published as Figure 2A in Lee et al., 2005. Further reproduction of this panel would need permission from the copyright holder.
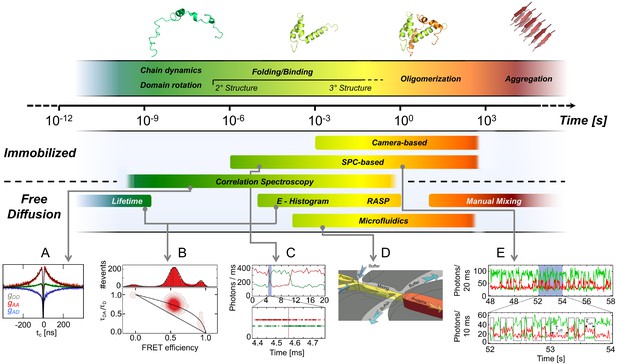
Exemplary methods for following smFRET dynamics on different timescales.
Top: Biomolecular dynamics cover a wide range of timescales. Biomolecular rotations occur in the pico- to nanosecond range, while conformational changes take place in nano- to microseconds (ns-µs), as in chain dynamics of disordered proteins, and protein folding in microseconds to minutes. Transitions along energetically unfavorable pathways can take up to hours or longer, as in protein misfolding (Borgia et al., 2011; Tosatto et al., 2015). (2013 Elsevier Ltd. All rights reserved. The figure was originally published as Figure 1 in Schuler and Hofmann, 2013. Current Opinion in Structural Biology, 23(1): 36–47. Further reproduction of this panel would need permission from the copyright holder.) Bottom: (A) Picosecond (ps) to millisecond (ms) processes are typically examined with confocal methods such as polarization-resolved fluorescence lifetime measurements and Fluorescence Correlation Spectroscopy (FCS). Example shown: chain dynamics of an IDP from nsFCS. (B) Conformational states are identified by individual populations with characteristic positions in the FRET efficiency - lifetime diagrams as discussed in the sections Detection and characterization of intra-state dynamics and Future of smFRET (adapted from Soranno et al., 2012). (C) Fast transitions measured using confocal microscopy can be analyzed using the photon trajectory and applying a photon-by-photon maximum likelihood approach (2018 Elsevier Ltd. All rights reserved. The figure was originally published as Figures 2 and 3 in Chung and Eaton, 2018. Current Opinion in Structural Biology, 48: 30–39. Further adaptation of this panel would need permission from the copyright holder.) The timescale over which kinetics can be measured can be extended for diffusing molecules at low concentrations by using a recurrence analysis of single particles (RASP, Hoffmann et al., 2011). (D) Non-equilibrium experiments over extended periods of time can be performed with microfluidic mixing devices. (Copyright 2011, Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. Reproduced from Gambin et al., 2011, with permission. Nature Methods 8:239–241. Further reproduction of this panel would need permission from the copyright holder.) (E) Slow changes in conformations over a broad range of timescales can be followed in smFRET efficiency trajectories registered by single-photon counting (SPC) or cameras over minutes to many hours when the sample is immobilized (adapted from Figure 1 of Zosel et al., 2018).
© 2013, Elsevier Ltd. All rights reserved. Figure 3 (top) and panel A was originally published as Figure 1 in Schuler and Hofmann, 2013. Further reproduction of this panel would need permission from the copyright holder.
© 2018, Elsevier Ltd. All rights reserved. Panel C was originally published as Figures 2 and 3 in Chung and Eaton, 2018. Further adaptation of this panel would need permission from the copyright holder.
© 2011, Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. Panel D was originally published as Figure 1f in Gambin et al., 2011. Further reproduction of this panel would need permission from the copyright holder.
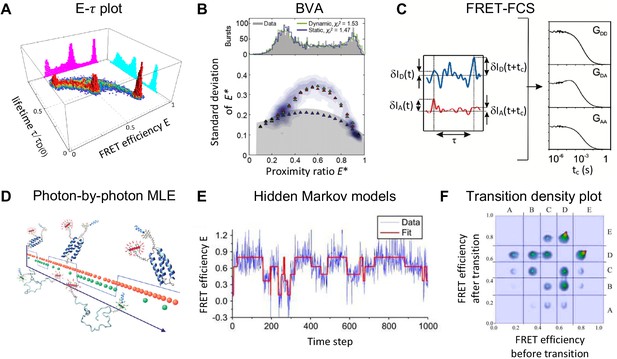
Exemplary selection of approaches to detect and quantify conformational dynamics in smFRET.
(A) Dynamics in a three-state system are detected using the two-dimensional distribution of the FRET efficiency and donor fluorescence lifetime (Reproduced from Gopich and Szabo, 2012. Further reproduction of this panel would need permission from the copyright holder.) (B) The two-state dynamics of a DNA hairpin are revealed from the standard deviation of the proximity ratio that is higher than expected for photon counting statistics alone in the burst variance analysis (BVA). (2011 The Biophysical Society. Published by Elsevier Inc All rights reserved. The figure was originally published as Figure 4C in Torella et al., 2011. Biophysical Journal, 100(6): 1568–1577. Further reproduction of this panel would need permission from the copyright holder.) (C) In fluorescence correlation spectroscopy (FCS), the dynamics show up as a positive correlation in the autocorrelation functions and and an anti-correlation in the cross-correlation function (2010 American Chemical Society Ltd. All rights reserved. The figure was originally published as Figure 1B and C in (Gurunathan and Levitus, 2010, reproduced with permission). Copyright 2010 ACS Publications. Further reproduction of this panel would need permission from the copyright holder.) (D) Photon-by-photon maximum likelihood estimation (MLE) infers the kinetic parameters directly from the photon arrival times (2011 American Chemical Society Ltd. All rights reserved. The figure was originally published as the Abstract Figure in Chung et al., 2011, reproduced with permission. Copyright 2011 ACS Publications. Further reproduction of this panel would need permission from the copyright holder.) (E–F) A hidden Markov model (HMM) is applied to the time traces of the FRET efficiency to estimate the states and interconversion rates (E). From the transition density plot (F), the connectivity of the FRET states is revealed. Displayed data in E and F are simulated. (Panels E and F: 2006 The Biophysical Society. Published by Elsevier Inc All rights reserved. The figures were originally published as Figure 4A and D in McKinney et al., 2006. Biophysical Journal, 91(5): 1941–1951. Further reproduction of this panel would need permission from the copyright holder.)
© 2012, Gopich and Szabo. Panel A was originally published as Figure 1C in Gopich and Szabo, 2012. Further reproduction of this panel would need permission from the copyright holder.
© 2011, The Biophysical Society. Published by Elsevier Inc. All rights reserved. Panel B was originally published as Figure 4C in Torella et al., 2011. Further reproduction of this panel would need permission from the copyright holder.
© 2010, American Chemical Society Ltd. All rights reserved. Panel C was originally published as Figures 1B and 1C in Gurunathan and Levitus, 2010. Further reproduction of this panel would need permission from the copyright holder.
© 2011, American Chemical Society Ltd. All rights reserved. Panel D was originally published as the abstract figure in Chung et al., 2011. Further reproduction of this panel would need permission from the copyright holder.
© 2006, The Biophysical Society. Published by Elsevier Inc. All rights reserved. Panels E and F were originally published as Figures 4A and 4D in McKinney et al., 2006. Further reproduction of this panel would need permission from the copyright holder.
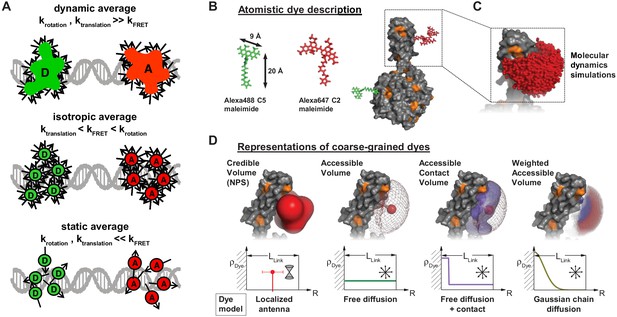
Dye models for FRET.
(A) The different kinetic averaging regimes for rotation and diffusion are shown schematically. In the dynamic averaging regime, rotation and diffusion happen on a timescale faster than the FRET process. In the isotropic averaging regime, translation is slower than the FRET process, but rotation is fast. The static average applies if both rotation and diffusion are slow compared to the rate of energy transfer. For most experimental situations, the isotropic average is most appropriate. (2008 National Academy of Sciences. Reproduced from Wozniak et al., 2008. Further reproduction of this panel would need permission from the copyright holder.) (B) A schematic of a donor/acceptor fluorophore pair (Alexa488, Alexa647) attached to the protein Atlastin-1. (C) The accessible volume that the acceptor fluorophore can explore is determined from a molecular dynamics simulation. (D) Different coarse-grained dye models are used in the community to describe the three-dimensional dye density ρDye (see main text). (Panels B, C, and D were reproduced from Dimura et al., 2016, Current Opinion in Structural Biology with permission, published under the Creative Commons Attribution 4.0 International Public License (CC BY 4.0; https://creativecommons.org/licenses/by/4.0/).)
© 2008, National Academy of Sciences. Panel A was originally published as Figure S2 in Wozniak et al., 2008. Further reproduction of this panel would need permission from the copyright holder.
© 2016, Dimura et al. Panels B, C and D were originally published as Figure 1a-d in Dimura et al., 2016, published under the Creative Commons Attribution 4.0 International Public License.
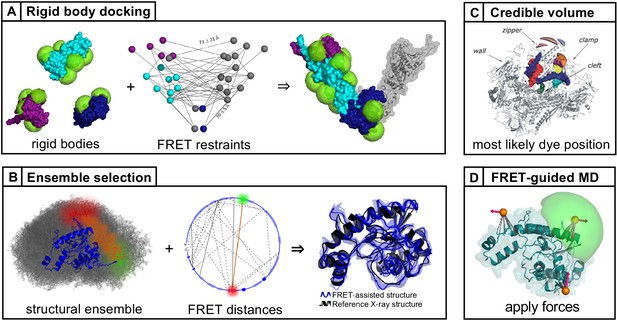
Approaches to integrative modeling using FRET restraints.
(A) Rigid bodies, representing either different domains of a protein or different proteins within a complex, are arranged by translation and rotation to satisfy the FRET restraints. Adapted from Figure 1 of Hellenkamp et al., 2017. (B) The structure that best satisfies all FRET restraints is selected from a prior ensemble. Adapted from Figure 2 of Dimura et al., 2020. (C) Using triangulation, the most likely dye position is estimated from the FRET distances with respect to known reference positions on the structure. (Reproduced from Andrecka et al., 2009, Nucleic Acid Research with permission, published under the Creative Commons Attribution-NonCommercial 4.0 International Public License (CC BY NC 4.0; https://creativecommons.org/licenses/by-nc/4.0). Further reproduction of this panel would need permission from the copyright holder.) (D) Starting from a known structure, a molecular dynamics simulation is guided using the FRET information by applying forces based on the FRET distances. (Reproduced from Dimura et al., 2020. Further reproduction of this panel would need permission from the copyright holder.)
© 2009, Andrecka et al. Panel C was originally published as Figure 2A in Andrecka et al., 2009, published under the Creative Commons Attribution-NonCommercial 4.0 International Public License.
© 2020, Dimura et al. Panel D was originally published as Figure 2A in Dimura et al., 2020. Published under the Creative Commons Attribution 4.0 International Public License.
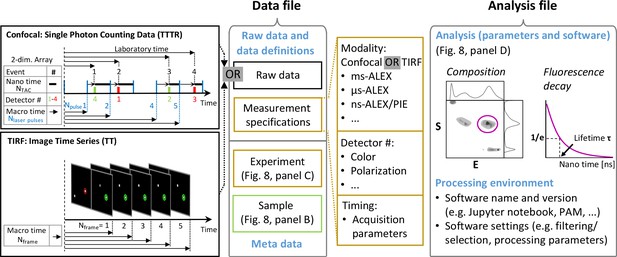
Concept for data storage following the FAIR principle.
All essential information should be contained in two data files, one for raw data and a second with the essential information regarding the associated analysis. (Left) Structure of proposed data file formats containing confocal or TIRF raw data in a time-tagged, TT, or time-tagged time-resolved, TTTR, format together with sample- and experiment-specific metadata (for details, see Figure 8). The measurement specifications are needed by the reading routine to reconstruct the photon trace from the stored data, for example, for timing in the TT format (sampling frequencies expressed as time bins or frame rates) or in the TTTR format (laser repetition rates, time binning in time-correlated single-photon counting). Moreover, the detector that measured the signal is noted (detector #) along with the detection time with a given time resolution. For representing the detection time of single photons in time-resolved smFRET studies, the TTTR format is used where the time corresponds to the sum of the macro time and the nano time (upper left panel). The macro time comprises multiple cycles of excitation laser pulses (blue vertical lines) and the nano time is determined by time-correlated single-photon counting with picosecond resolution. The TTTR format is the most compact data format for single-molecule fluorescence data for detection times with picosecond time resolution and macro-times of hours (Felekyan et al., 2005). The representation of the photon detection of intensity-based and imaging-based smFRET studies is in the TT format, where the macro-time comprises multiples of the external clock pulse or readout cycles, where single or several photons were detected. The stack of TIFF images acquired in TIRF measurements (lower left panel) is transformed into the TT format for analyzing photon time traces for selected spots. (Middle) For the corresponding data file, a metadata system as implemented in the Photon-HDF5 file format (Ingargiola et al., 2016a) is suggested. (Right) The analysis file should contain the determined parameters obtained by a quantitative analysis together with analysis metadata that assure evaluation, reproducibility, and re-usability of the analysis. The processed data should be documented as outlined in Figure 8D.
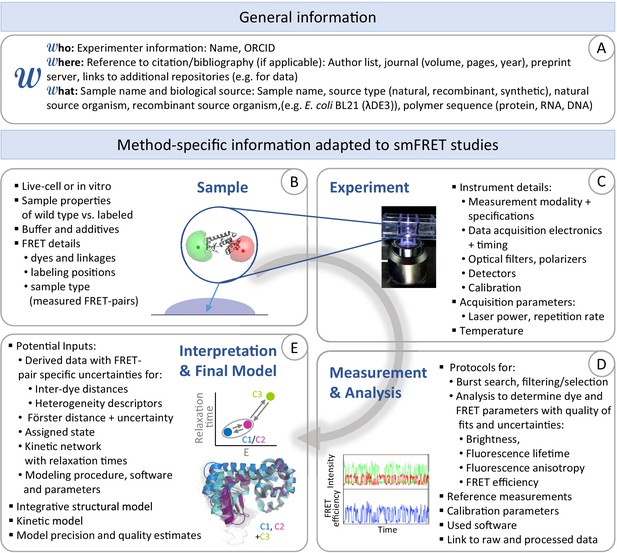
Disseminating the results of smFRET studies.
Recommended categories for data and method-specific information (metadata), which are needed for documentation of smFRET studies where the authors want to archive their obtained kinetic/structural models. (A) General information. (B) Information on the sample and FRET-specific properties. (C) Information on the experiment and data acquisition. (D) Information on processing and analysis procedures. (E) Information on the interpretation of the data and the final kinetic network or structural model.
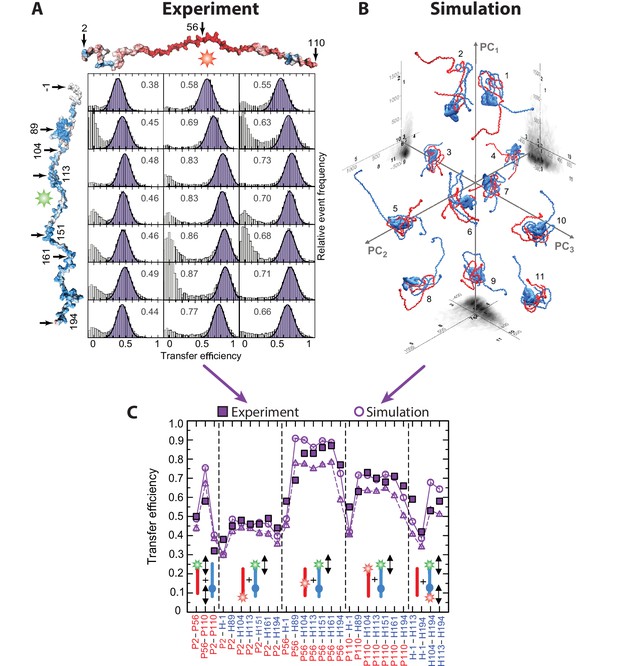
Using smFRET to investigate the structure and dynamics of ultrahigh-affinity IDP complexes.
(A) SmFRET efficiency histograms for FRET between a donor label (Alexa488) attached at various positions to the linker histone H1 (shown in blue) with the IDP ProTα (shown in red) labeled at different positions with the acceptor fluorophore (Alexa594). (B) For structural calculations of the H1-ProTα complex, coarse-grained MD simulations were performed. From the MD simulations, an ensemble of structures was determined. Eleven examples of configurations are shown and projected onto the first three principle components (PC1, PC2, and PC3) of the inter-residue distance map. 2D projections of the full ensemble are shown in gray (axes are labeled in Å). (C) A comparison of the experimental FRET efficiencies (filled squares) and the FRET efficiencies estimated from simulated structures (open circles) shows good agreement between the measured and simulated values. Pictograms indicate the variations of dye positions studied. (Panels A, B, and C: Copyright 2018, Nature Publishing Group, a division of Macmillan Publishers Limited. All rights reserved. Reproduced from Borgia et al., 2018, with permission. Further reproduction of this panel would need permission from the copyright holder.)
© 2018, Macmillan Publishers Limited, part of Springer Nature. All rights reserved. Panels A-C were originally published as Figure 3i, 4c and 4a in Borgia et al., 2018. Further reproduction of this panel would need permission from the copyright holder
Tables
List of available software for smFRET analysis.
| Software | Type | Description | URL |
|---|---|---|---|
| ALiX | Confocal | ALiX is developed for basic research on diffusing two-color smFRET in single or multiple spot geometries (Ingargiola et al., 2017). | https://sites.google.com/a/g.ucla.edu/alix/ |
| Fretica | Confocal | Fretica, a Mathematica package with a backend written in C++, is a user-extendable toolbox that supports MFD, PIE/ALEX, PCH (Huang et al., 2004; Müller et al., 2000), FIDA (Gopich and Szabo, 2005b; Kask et al., 1999), PDA (Antonik et al., 2006; Ernst et al., 2020), recurrence analysis (Hoffmann et al., 2011), fluorescence lifetime fitting, FLIM, FCS, FLCS (Arbour and Enderlein, 2010; Dertinger et al., 2007), dual-focus FCS, nsFCS (Nettels et al., 2007; Schuler and Hofmann, 2013), maximum likelihood estimation from photon-by-photon (Gopich and Szabo, 2009) and binned trajectories, simulation of confocal experiments and more (Nettels and Schuler, 2020; https://schuler.bioc.uzh.ch/programs/). | https://schuler.bioc.uzh.ch/programs/ |
| FRET_3colorCW | Confocal | C++ and MATLAB GUI-based CPU-GPU co-parallelization software package for an enhanced maximum likelihood analysis of two- and three-color fluorescence photon trajectories generated by continuous-wave donor excitation (Yoo et al., 2020). | https://github.com/hoisunglab/FRET_3colorCW |
| gSMFRETda | Confocal | gSMFRETda is a GPU-capable program for Monte Carlo simulations of PDA. It can sample dwell time and other parameters in fine grids, thus allowing the analysis of rapid dynamic interconversions (Liu et al., 2019). | https://github.com/liu-kan/gSMFRETda |
| H2MM | Confocal | H2MM is a maximum likelihood estimation algorithm for a photon-by-photon analysis of smFRET experiments (Pirchi et al., 2016). | http://pubs.acs.org/doi/suppl/10.1021/acs.jpcb.6b10726/suppl_file/jp6b10726_si_002.zip |
| MFD Spectroscopy and Imaging | Confocal | A modular software package for confocal fluorescence spectroscopy and imaging experiments using multiparameter fluorescence detection (MFD) with all tools (FCS, fFCS, PDA, seTCSPC, trace analysis, 2D simulation of MFD diagrams, Burbulator) and multiparameter fluorescence image spectroscopy (MFIS) (Antonik et al., 2006; Felekyan et al., 2005; Kühnemuth and Seidel, 2001; Widengren et al., 2006; Felekyan et al., 2020). | https://www.mpc.hhu.de/software/3-software-package-for-mfd-fcs-and-mfis |
| OpenSMFS | Confocal | A collection of tools (Ingargiola et al., 2016b) for solution-based single-molecule fluorescence spectroscopy, including smFRET, FCS, MC-DEPI (Ingargiola et al., 2018b). | https://github.com/OpenSMFS |
| smfBox | Confocal | A confocal smFRET platform, providing build instructions and open-source acquisition software (Ambrose et al., 2020). | https://craggslab.github.io/smfBox/ |
| rFRET | Confocal Imaging | rFRET is a comprehensive, MATLAB-based program for analyzing ratiometric microscopic FRET experiments (Nagy et al., 2016). | https://peternagy.webs.com/fret#rfret |
| ChiSurf | Confocal Imaging Ensemble | ChiSurf is a fluorescence analysis platform for the analysis of time-resolved fluorescence decays (Peulen et al., 2017). | https://github.com/Fluorescence-Tools/ChiSurf/wiki |
| PAM - PIE Analysis with MATLAB | Confocal Imaging Ensemble | PAM (PIE analysis with MATLAB) is a GUI-based software package for the analysis of fluorescence experiments and supports a large number of analysis methods ranging from single-molecule methods to imaging (Schrimpf et al., 2018; Barth et al., 2020). | RRID:SCR_020966, https://www.cup.uni-muenchen.de/pc/lamb/software/pam.html |
| AutoSiM | TIRF | AutoSiM is a deep-learning developed MATLAB program for automatically selecting and sorting smFRET traces (Li et al., 2020a). | https://doi.org/10.7302/ck2m-qf69 |
| BIASD | TIRF | BIASD uses Bayesian inference to infer transition rates that are more than three orders of magnitude larger than the acquisition rate of the experimental smFRET data (Kinz-Thompson and Gonzalez, 2018). | http://github.com/ckinzthompson/biasd |
| DeepFRET | TIRF | smFRET software based on deep-learning for automatic trace selection and classification. It includes all common features: image analysis, background-corrected trace-extraction, hidden Markov analysis, correction factor application, and data visualization (Thomsen et al., 2020). | https://github.com/hatzakislab/DeepFRET-GUI |
| ebFRET | TIRF | ebFRET performs combined analysis on multiple smFRET time-series to learn a set of rates and states (van de Meent et al., 2014). | https://ebfret.github.io/ |
| FRETboard | TIRF | smFRET data preprocessing and analysis using algorithms of choice and user supervision. Also offered as a web-based user interface (de Lannoy et al., 2020). | https://github.com/cvdelannoy/FRETboard |
| HaMMy | TIRF | smFRET analysis and hidden Markov modeling (McKinney et al., 2006). | http://ha.med.jhmi.edu/resources/ |
| hFRET | TIRF | hFRET uses variational Bayesian inference to estimate the parameters of a hierarchical hidden Markov model, thereby enabling robust identification and characterization of kinetic heterogeneity (Hon and Gonzalez, 2019). | https://github.com/GonzalezBiophysicsLab/hFRET |
| iSMS | TIRF | iSMS is a user-interfaced software package for smFRET data analysis. It includes extraction of time-traces from movies, traces grouping/selection tools according to defined criteria, application of corrections, data visualization and analysis with hidden Markov modeling, and import/export possibilities in different formats for data-sharing (Preus et al., 2015; Preuss et al., 2020). | http://isms.au.dk/ |
| MASH-FRET | TIRF | MASH-FRET is a MATLAB-based software package for the simulation (Börner et al., 2018) and analysis of single-molecule FRET videos and trajectories (video processing [Hadzic et al., 2016], histogram analysis [König et al., 2013], and transitions analysis [Hadzic et al., 2018; König et al., 2013]). | https://rna-fretools.github.io/MASH-FRET/ |
| miCUBE | TIRF | TIRF smFRET platform, providing detailed build instructions and open-source acquisition software (Martens et al., 2019). | https://hohlbeinlab.github.io/miCube/index.html |
| SMACKS | TIRF | SMACKS (single-molecule analysis of complex kinetic sequences) is a maximum-likelihood approach to extract kinetic rate models from noisy single-molecule data (Schmid et al., 2016). | https://www.singlemolecule.uni-freiburg.de/software/smacks |
| smCamera | TIRF | smFRET data acquisition (Windows. exe) and analysis (IDL, MATLAB) with example data (Roy et al., 2008). | http://ha.med.jhmi.edu/resources/ |
| SPARTAN | TIRF | Automated analysis of smFRET multiple single-molecule recordings. It includes extraction of traces from movies, trace selection according to defined criteria, application of corrections, hidden Markov modeling, simulations, and data visualization (Juette et al., 2016). | https://www.scottcblanchardlab.com/software |
| STaSI | TIRF | STaSI uses the Student’s t-test and groups the segments into states by hierarchical clustering (Shuang et al., 2014). | https://github.com/LandesLab/STaSI |
| TwoTone | TIRF | A TIRF-FRET analysis package for the automatic analysis of single-molecule FRET movies (Holden et al., 2011). | https://groups.physics.ox.ac.uk/genemachines/group/Main.Software.html |
| vbFRET | TIRF | vbFRET uses variational Bayesian inference to learn hidden Markov models from individual, smFRET time trajectories (Bronson et al., 2009). | http://www.columbia.edu/cu/chemistry/groups/gonzalez/software.html |
| Fast NPS | Modeling | A nano-positioning system for macromolecular structural analysis (Eilert et al., 2017). | http://dx.doi.org/10.17632/7ztzj63r68.1 |
| Fluordynamics | Modeling | Fluordynamics is a PyMOL plugin to label biomolecules with organic fluorophores for all-atom molecular dynamics simulations. It builds on AMBERDYES (Schepers and Gohlke, 2020) and extends the force field to common nucleic acid linker chemistries (Steffen et al., 2016). | https://github.com/RNA-FRETools/fluordynamics |
| FPS | Modeling | A toolkit for FRET restrained modeling of biomolecules and their complexes for quantitative applications in structural biology (Kalinin et al., 2012). | https://www.mpc.hhu.de/software/1-fret-positioning-and-screening-fps |
| FRETraj | Modeling | FRETraj is a Python API to the LabelLib package, which integrates into PyMOL to interactively calculate accessible-contact volumes and predict FRET efficiencies (Steffen et al., 2016). | https://github.com/RNA-FRETools/fretraj |
| FRETrest in Amber20 | Modeling | FRETrest is a set of helper scripts for generating FRET-restraints for Molecular Dynamics (MD) simulations performed with the AMBER Software Suite (Dimura et al., 2020). | http://ambermd.org/doc12/Amber20.pdf |
| LabelLib | Modeling | LabelLib is a C++ library for the simulation of the accessible volume (AV) of small probes flexibly coupled to biomolecules (Dimura et al., 2016; Kalinin et al., 2012). | https://github.com/Fluorescence-Tools/LabelLib |
A list of repositories for citable data storage.
| Repository name | URL | Size limits | Fee/Costs |
|---|---|---|---|
| Dryad Digital Repository | https://datadryad.org/stash | No | $120 USD for first 20 GB, and $50 USD for each additional 10 GB |
| figshare | https://figshare.com/ | 1 TB per dataset | It varies with publishers |
| Harvard Dataverse | https://dataverse.harvard.edu/ | 2.5 GB per file, 10 GB per dataset | Free |
| Open Science Framework | https://osf.io/ | 5 GB per file, multiple files can be uploaded | Free |
| Zenodo | https://zenodo.org/ | 50 GB per dataset | Donation |
| Mendeley Data | https://data.mendeley.com/ | 10 GB per dataset | Free |





