Dynamic Na+/H+ exchanger 1 (NHE1) – calmodulin complexes of varying stoichiometry and structure regulate Ca2+-dependent NHE1 activation
Figures
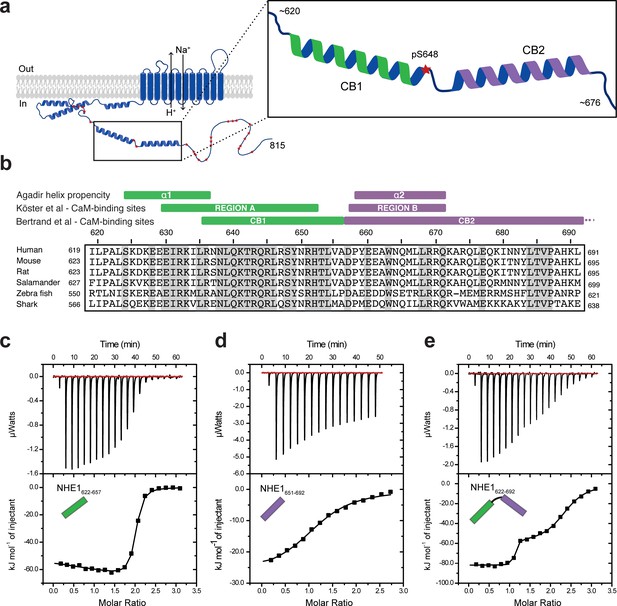
Na+/H+-exchanger:calmodulin (NHE1:CaM) interactions in vitro adopt different stoichiometries.
(a) Sketch of NHE1 (blue) in the membrane highlighting helical elements in the cytoplasmic tail and known phosphorylation sites (red stars). Magnified region depicts the two previously described CaM-binding (CB) α-helices CB1 (green) and CB2 (purple) with the phosphorylated S648. (b) Alignment of the sequences of human, mouse, rat, salamander, zebra fish, and shark NHE1 is highlighted in gray identical residues. Indicated above are structural and functional regions based on either an Agadir α-helix prediction, the CaM-binding region A (green) and region B (purple), as defined by Köster et al., 2011 or the CaM-binding region I (CB1) and II (CB2), as defined by Bertrand et al., 1994. (c)-(e). Binding NHE1 to CaM by isothermal titration calorimetry (ITC). In representative ITC experiments, we injected (c) NHE1622-657 or (d) NHE1651-692 or (e) NHE1622-692 into CaM. The upper parts show baseline-corrected raw data from the titrations, and the lower parts the normalized integrated binding isotherms with the fitted binding curves assuming a single binding event in (d) and two binding events in (c) and (e). The NHE1 peptides titrated into CaM are shown in cartoon.
-
Figure 1—source data 1
Isothermal titration calorimetry raw data and fits of triplicate experiments for the titration of calmodulin with H1, H2, and H1H2 (relating to Figure 1c–e).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig1-data1-v2.xlsx
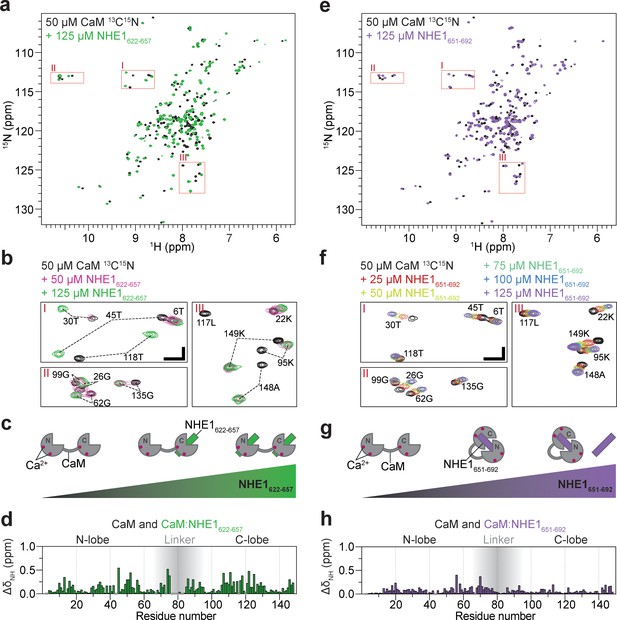
Calmodulin (CaM) interaction with NHE1622-657 (H1) and NHE1651-692 (H2).
(a) 1H,15N HSQC spectra of 50 μM 15N-CaM alone (black) and in the presence of 125 μM unlabeled NHE1622-657 (H1, green). (b) Zoom on the three highlighted regions from the spectrum in (a); 1H,15N HSQC spectra of 50 μM 15N-CaM in the presence of 0 μM (black), 50 μM (dashed red), and 125 μM (green) unlabeled NHE1622-657. The horizontal and vertical scale bars correspond to 0.1 ppm δ1H and 0.5 ppm δ15N, respectively. (c) Cartoon representation of CaM interaction with increasing amounts of NHE1622-657. (d) Chemical shift perturbations (CSP; ΔδNH) of CaM upon interaction with NHE1622-657 at saturation (125 μM NHE1622-657, 50 μM 15N-CaM, denoted 2:1 NHE1:CaM). (e) 1H,15N HSQC spectra of 50 μM 15N-CaM alone (black) and in the presence (purple) of unlabeled NHE1651-692. (f) Zoom on the three highlighted regions from the spectrum in (e); 1H,15N HSQC spectra of 15N-CaM titrated from 0 to 125 μM (black to purple) with unlabeled NHE1651-692 (H2). The horizontal and vertical scale bars correspond to 0.1 ppm δ1H and 0.5 ppm δ15N, respectively. (g) Cartoon representation of CaM interaction with increasing amounts of NHE1651-692. (h) CSP (ΔδNH) of CaM upon interaction with NHE1651-692 at saturation (125 μM NHE1651-692, 50 μM 15N-CaM). HSQC - Heteronuclear Single Quantum Coherence; DMSO - Dimethylsulfoxide; RMSD - Root-mean-square deviation; NOE - Nuclear overhauser effect; BSA - Bovine serum albumine; SDS - Sodium dodecylsulfate; IPTG - Isopropyl-β-D-thiogalactoside; DSS -2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt.
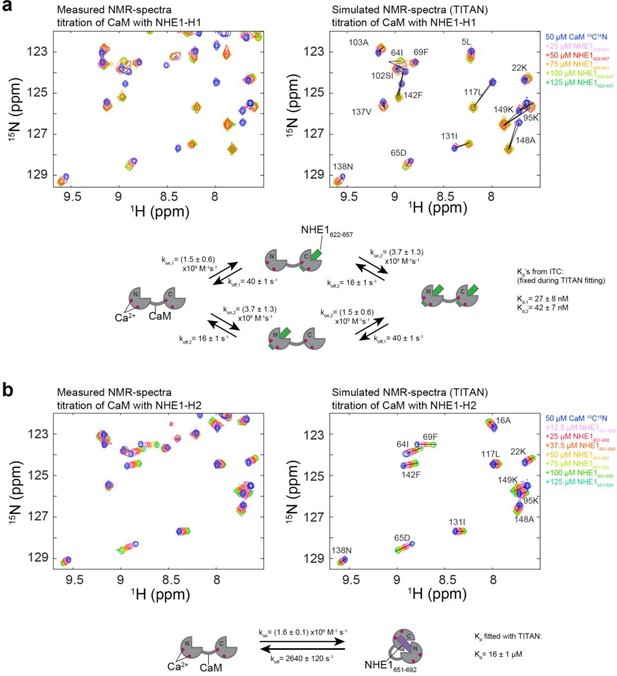
TITAN 2D nuclear magnetic resonance lineshape analysis for extraction of binding kinetics.
(a) Side-by-side comparison of measured (left) and simulated (right) 1H,15N HSQC spectra of 50 μM calmodulin (CaM) (13C,15N) titrated with up to 125 μM unlabeled NHE1622-657. The simulated spectrum was obtained by fitting the data to a four-state model as depicted in the cartoon below with the parameters for koff obtained from the fit and the Kds taken from the isothermal titration calorimetry analysis. (b) Side-by-side comparison of measured (left) and simulated (right) 1H,15N HSQC spectra of 50 μM CaM (13C,15N) titrated with up to 125 μM unlabeled NHE1651-692. The simulated spectrum was obtained by fitting the data to a two-state model as depicted in the cartoon below with the parameters koff and Kd obtained from the fit. The black lines in the simulated spectra connect the fitted signal positions of the four or two states in (a) and (b), respectively.
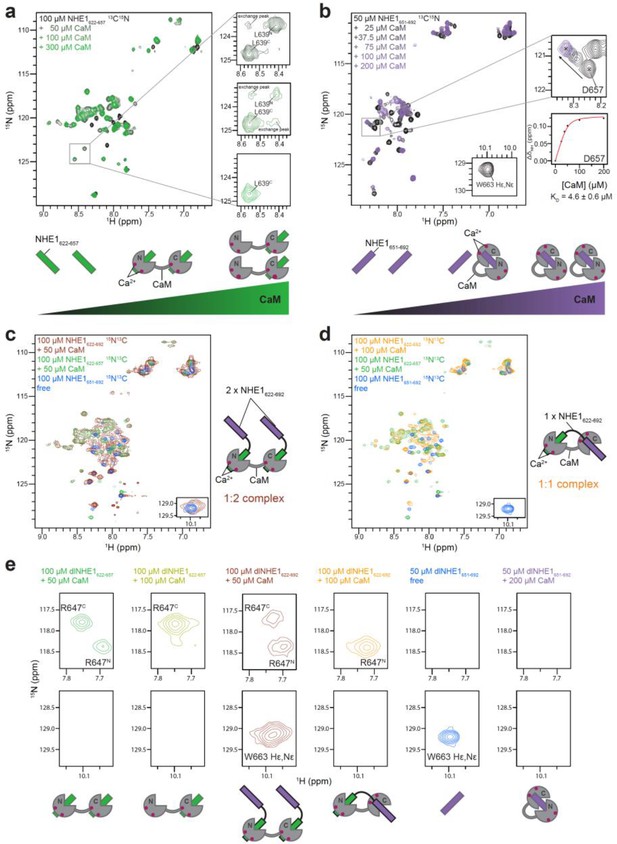
Titration of 15N-labeled H1, H2, and H1H2 with unlabeled calmodulin (CaM).
(a) 1H,15N HSQC spectra of 100 μM 15N-NHE1622-657 titrated with up to 300 μM (black to green) unlabeled CaM. The nuclear magnetic resonance (NMR) signals of L639 at three different NHE1622-657:CaM ratios; 1:1 (top), 1:2 (middle), and 1:3 (bottom), are shown to the right. N and C indicate if the highlighted residue is bound to the N- or C-lobe of CaM. Weak exchange peaks indicate intermediate/slow exchange (~0.1 s−1) of NHE1622-657 between the two CaM lobes during the experiment. The cartoon representation depicts the most prevalent NHE1622-657:CaM states as a function of increasing [CaM]. (b) 1H,15N HSQC spectra of 50 μM 15N-NHE1651-692 titrated with up to 200 μM (black to purple) with unlabeled CaM. NMR shifts of D657 shown to the right with a fit of the chemical shift perturbations as a function of CaM concentration, yielding a KD of 4.6 ± 0.6 μM. The cartoon representation depicts the most prevalent NHE1651-692:CaM states as a function of increasing [CaM]. (c) 1H,15N HSQC spectra of 100 μM 15N-NHE1622-692 in the presence of 50 μM unlabeled CaM. The signals are severely broadened and overlapping, but the resonances closely resemble signals from the spectrum of H1 bound to both lobes of CaM (green) or free H2 (blue), confirming the 1:2 (CaM:H1) complex as depicted in the cartoon to the right. (d) 1H,15N HSQC spectra of 100 μM 15N-NHE1622-692 in the presence of 100 μM unlabeled CaM. The signals closely resemble resonances from the spectrum of H1 bound to the N-lobe of CaM (green), and few signals resemble resonances from the far C-terminal region of free H2 (blue), confirming the 1:1 (CaM:H1) complex as depicted in the cartoon to the right. (e) Zooms of the 1H,15N HSQC spectra highlighting a representative resonance of H1 (R647, top row) and H2 (HεNε W663, bottom row). The cartoons depict the most prevalent NHE1:CaM state at the given conditions. dl denotes double-labeled (13C,15N).
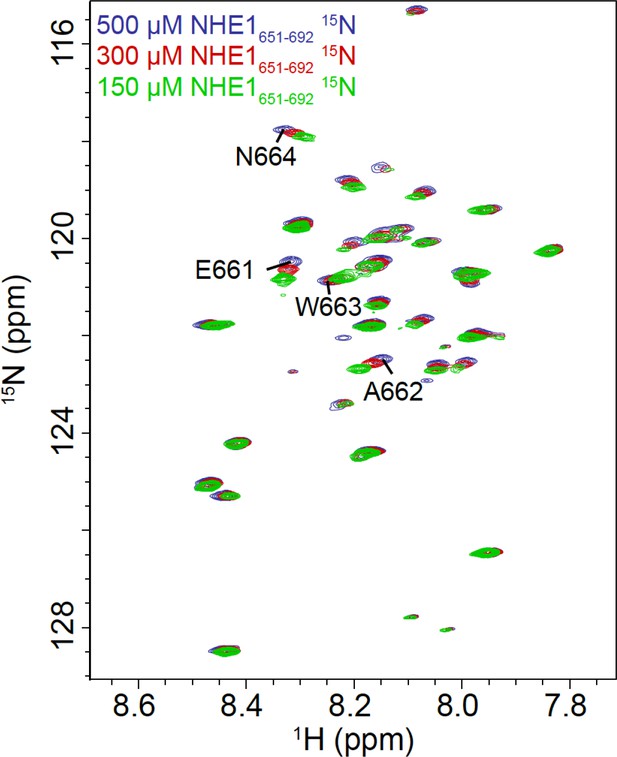
Concentration dependence of the nuclear magnetic resonance chemical shift of Na+/H+-exchanger (NHE1) H2.
1H,15N HSQC spectra of NHE1 H2 at different concentrations show concentration-dependent changes in chemical shifts, indicating a weak self-association of the peptide at high concentrations. The most affected resonances are highlighted and correspond to the stretch of residues E661–N664 of NHE1.
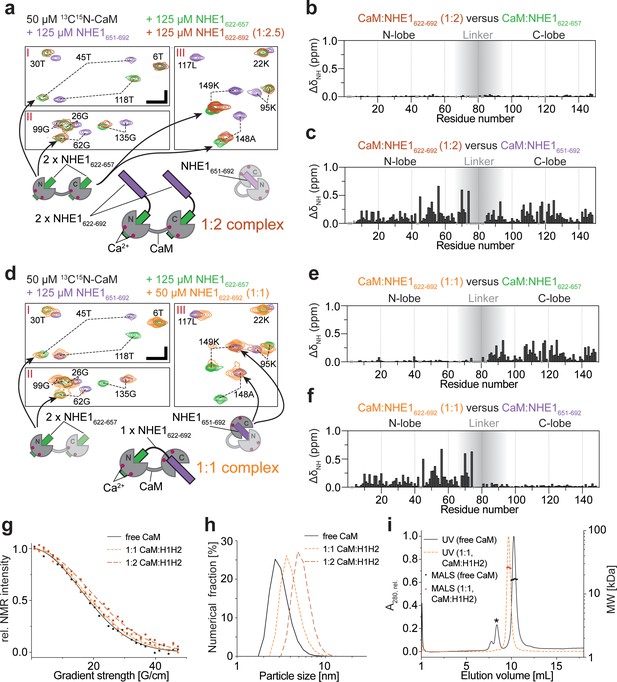
Calmodulin (CaM) interaction with NHE1622-692 (H1H2) at two different stoichiometries.
(a) The 1:2 CaM:NHE1622-692 (H1H2) complex (molar ratio 1:2). Three regions of the 1H,15N HSQC spectra of 50 μM 15N-CaM with 125 μM NHE1622-657 (green), 125 μM NHE1651-692 (purple), or 125 μM NHE1622-692 (orange) are shown. The horizontal and vertical scale bars correspond to 0.1 ppm δ1H and 0.5 ppm δ15N, respectively. The complexes giving rise to the indicated peaks are depicted in the cartoons below (right and left), with the 1:2 complex in the center. (b) Chemical shift perturbations (CSPs) (ΔδNH) between the 1:2 CaM:NHE1622-692 complex and the 1:2 CaM:NHE1622-657 complex (dark red and green spectra in a). The gray shade illustrates the linker region of CaM. (c) CSPs (ΔδNH) between 1:2 CaM:NHE1622-692 and 1:1 CaM:NHE1651-692 (dark red and purple spectra in a). (d) The 1:1 CaM:NHE1622-692 complex. Three regions of the 1H,15N HSQC spectra of 50 μM 15N-CaM in the presence of 125 μM NHE1622-657 (green), 125 μM NHE1652-692 (purple), or 50 μM NHE1622-692 (orange). The horizontal and vertical scale bars correspond to 0.1 ppm δ1H and 0.5 ppm δ15N, respectively. Smaller cartoons represent the 1:2 CaM:NHE1622-657 complex (left) and 1:1 CaM:NHE1651-692 complex (right) and the 1:1 CaM:NHE1622-692 complex identified from overlapping peaks in the 1H,15N HSQC spectra. (e) CSPs (ΔδNH) between 1:1 CaM:NHE1622-692 and 1:2 CaM:NHE1622-657 (orange and green spectrum in d). (f) CSPs (ΔδNH) between 1:1 CaM:NHE1622-692) and 1:1 CaM:NHE1651-692 (orange and purple spectrum in (d). The chemical shifts are available in BMRB accession code 34521. (g) Pulsed field gradient nuclear magnetic resonance diffusion measurements of 40 μM 15N-CaM without ligand (black), with 40 μM NHE1622-692 (1:1, orange), and with 80 μM NHE1622-692 (1:2, red). (h) Number-averaged particle size distribution from dynamic light scattering at a total protein concentration of 1 mg/mL in all three cases. (i) Size exclusion chromatography coupled to multiple angle light scattering elution profiles of free CaM (black, *bovine serum albumin from calibration run) and the 1:1 complex of CaM:H2 (orange).
-
Figure 3—source data 1
Pulsed field gradient nuclear magnetic resonance raw data (relating to Figure 3g).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Raw dynamic light scattering correlation data (relating to Figure 3h).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Raw size exclusion chromatography coupled to multiple angle light scattering data (relating to Figure 3i).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig3-data3-v2.xlsx
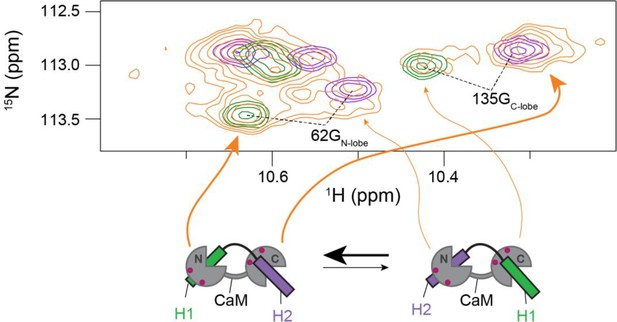
Minor population at a stoichiometry of 1:1 (CaM:H1H2).
Zoom on a representative region of the 1H,15N HSQC spectrum of calmodulin (CaM) in complex with either H1 (green), H2 (purple), or H1H2 at a stoichiometry of 1:1 (orange). The signals are drastically broadened in the case of H1H2, indicating conformational exchange between different states under these conditions. From the signal intensity distribution between these states, the main state in solution has H1 bound to the N-Lobe (overlap between orange and green peaks for 62G) and H2 bound to the C-lobe (overlap between orange and purple peaks for 135G). However, the minor population present agrees with the opposite architecture, where H1 is bound to the C-Lobe and H2 bound to the N-lobe. The two 1:1 complexes that are in accordance with these architectures are depicted in the cartoon in the bottom.
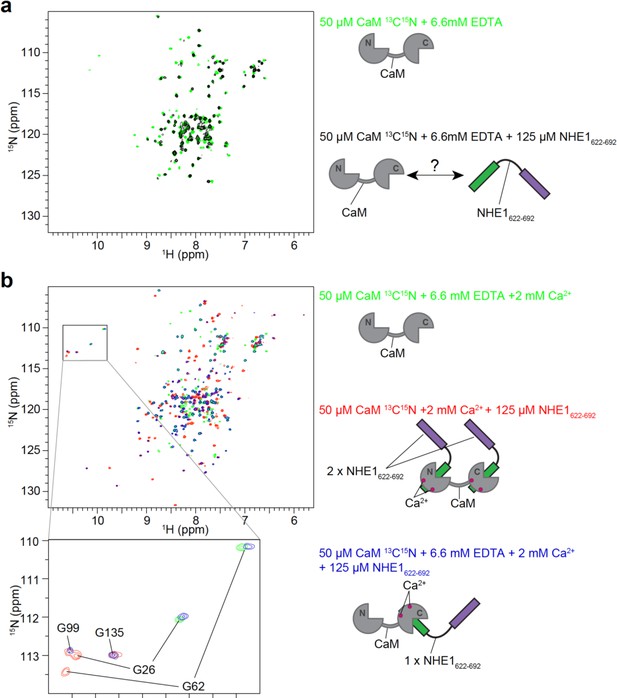
Influence of ethylenediaminetetraacetic acid (EDTA) on calmodulin (CaM) interaction with NHE1622-692.
(a) In the absence of Ca2+, small chemical shift perturbations and line broadening of apo-CaM were observable after addition of excess NHE1 H1H2, showing that NHE1:CaM complex formation can also occur in the absence of Ca2+. (b) In the presence of Ca2+ and excess EDTA, free CaM is stripped from Ca2+ as can be seen from the similarity of the green 1H,15N HSQC spectra in (a) and (b). NHE1 H1H2 increases the Ca2+ affinity of the C-lobe of CaM, so that the main observable state in solution has resonances of the N-lobe closely resembling the apo state (G26 and G62), while resonances of the C-lobe resemble the state where the C-lobe is bound to Ca2+ and H1 (G99 and G135).
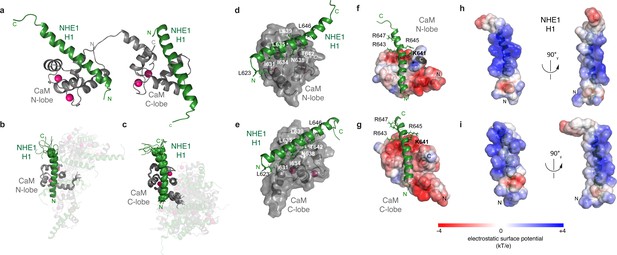
The ternary complex between calmodulin (CaM) and two Na+/H+-exchanger (NHE1) H1s.
(a) Cartoon of the complex showing the two lobes of CaM in gray and the two NHE1 H1 helices in green. Ca2+ ions are shown in magenta. (b, c) Superimposition of the 10 lowest energy structures of the calculated ensemble aligned by (b) the CaM N-lobe with NHE1 H1 bound or (c) the CaM C-lobe with the second NHE1 H1 bound, showing mobility around the CaM linker. (d, e) Hydrophobic residues of NHE1 H1 forming contacts to CaM in the (d) N-lobe and (e) C-lobe, respectively. Electrostatic complementarity between Ca2+-CaM and NHE1-H1 in the (f) CaM N-lobe and the (g) CaM C-lobe shown by the surface potential of CaM and (h, i) for the two correspondingly bound H1 helices (PDB accession code 6zbi).
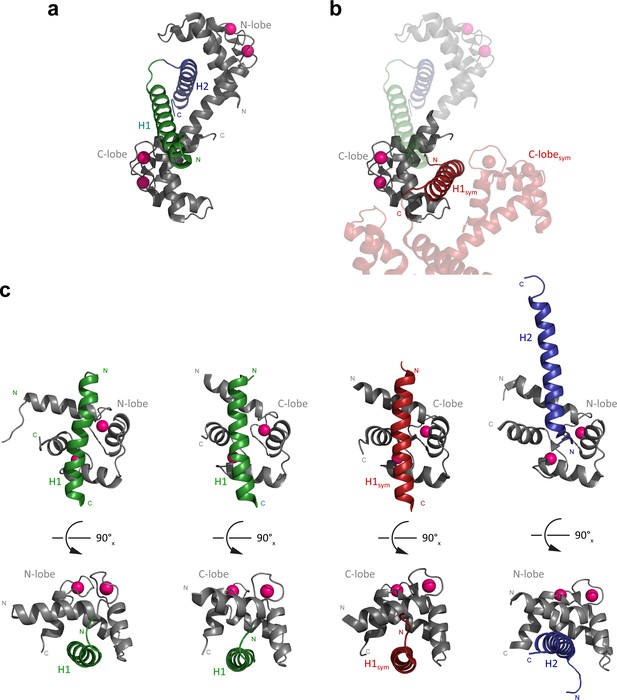
Comparison of the crystal structure (PDB: 2ygg) and the nuclear magnetic resonance (NMR) structure presented here (PDB: 6zbi).
(a) Crystal structure of the 1:1 Na+/H+-exchanger:calmodulin (NHE1:CaM) complex (PDB code: 2ygg) with the N-lobe bound to H2 and the backside of the C-lobe bound to H1 (PBD code: 6zbi). (b) Same as in (a) with a symmetry-related molecule of the crystal lattice included (red), showing that the hydrophobic cleft of the CaM C-lobe is occupied with H1 of this symmetry-related complex (H1sym). (c) Side-by-side comparison of the structures of CaM N-lobe bound to H1 (NMR structure, left), CaM C-lobe bound to H1 (NMR structure, middle left), CaM C-lobe bound to H1sym (crystal structure, middle right), and the CaM N-lobe bound to H2 (crystal structure, right). The three structures with H1 bound to the hydrophobic cleft of a CaM lobe are very similar to Cα-RMSDs of 2.3 Å (N-lobe + H1NMR:C-lobe + H1NMR), 1.8 Å (N-lobe + H1NMR:C-lobe + H1sym,X-ray), and 1.1 Å (C-lobe + H1NMR:C-lobe + H1sym,X-ray), respectively.
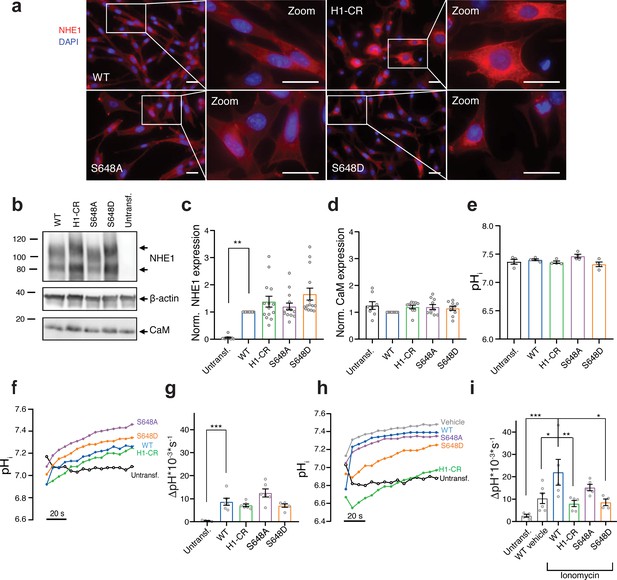
S648 phosphorylation status regulates Ca2+-dependent Na+/H+-exchanger (NHE1) activation.
(a) Representative immunofluorescence (IF) images of PS120 cells stably expressing wild-type (WT) or variant hNHE1 and stained for NHE1 (red) and DAPI (blue) to evaluate NHE1 expression and localization. Images were taken at ×60 magnification on an inverted Olympus Cell Vivo IX83 microscope, and areas marked with white squares are magnified. Scale bars represent 20 μm (n = 5). IF images of calmodulin (CaM) in all cell lines are shown in Figure 5—figure supplement 1. (b) Representative western blot of the four variant PS120 cell lines as well as untransfected PS120 cells blotted for NHE1 and endogenous CaM. For NHE1, the lower band corresponds to the immature protein and the upper band to fully glycosylated NHE1. β-actin was used as loading control (n = 8–14). (c, d) Total NHE1 (n = 11–14) or CaM (n = 8–10) expression was quantified using ImageJ and normalized for each variant to the loading control and then to the expression in the WT NHE1 cell line. (e) Steady-state pHi measured in Ringer solution in the absence of HCO3- for each cell line. (n = 4). (f–i) Representative traces of BCECF-AM fluorescence converted to pHi for the four NHE1 variant cell lines and untransfected PS120 cells illustrating the time course of pHi recovery in Ringer in the absence of HCO3- after an NH4Cl-prepulse-induced acid load without (f, g) or with (h, i) the addition of 5 μM ionomycin or vehicle (EtOH). The recovery rates as ∆pH*10−3*s−1 of each cell line are depicted in (g) (n = 4–6) and (i) (n = 4–6). The rate of pHi recovery is expressed as ΔpH/s because steady-state pHi and acidification were similar between cell lines, allowing direct comparison. It should be noted that the variant NHE1 proteins 1K3R1D3E, S648A, and S648D harbor a spontaneous F395Y mutation. The expression and pHi recovery rates of two WT NHE1s (two different clones) did not differ from that of F395Y (Figure 5—figure supplement 2). Data represent mean ± SEM. For (d, g, i), one-way analysis of variance with Dunnett’s post-test comparing all cells to WT was carried out. For (c, e), Kruskal–Wallis non-parametric test comparing all cells to WT was carried out. Exact p-values stated for each group compared to WT were as follows: (c) untransf. p = 0.002, H1-CR p > 0.9999, S648A p > 0.9999, S648D p = 0.1054; (d) untransf. p = 0.8894, H1-CR p = 0.6035, S648A p = 0.5309, S648D p = 0.8852; (e) untransf. p > 0.9999, H1-CR p > 0.9999, S648A p > 0.9999, S648D p = 0.6911; (g) untransf. p = 0.0009, H1-CR p = 0.7781, S648A p = 0.1097, S648D p = 0.7652; (i) untransf. p = 0.0007, WT vehicle p = 0.0254, H1-CR p = 0.0089, S648A p = 0.3367, S648D p = 0.0195. *, ** and *** denote p < 0.05, p < 0.01 and p < 0.001, respectively.
-
Figure 5—source data 1
Raw quantification data for Na+/H+-exchanger western blots (relating to Figure 5b, c).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Raw quantification data for calmodulin western blots (relating to Figure 5b, d).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Raw western blots (relating to Figure 5b).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig5-data3-v2.pptx
-
Figure 5—source data 4
pHi data summaries (relating to Figure 5e–i).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig5-data4-v2.xlsx
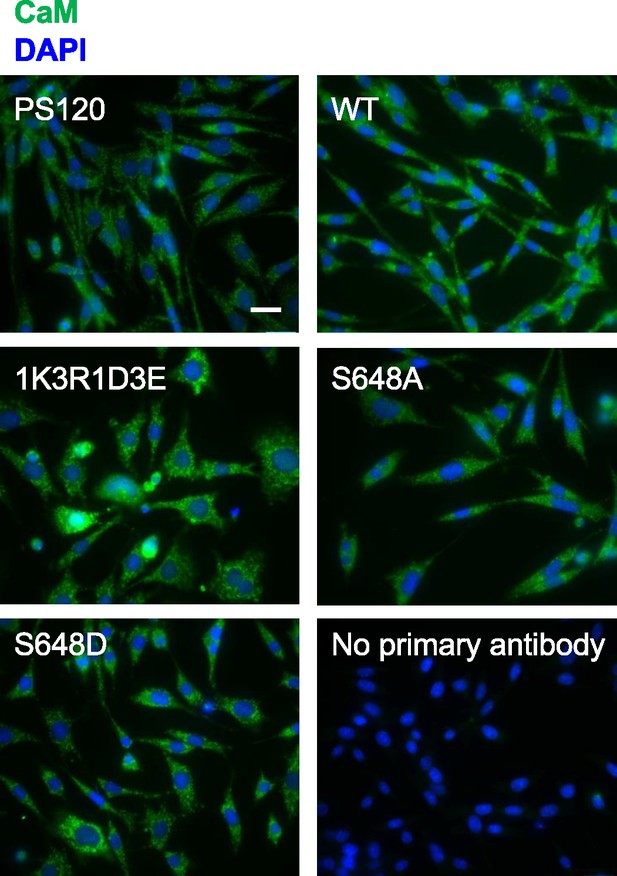
Immunofluorescence staining of endogenous calmodulin (CaM) in PS120 cell lines stably expressing wildtype (WT) or variant hNHE1.
Representative immunofluorescence images of PS120 cells stably expressing WT or variant hNHE1 and stained for CaM (green) and DAPI (blue) to evaluate endogenous CaM expression and localization. Images were taken at ×60 magnification on an Olympus Cell Vivo IX83 microscope and are from the same cell passages as the pictures shown in Figure 5a (main text). Scale bars represent 20 μm (n = 5).
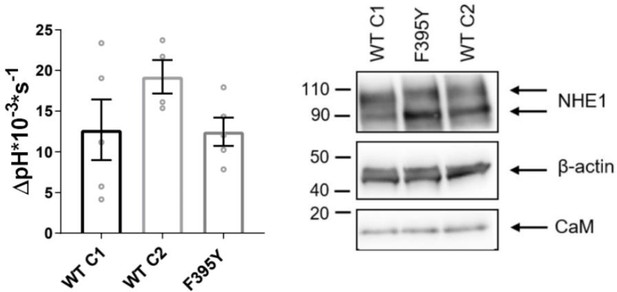
Comparison of expression levels and pHi recovery rates of PS120 cells expressing two wildtype (WT) and F395Y hNHE1.
Left: Recovery rates as ∆pH * 10−3 * s−1 of three PS120 cell lines stably expressing WT hNHE1 (WT C1 and C2) or hNHE1 with a spontaneous F395Y mutation, which is present in the cell lines 1K3R1D3E, S648A, and S648D. The WT C1 cell line was used as ‘WT’ in all experiments in the main text. pHi recovery rate was measured in Ringer solution after an NH4Cl-prepulse-induced acid load (n = 4–5). One-way analysis of variance with Dunnett’s post-test comparing cell lines to WT C1 was carried out. Right: Representative western blot of these PS120 cell lines blotted for Na+/H+-exchanger (NHE1) and endogenous CaM. β-Actin was used as loading control (n = 5).
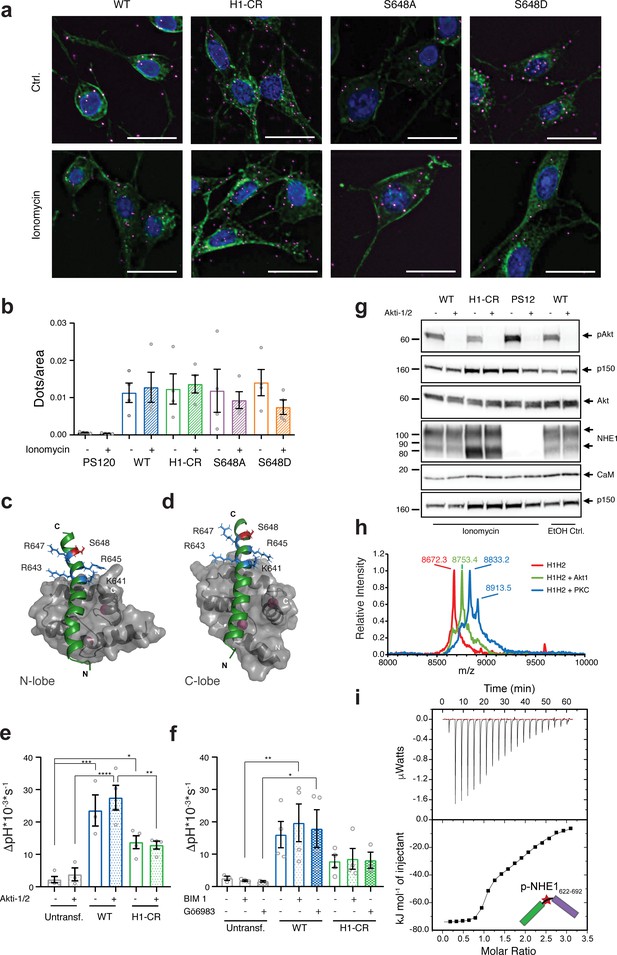
Cellular Na+/H+-exchanger:calmodulin (NHE1:CaM) proximity is resistant to H1-CR and S648 NHE1 mutations, and Akt and PKC activity does not significantly impact ionomycin-induced NHE1 activity.
(a) Proximity ligation assay (PLA) of wildtype (WT) and variant hNHE1 in PS120 cells, with addition of vehicle (EtOH) or 5 μM ionomycin as shown (n = 4–7). Negative controls are shown in Figure 6—figure supplement 1 PLA signal is magenta, co-staining for NHE1 is green, and nuclei (DAPI) are blue. Scale bars represent 20 μm. (b) Quantification of PLA signal in ImageJ after background subtraction and normalization to cell area. Approximately 400 cells were counted per condition per n (n = 4). Two-way analysis of variance (ANOVA) with Tukey’s post-test comparing all conditions to one another showed no significant differences between groups. As there are 45 exact p-values for this figure, these are available upon request. (c, d) Structures of the individual lobes of the (2:1) NHE1 H1-CaM complex highlighting the position of the mutated residues and S648 in (c) the N-lobe and (d) the C-lobe. (e) Recovery rates as ∆pH * 10−3 * s−1 of each cell line with addition of 10 μM Akti-1/2 or vehicle (DMSO) (n = 4). Two-way ANOVA with Tukey’s post-test was used to compare cell types to one another within each treatment group. Exact p-values were as follows: (e) Ctrl group: untransf. WT p = 0.0001, untransf. H1-CR p = 0.0141, WT H1-CR p = 0.0559; Akti-1/2 group: untransf. WT p < 0.0001, untransf. H1-CR p = 0.0792, WT H1-CR p = 0.0024. (f) As in (e), except that where indicated cells were treated with 5 µM of bisindolylmaleimide I (BIM) or 3 µM of Gö6983 (Go) (n = 4) or DMSO (vehicle). Exact p-values were as follows: Ctrl group: untransf. WT p = 0.0501, WT H1-CR p = 0.2291; BIM I group: untransf. WT p < 0.0091, WT H1-CR p = 0.0844; Go group: untransf. WT p < 0.0370, WT H1-CR p = 0.1823. (g) Western blot assessing the inhibition of Akt activity by Akti-1/2, assessed as phosphorylation of Ser473 of Akt (pAkt) compared to total Akt, in PS120 cells expressing WT and variant NHE1 (n = 4). The blot also illustrates endogenous levels of CaM and exogeneous NHE1. p150 was used as a loading control. NHE1 and CaM were detected on the same membrane as Akt. (h) Matrix-assisted laser desorption/ionization time of flight mass spectrometry analyses of NHE1-H1H2 before (red) and after addition of Akt (green) or protein kinase C (blue). (i) Representative binding experiments of Akt-phosphorylated NHE1-H1H2 titrated into CaM using isothermal titration calorimetry. The upper part shows baseline-corrected raw data from the titrations, and the lower part the normalized integrated-binding isotherms with the fitted binding curves assuming a two-site binding event. The peptide titrated into CaM is shown in cartoon, and the star indicates the phosphorylated S648. Data represent mean ± SEM. *, **, *** and **** denote p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively.
-
Figure 6—source data 1
Raw Na+/H+-exchanger calmodulin proximity ligation assays data (relating to Figure 6a, b).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig6-data1-v2.xlsx
-
Figure 6—source data 2
pHi data summaries (relating to Figure 6e, f).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Raw western blots (relating to Figure 6g).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig6-data3-v2.pptx
-
Figure 6—source data 4
Isothermal titration calorimetry raw data and fits of triplicate experiments for the titration of calmodulin with pS648 H1H2 (Figure 6h).
- https://cdn.elifesciences.org/articles/60889/elife-60889-fig6-data4-v2.xlsx
Lack of proximity ligation assays (PLA) signal in PS120 cells lacking Na+/H+-exchanger (NHE1).
Representative images of PS120 control cells lacking NHE1 and subjected to PLA with both NHE1 and calmodulin antibodies, and addition of vehicle (EtOH) or 5 μM ionomycin as indicated (n = 4–5). Images correspond to those shown in Figure 6A. The PLA signal is magenta, co-staining for NHE1 is green, and nuclei (DAPI) are blue. As seen, negligible PLA signal is observed in cells lacking NHE1, confirming assay specificity. Scale bars represent 20 μm.
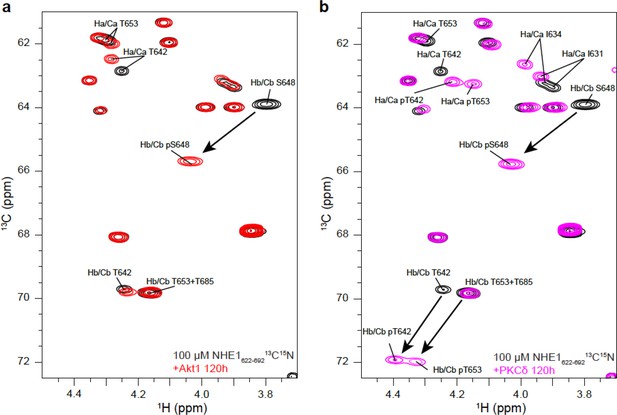
Identification of phosphorylated residues in Na+/H+-exchanger (NHE1) H1H2 by nuclear magnetic resonance.
13C,1H HSQC spectra of NHE1-H1H2 before (black) and after 120 hr of phosphorylation with Akt1 (a, red) or protein kinase C (PKC)δ (b, pink). The arrows highlight the large shift of the HβCβ peaks upon phosphorylation of residue S648 by (a) Akt1 and residues T642, S648, and T653 by (b) PKCδ.
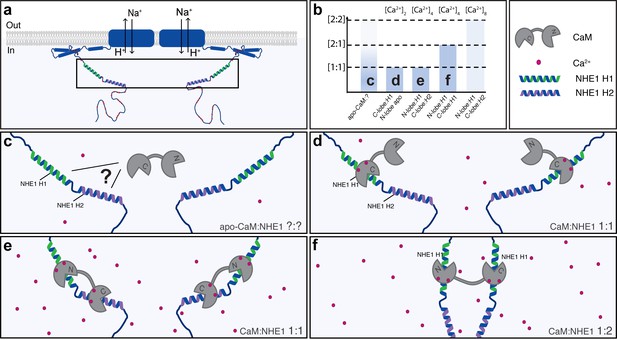
A dynamic view on Na+/H+-exchanger:calmodulin (NHE1:CaM) interaction.
Multiple possible complexes exist depending on the [NHE1], [CaM], and [Ca2+], including ternary complexes, suggesting that CaM may also contribute to NHE1 dimerization. (a) Schematic of NHE1 with a zoom on the CaM-binding region. The orientation of the monomers (N-terminal to N-terminal) in the NHE1 dimer is arbitrarily chosen. (b) Overview of the multiple complexes between CaM and NHE1 and their dependence on Ca2+ (top). The indices (c–f) refer to the states shown in cartoon below. (c) Interaction between apo-CaM and NHE1 is seen, but its structure is unknown (indicated by a question mark). (d) At low Ca2+ levels, NHE1-H1 is bound to the Ca2+-loaded C-lobe of CaM, whereas the N-lobe is in its apo-state. (e) At high Ca2+ levels, and high NHE1:Ca2+-CaM ratio, a 1:1 complex is formed with H1 bound to the N-lobe and H2 bound to the C-lobe of CaM. (f) At high Ca2+ levels, and low NHE1:Ca2+-CaM ratio, a 1:2 complex is formed, where NHE1-H1 is bound to both the N-lobe and the C-lobe of CaM.
Tables
Thermodynamic parameters of NHE1:CaM interactions.
| ΔH1 (kJ/mol) | TΔS1 (kJ/mol) | Kd,1 (nM) | n1 | ΔH2 (kJ/mol) | TΔS2 (kJ/mol) | Kd,2 (nM) | n2 | |
|---|---|---|---|---|---|---|---|---|
| H1 (one site) | −60 ± 3 | −17 ± 2 | 23 ± 5 | 1.92 ± 0.06 | – | – | – | – |
| H1 (two site) | −39 ± 11 | 4 ± 10 | 27 ± 8 | 0.92 ± 0.05 | −83 ± 8 | −41 ± 8 | 42 ± 6 | 0.98 ± 0.01 |
| H2 (one site) | −29 ± 4 | 0 ± 3 | 8900 ± 600 | 1.15 ± 0.08 | – | – | – | – |
| H1H2 (two site) | −84 ± 5 | −29 ± 5 | 0.27 ± 0.07 | 1.04 ± 0.05 | −62 ± 3 | −26 ± 3 | 560 ± 10 | 1.22 ± 0.06 |
| H1H2 pS648 (two site) | −75 ± 3 | −27 ± 2 | 5 ± 4 | 0.93 ± 0.02 | −58 ± 7 | −26 ± 8 | 2700 ± 700 | 1.16 ± 0.06 |
| NHE1: Na+/H+-exchanger; CaM: calmodulin. | ||||||||
Exchange kinetics from 2D NMR lineshape analysis.
| kon,1 (M−1 s−1) | koff,1 (s−1) | Kd,1 (nM) | kon,2 (M−1 s−1) | koff,2 (s−1) | Kd,2 (nM) | |
|---|---|---|---|---|---|---|
| H1 (two site) | (1.5 ± 0.6) × 109 | 40 ± 5 | 27* | (3.7 ± 1.3) × 108 | 16 ± 3 | 42* |
| H2 (one site) | (1.6 ± 0.1) × 108 | 2600 ± 100 | 16000 ± 1000 | – | – | – |
-
*Kds taken from ITC analysis, kept constant during fitting.
NMR: nuclear magnetic resonance; ITC: isothermal titration calorimetry.
Size evaluation of CaM-H1H2 complexes at different stoichiometries.
| DNMR (1010 m2 s−1) | Rh,NMR (Å) | Number; volume; intensity averaged Rh, DLS (Å) | Elution volumeSEC (mL) | MWMALS (kDa) | |
|---|---|---|---|---|---|
| Free CaM | 1.57 ± 0.05 | 20.8 ± 0.8 | 16 ± 1; 20 ± 1; 27 ± 2 | 10.27 ± 0.05 | 17.0 ± 0.9 |
| CaM:H1H2 (1:1) | 1.43 ± 0.04 | 22.8 ± 0.7 | 21 ± 2; 26 ± 2; 34 ± 2 | 9.68 ± 0.05 | 25.0 ± 1.6 |
| CaM:H1H2 (1:2) | 1.20 ± 0.05 | 27.2 ± 0.8 | 27 ± 2; 31 ± 2; 37 ± 2 | –* | –* |
-
*Not obtainable due to interaction of H1H2 with the SEC column material.
CaM: calmodulin; SEC: size exclusion chromatography; MALS: Multi-angle light scattering.
Structural statistics (PDB accession code 6zbi).
| Nuclear magnetic resonance restraints | |
|---|---|
| Distance, short-range |i–j| ≤ 1 | 1019 |
| Distance, medium-range 1 < |i–j| < 5 | 361 |
| Distance, long-range |i–j| ≥ 5 | 224 |
| Distance, intermolecular | 213 |
| Dihedral restraints (Talos-N): | 336 |
| Average pairwise RMSD | |
| N-lobe + H1N 8.73 (chain A), 623.644 (chain C) | Backbone: 0.77 ± 0.16 Å |
| Heavy atoms: 1.42 ± 0.14 Å | |
| C-lobe + H1C 83.148 (chain A), 623.644 (chain B) | Backbone: 0.83 ± 0.18 Å |
| Heavy atoms: 1.57 ± 0.17 Å | |
| Ramachandran statistics | |
| Most favored regions | 95.7% |
| Additionally allowed regions | 3.7% |
| Generously allowed regions | 0.3% |
| Disallowed regions | 0.3% |





