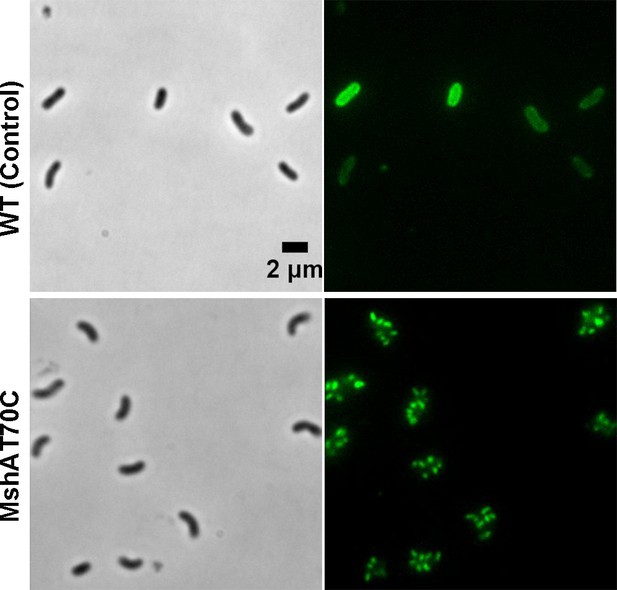
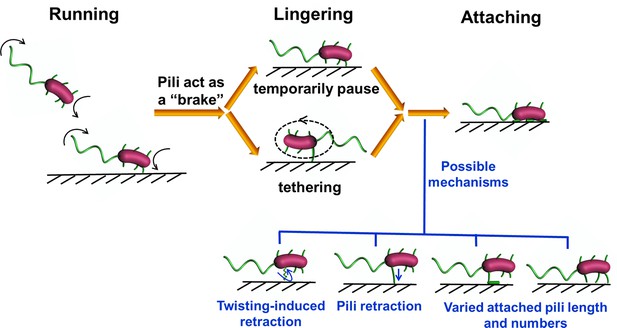
Crash landing of Vibrio cholerae by MSHA pili-assisted braking and anchoring in a viscoelastic environment
Figures
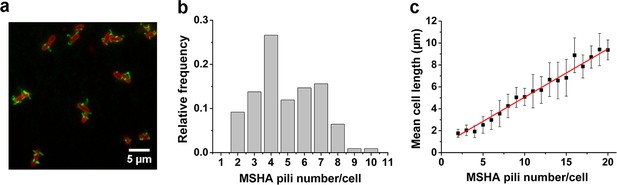
MSHA pili are evenly distributed along cell length with a constant length density.
(a) Examples of labeled MSHA pili observed on cell bodies. Green fluorescence showing the AF488-mal labeled MSHA pili, red fluorescence showing the FM4-64 labeled plasma membrane. (b) Distribution of pili number per cell cultivated in LB medium. Ncell=110. (c) The MSHA pili number per cell is linearly correlated with the cell length. Cells with longer length were obtained by 30–50 min treatment using 10 μg/mL cephalexin. Ncell=368. LB, Luria-Bertani; MSHA, mannose-sensitive hemagglutinin.
-
Figure 1—source data 1
Source data for Figure 1b.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Source data for Figure 1c.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig1-data2-v2.xlsx
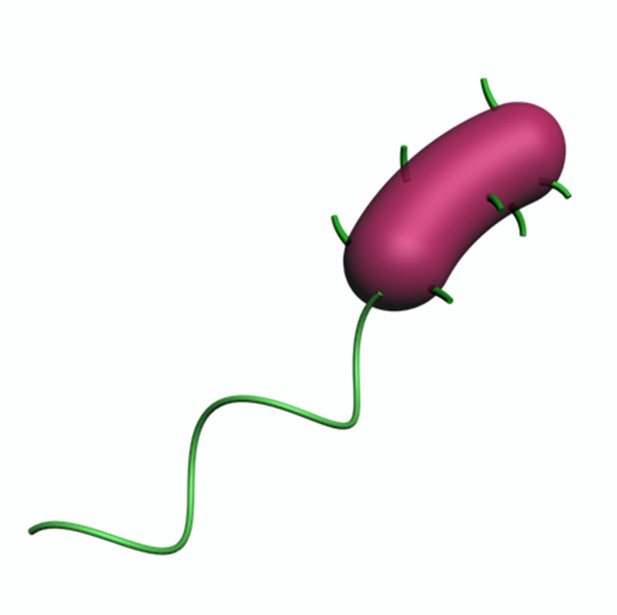
A 3D view of a typical Vibrio cholerae cell showing the whole-body distribution of MSHA pili; this cell has six pili. MSHA, mannose-sensitive hemagglutinin.
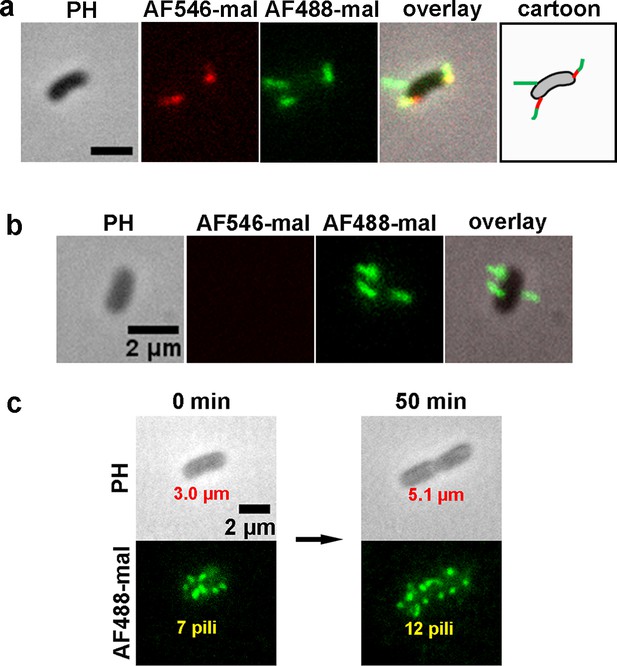
MSHA pili labeling of the prepared samples.
To evaluate changes to the MSHA pilus during cell growth, the MSHA pili were labeled with two different colored dyes, AF546-mal (red) and AF488-mal (green), at 0 and 40 min, respectively. (a) Representative double-color labeling image of MshAT70C cell, showing the new separate pilus (top left, green) and the secondary segments (lower left, green) at the end of the primary segments (lower left, red). Scale bar, 2 μm. (b) Representative double-color labeling image for a newly dividing cell, which is only labeled with AF488-mal. (c) In situ observation of MSHA pili growth stained at 0 and 50 min with AF488-mal. The results show that during a period of 50 min, the length of the cell changes from 3.0 to 5.1 μm, while the number of pili increases from 7 to 12. MSHA, mannose-sensitive hemagglutinin.
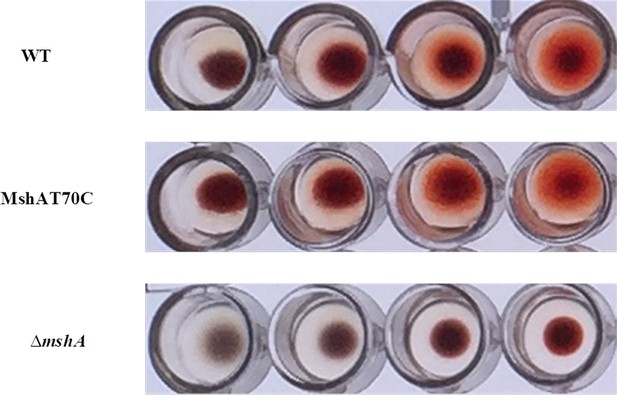
Hemagglutination assays.
MshAT70C point mutation does not affect MSHA pilus function. Vibrio cholerae strains were grown in LB medium and assayed for MSHA production by hemagglutination. Twofold dilutions of mid-log cultures of bacteria (left to right) were assayed for their ability to agglutinate sheep erythrocytes. The assay was repeated three times, and representative results are shown. LB, Luria-Bertani; MSHA, mannose-sensitive hemagglutinin.
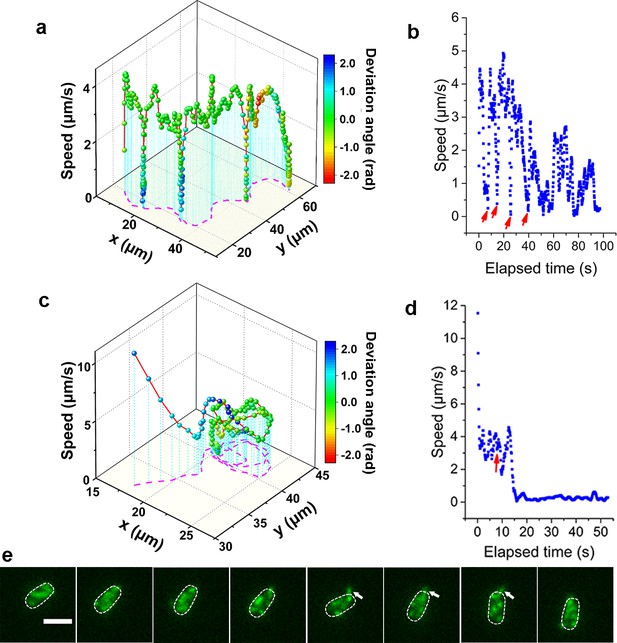
Analysis of roaming and orbiting, using cells of MSHA labeled MshAT70C.
The 3D plot and speed changes over time of representative (a, b) roaming and (c, d) orbiting cells, respectively. The magenta dashed lines in panels (a) and (c) are the trajectories of cells and the color maps indicate the deviation angle. The arrows in panel (b) represent temporary attachment between MSHA pili and surface, where the speeds are close to 0. (e) Time-lapse images of the orbiting cell in panels (c, d) at 130 ms intervals (see Video 2 for more details). The arrowheads show the stretched pilus, which corresponds to the red arrow in panel (d), indicating temporary attachment and stretching of pilus on the surface. Dashed lines indicate the estimated envelope of the cell body. Scale bar, 2 μm. MSHA, mannose-sensitive hemagglutinin.
-
Figure 2—source data 1
Source data for Figure 2a.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Source data for Figure 2b.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Source data for Figure 2c.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Source data for Figure 2d.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig2-data4-v2.xlsx
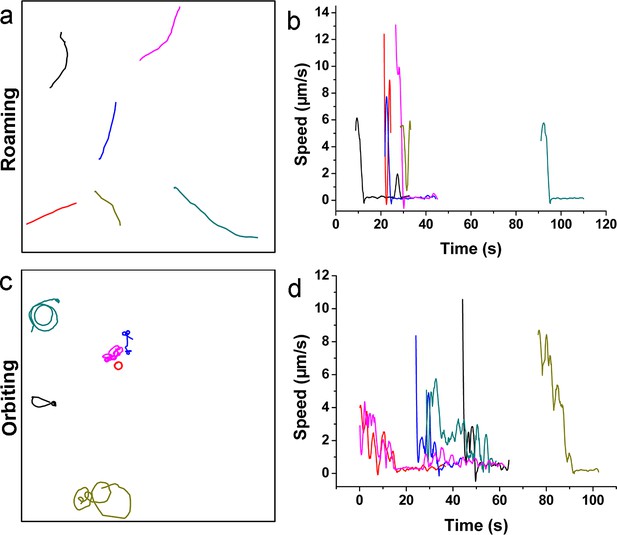
Quantitative analysis of roaming and orbiting by MSHA labeled MshAT70C in 2% LB with 1% MC.
(a) Trajectories and (b) speed of typical roaming cells; (c) Trajectories and (d) speed of typical orbiting cells. LB, Luria-Bertani; MC, methylcellulose; MSHA, mannose-sensitive hemagglutinin.
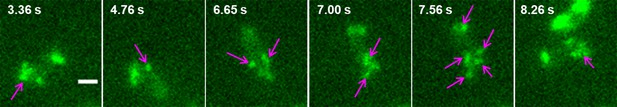
Switch of temporarily attached pili.
When transient pauses happened, the attached pilus could be switched from one to another or more. The arrows show the apparent pili attached to the surface. Scale bar, 1 μm.
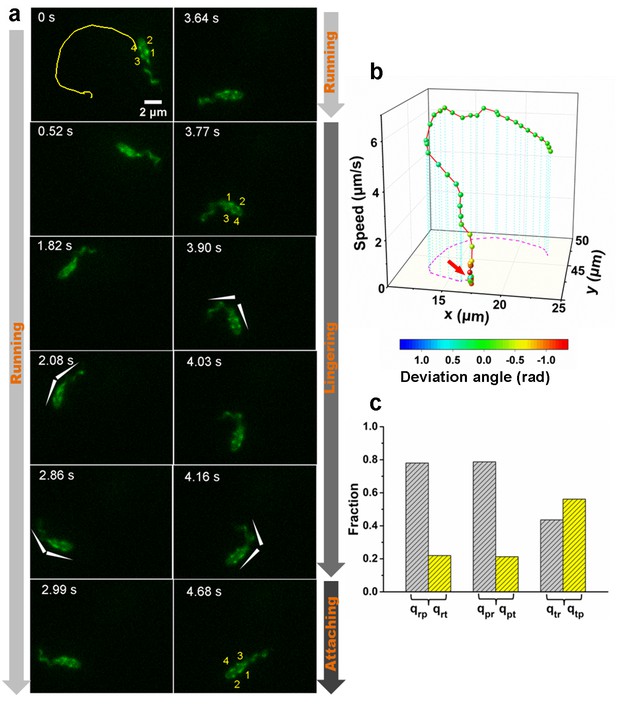
An example of a typical landing sequence of a Vibriocholerae cell with MSHA pili and flagella both labeled (MshAT70CFlaAA106CS107C).
(a) Representative image sequences showing the behavior of MSHA pili and flagellum. For easy identification, four pili of the example cell in (a) are numbered from 1 to 4, which revolve around the major axis of the cell periodically as the cell swims. The white arrowheads indicate the orientation of the cell body and flagellum. (b) A 3D plot of speed and deviation angle of the representative cell in panel (a) over its trajectory. The red arrow in panel (b) represents the position, where the pili touch surface, causing a deflection. (c) The conditional probabilities qij that the bacterium transitions from state i to j. The number of transition events used for estimating these conditional probabilities is 666. r: running state, t: tethered state, p: paused state. MSHA, mannose-sensitive hemagglutinin.
-
Figure 3—source data 1
Source data for Figure 3b.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for Figure 3c.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig3-data2-v2.xlsx
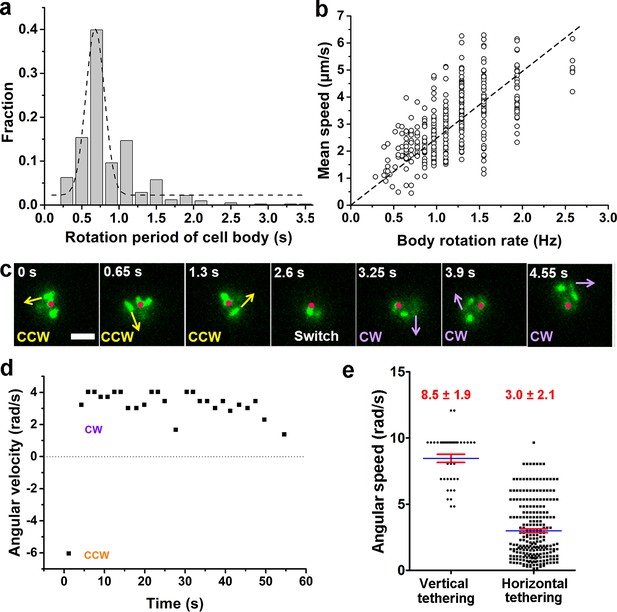
Characterization of running and tethered cells.
(a) Distribution of the rotation period of the cell body. The dashed line represents Gaussian fitting. A total of 416 rotation events from 54 cells were used for statistical analysis. (b) Measured relation between the rotation rate of the cell body and the mean swimming speed of the cell. The dotted line represents linear fitting result. Ncell=47. (c) An example of a typical tethered motion, showing a cell performing a circular motion around a center point (the red dot) with the direction of motion (noted by arrows) switched from CCW to CW. Scale bar, 2 μm. (d) The angular velocity of the tethered cell in panel (c) over a short duration showing a pair of CW (positive angular velocity) and CCW (negative angular velocity) intervals; (e) Distribution of angular speed of circular motion for horizontal (241 intervals from 25 cells) and vertical (38 intervals from 5 cells) tethered cells. CCW, counterclockwise; CW, clockwise.
-
Figure 4—source data 1
Source data for Figure 4a.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Source data for Figure 4b.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Source data for Figure 4d.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Source data for Figure 4e.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig4-data4-v2.xlsx
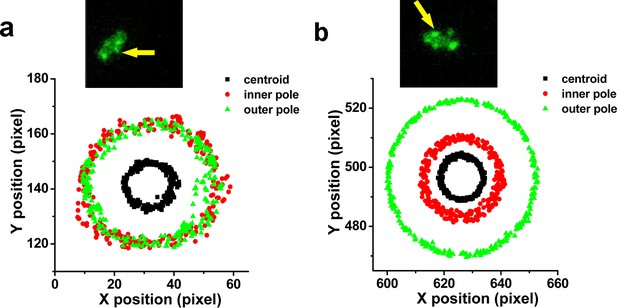
Examples show positions of two poles and centroid of tethered motility.
The contributing MSHA pili were indicated by yellow arrows. (a) ~1/2 position; (b) 1/3 or 2/3 position. MSHA, mannose-sensitive hemagglutinin.
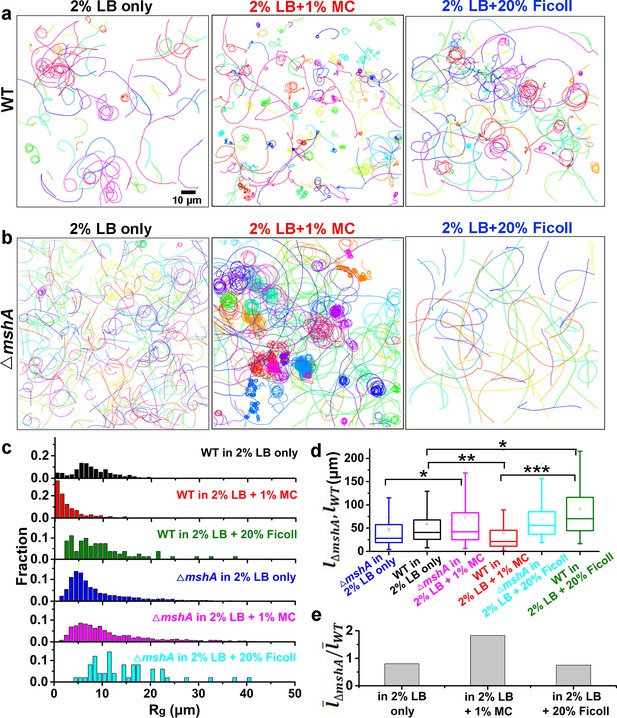
Role of MSHA pili in cell landing is more apparent in viscoelastic non-Newtonian solutions than viscous Newtonian fluids.
(a) Examples of WT cell trajectories showing both roaming and orbiting motilities in 2% LB only, 2% LB with 1% MC, and 2% LB with 20% Ficoll; (b) Examples of cell trajectories of △mshA; (c) Histograms of Rg of WT and △mshA in different solutions; (d) A box plot summary of path lengths of WT and △mshA. Statistical significance was determined with one-way ANOVA followed by Tukey’s multiple comparison test comparing the different groups (*p<0.05; **p<0.01; ***p<0.001). The data were analyzed using the Prism 5.0 software program (GraphPad Software, La Jolla, CA, USA). (e) The ratio of mean path length between △mshA and WT, . LB, Luria-Bertani; MC, methylcellulose; MSHA, mannose-sensitive hemagglutinin.
-
Figure 5—source data 1
Source data for Figure 5c.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Source data for Figure 5d.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Source data for Figure 5e.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig5-data3-v2.xlsx
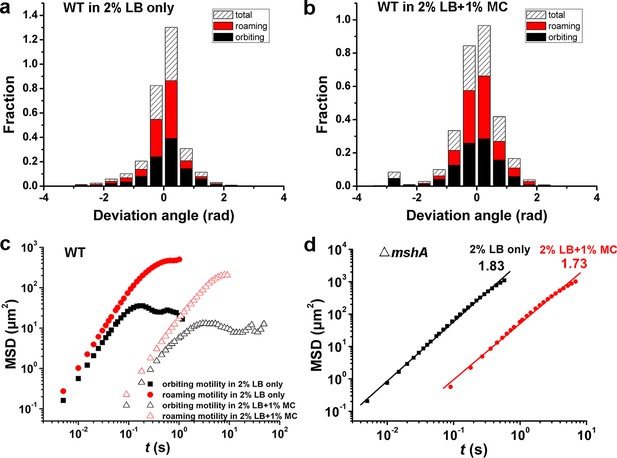
Motility characterization of WT and △mshA cells in 2% LB only and in 2% LB with 1% MC.
(a) Histograms of deviation angle for WT in 2% LB only. (b) Histograms of deviation angle for WT in 2% LB+1% MC viscous solution. Black represents orbiting motility and red represents roaming motility. (c, d) Mean square displacements (MSDs) of WT (c), △mshA (d) in 2% LB only and 2% LB+1% MC viscous solution. LB, Luria-Bertani; MC, methylcellulose; MSHA, mannose-sensitive hemagglutinin.
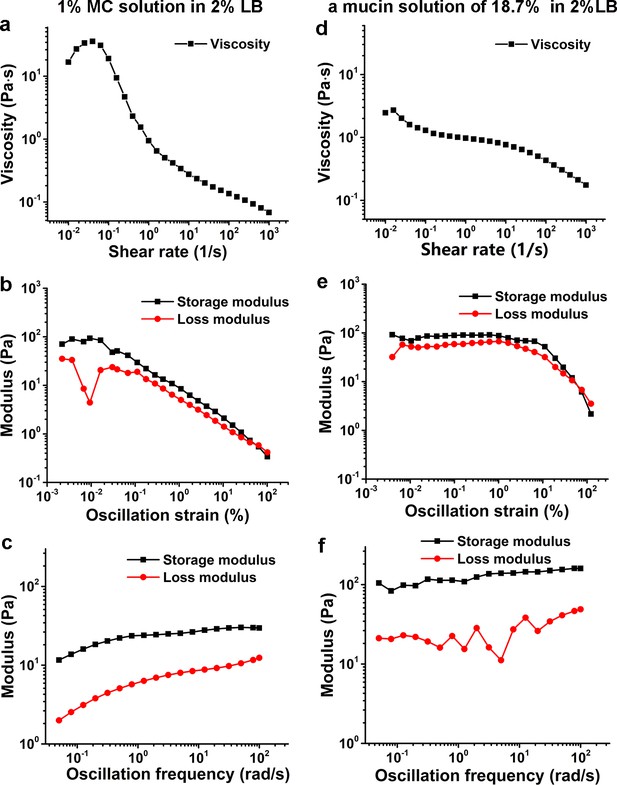
Characterization of viscoelasticity of the 1% MC solution in 2% LB (a–c) and a mucin solution of 18.7% (w/w) in 2% LB (d–f) at 26°C.
(a) and (d) show viscosity as a function of shear rate. (b) and (e) display modulus as a function of the oscillation strain, using cone-plate geometry. (c) and (f) show modulus as a function of the oscillation frequency under an oscillation strain of 0.1%, using cone-plate geometry. LB, Luria-Bertani; MC, methylcellulose.
-
Figure 6—source data 1
Source data for Figure 6a.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Source data for Figure 6b.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Source data for Figure 6c.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Source data for Figure 6d.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data4-v2.xlsx
-
Figure 6—source data 5
Source data for Figure 6e.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data5-v2.xlsx
-
Figure 6—source data 6
Source data for Figure 6f.
- https://cdn.elifesciences.org/articles/60655/elife-60655-fig6-data6-v2.xlsx
Videos
Time-lapse fluorescence imaging showing a typical roaming cell (indicated by the arrowhead) with labeled MSHA pili in 2% LB+1% MC viscoelastic medium.
This video was recorded every 390 ms for 98 s and displayed at 20 frames per second (fps).
Time-lapse fluorescence imaging showing a typical orbiting cell with labeled MSHA pili in 2% LB+1% MC medium.
This video was recorded every 130 ms for 15 s and displayed at 10 fps.
Time-lapse fluorescence imaging showing switch of pili.
When transient pauses happened, the attached pilus could be switched from one to another or more. See also Figure 2—figure supplement 2. This video was recorded every 70 ms for 10 s and displayed at 5 fps.
Time-lapse fluorescence imaging showing linear motion bent into a circular motion that is centered around the attachment point between MSHA pili and the surface, which can act as an anchor point.
This video was recorded every 130 ms for 8 s and displayed at 10 fps.
Time-lapse fluorescence imaging showing dynamic movement of a MshAT70CFlaAA106CS107C cell with the labeled flagellum and MSHA pili in 2% LB+1% MC medium.
This video was recorded every 130 ms for 13 s and displayed at 10 fps.
Time-lapse fluorescence imaging showing five MSHA pili of a WT cell stuck to the surface and kept still or fluctuated frequently.
This video was recorded every 460 ms for 25 s and displayed at 10 fps.
Time-lapse fluorescence imaging showing a typical tethered cell performing a circular motion around a fixed point with the direction of motion switched from CCW to CW.
See also Figure 4c. This video was recorded every 130 ms for 6 s and displayed at 5 fps.
Time-lapse fluorescence imaging showing different adhesion points of a pilus.
When the tip of the pilus was free (~3.5 s), the upper part of the pilus was still capable of keeping the cell adhered. This video was recorded every 130 ms for 13 s and displayed at 10 fps.
Time-lapse fluorescence imaging showing the motion evolution of the flagellum from rotating to stopping eventually.
This video was recorded every 130 ms for 10 s and displayed at 10 fps.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Parent strain (Vibrio cholerae) | C6706 | Joelsson et al., 2006 | ||
| Plasmids (DH5α λpir) | pWM91 | Metcalf et al., 1996 | Suicide vector | |
| Chemical compound, drug | Alexa Fluor 488 C5 Maleimide | Thermo Fisher Scientific | Cat. #: A10254 | |
| Chemical compound, drug | Alexa Fluor 546 C5 Maleimide | Thermo Fisher Scientific | Cat. #: A10258 | |
| Chemical compound, drug | Methyl cellulose (MC) | Solarbio | Cat. #: M8070 | |
| Chemical compound, drug | Ficoll 400 | Yuanye Bio-Technology | Cat. #: 26873-85-8 | MW=400 kDa |
| Software | GraphPad Prism software | RRID:SCR_002798 | ||
| Software | Matlab | RRID:SCR_001622 | matlab R2015a | |
| Software | Leica LAS-X | RRID:SCR_013673 |
Strains used in this study.
| Strain | Description | Source or reference | |
|---|---|---|---|
| Parent strain (Vibrio cholerae) | C6706 SmR | Joelsson et al., 2006 | |
| ∆mshA | C6706 SmR, VC1807::CmR, mshA knockout | This study | |
| ∆flaA | C6706 SmR, VC1807::CmR, flaA knockout | This study | |
| MshAT70C | C6706 SmR, VC1807::KmR, MshAT70C | Ellison et al., 2017 | |
| FlaAA106C | C6706 SmR, VC1807::CmR, FlaAA106C | This study | |
| FlaAS107C | C6706 SmR, VC1807::CmR, FlaAS107C | This study | |
| FlaAA106CS107C | C6706 SmR, VC1807::CmR, FlaAA106CS107C | This study | |
| FlaAE332C | C6706 SmR, VC1807::CmR, FlaAE332C | This study | |
| FlaAG23C | C6706 SmR, VC1807::CmR, FlaAG23C | This study | |
| FlaAN26C | C6706 SmR, VC1807::CmR, FlaAN26C | This study | |
| FlaAN83C | C6706 SmR, VC1807::CmR, FlaAN83C | This study | |
| FlaAS325C | C6706 SmR, VC1807::CmR, FlaAS325C | This study | |
| FlaAS87C | C6706 SmR, VC1807::CmR, FlaAS87C | This study | |
| FlaAS376C | C6706 SmR, VC1807::CmR, FlaAS376C | This study | |
| FlaAV117C | C6706 SmR, VC1807::CmR, FlaAV117C | This study | |
| MshAT70C, ∆flaA | C6706 SmR, VC1807::KmR, flaA knockout | This study | |
| MshAT70C, FlaAA106C | C6706 SmR, VC1807::KmR, FlaAA106C | This study | |
| MshAT70C, FlaAS107C | C6706 SmR, VC1807::KmR, FlaAS107C | This study | |
| MshAT70C, FlaAA106CS107C | C6706 SmR, VC1807::KmR, FlaAA106CS107C | This study | |
Additional files
-
Source code 1
Source code for Figure 5a and b trajectory plots.
- https://cdn.elifesciences.org/articles/60655/elife-60655-code1-v2.zip
-
Supplementary file 1
This supplementary information contains Table S1 which lists plasmids and primers used.
- https://cdn.elifesciences.org/articles/60655/elife-60655-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60655/elife-60655-transrepform-v2.docx