Physically asymmetric division of the C. elegans zygote ensures invariably successful embryogenesis
Figures
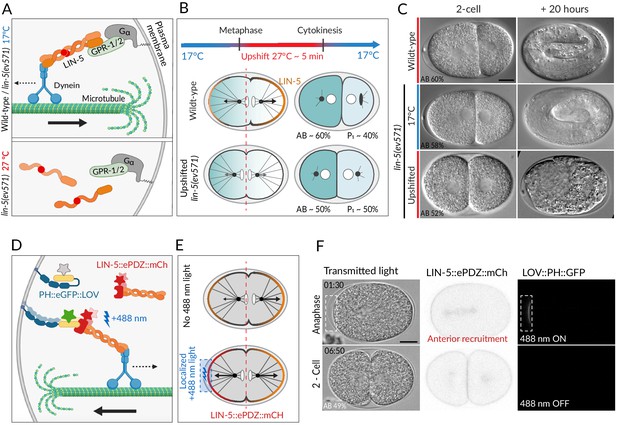
Two methods for equalizing first cell division in C. elegans embryos.
(A) The ternary complex comprising Gα, GPR-1/2, and LIN-5 dimers tethers a dynein complex (only the motor protein is represented here) to the cell cortex, thereby generating pulling forces on astral microtubules (top). The lin-5(ev571) temperature-sensitive mutant encodes a protein with a 3-amino acid insertion (red disc) in the coiled-coil domain; this mutant protein cannot dimerize or generate pulling forces at the restrictive temperature (bottom). (B) Schematic of transient temperature upshift during the first division of the C. elegans zygote to equalize AB and P1 sizes using lin-5(ev571). Such transient upshift has no impact on the asymmetry of the first unequal division in the wild type, yielding AB and P1 cells corresponding to ~60% and ~40% of embryo size, respectively (middle), due to asymmetric net cortical pulling forces acting on the spindle poles (arrows), stemming from the posterior enrichment of the ternary complex (orange). Cortical pulling forces are impaired upon transient upshift of lin-5(ev571) embryos, resulting in equalized first division (bottom). The red dashed line indicates the embryo center. Embryos are oriented with anterior to the left here and all figure panels. (C) Differential interference contrast (DIC) microscopy images from time-lapse recordings at the 2-cell stage (left) and 20 hr thereafter (right). Upshifted wild-type embryo (top) or lin-5(ev571) embryo kept at 17°C (middle) divide unequally and develop into 3-fold larvae that later hatch, while upshifting lin-5(ev571) embryos results in equalized division (bottom), which can lead to embryonic lethality. In this and other figure panels, scale bar is 10 µm and relative AB cell size at the 2-cell stage is indicated. See also Videos 1–3. (D) Schematic of optogenetic-mediated LIN-5::ePDZ::mCherry recruitment to the anterior cortex by localized activation of PH::eGFP::LOV interaction with a 488 nm laser. A helix in the LOV domain unfolds upon illumination, allowing binding of ePDZ. Star represents fluorescent protein fusion (GFP: gray to start with, green upon illumination; mCherry: red). (E) Embryos expressing LIN-5::ePDZ::mCherry and PH::eGFP::LOV divide unequally without 488 nm exposure due to asymmetric localization of LIN-5 (top). Optogenetic-mediated recruitment of LIN-5::ePDZ::mCherry to the anterior cortex during mitosis (blue rectangle) results in balanced net pulling forces on the two spindle poles and equal first division (bottom). (F) Images from time-lapse recording of optogenetic-mediated first division equalization. LIN-5::ePDZ::mCherry was recruited to the anterior cortex from anaphase onset until cytokinesis completion by scanning the 488 nm laser in the indicated rectangular region. Note that endogenous LIN-5 is tagged with ePDZ::mCherry, hence resulting in the fusion protein being present also on the spindle, the centrosomes, and the posterior cortex during anaphase. Note that that this strain also expresses GFP::TBB-2, which is not visible in the small illuminated portion of the cortex. Time indicated in min:s. See also Video 4.

Cell size measurement method and precision.
(A) The surfaces of AB and P1 were determined in the mid-plane image with the largest cross-sectional area. Cell boundaries were manually traced using the GFP::PH signal labeling the plasma membrane with the polygon tool in Fiji and the enclosed surface measured. Scale bar is 10 µm in this and all figure supplement panels. (B) Mean variation coefficient of repeated manual measurements of AB cell surface in 25 embryos, corresponding to the technical error, which is 0.66% ± 0.27 SD on average; each embryo was independently measured 3–5 times. (C) Cell volumes were segmented automatically using the Interactive Morphological 3D reconstruction plugin in Fiji, and the relative AB volume with respect to the sum of AB and P1 volumes calculated. (D) Correlation of relative AB surface and AB volume. Solid black line: linear model fitted to the data; gray dashed lines: 95% confidence interval for the fit. (E) Mean percentage error of relative AB size based on the difference between surface and volume measurements as a function of relative AB size, showing that there is no significant correlation between the two. Same embryos as in D. See also Figure 1—figure supplement 1—source data 1 and Figure 1—figure supplement 1—source code 1.
-
Figure 1—figure supplement 1—source code 1
Comparison of 3D and 2D cell size measurements.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig1-figsupp1-code1-v3.zip
-
Figure 1—figure supplement 1—source data 1
Matched 3D and 2D cell size measurements at the 2-cell stage.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig1-figsupp1-data1-v3.xlsx
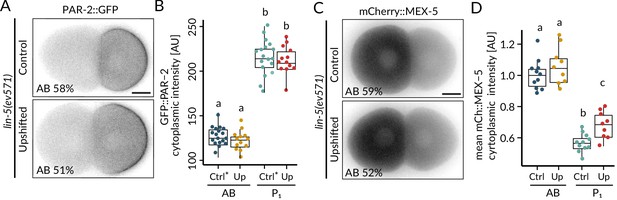
Upshifted lin-5(ev571) embryos are polarized.
Distribution (A) and corresponding quantification of normalized cytoplasmic intensity (B) of endogenously tagged PAR-2::GFP in control (Ctrl) and upshifted lin-5(ev571) embryos (Up). Note that in this case the control lin-5(ev571) condition included 13 non-upshifted embryos and 5 embryos upshifted in the early 2-cell stage (denoted with *). Distribution (C) and corresponding quantification of normalized cytoplasmic intensity (D) of endogenously tagged mCherry::MEX-5 in control (Ctrl) and upshifted lin-5(ev571) embryos (Up). See also Figure 1—figure supplement 2—source data 1 and Figure 1—figure supplement 2—source code 1.
-
Figure 1—figure supplement 2—source code 1
Analysis of PAR-2::GFP and mCherry::MEX-5 intensities.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig1-figsupp2-code1-v3.zip
-
Figure 1—figure supplement 2—source data 1
PAR-2::GFP and mCherry::MEX-5 intensities.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig1-figsupp2-data1-v3.xlsx
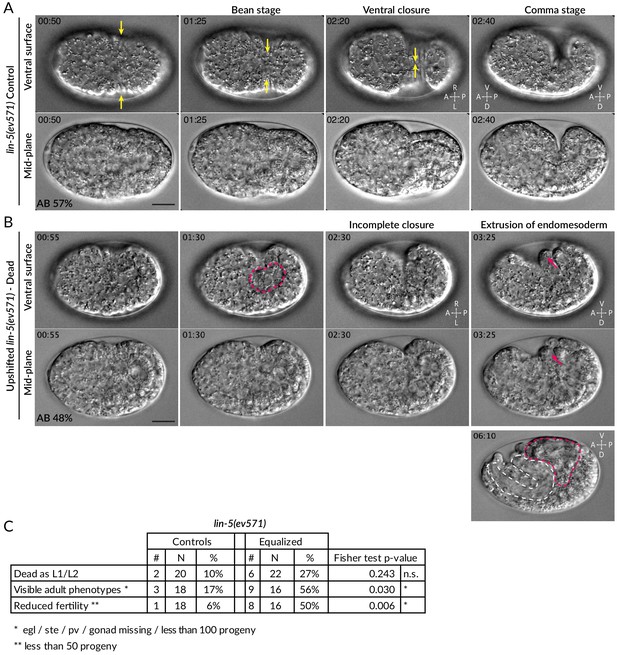
Ventral rupture in equalized embryos.
(A) Four time points from DIC recording of hypodermal ventral closure in control lin-5(ev571) embryo upshifted at the 2-cell stage. Hypodermal cells migrate from both sides towards the ventral midline (yellow arrows indicate movement), where they meet and seal the embryo in a continuous skin layer. The embryo flips on its side at the comma stage when the elongation phase of morphogenesis begins. In this and other figure supplement panels, relative AB cell size at 2-cell is indicated. See also Video 5. (B) Incomplete ventral closure (red dashed area) in upshifted lin-5(ev571) embryo that will later die, with extrusion of internal tissues (red arrows). The resulting phenotype at 06:10 shows pharyngeal (white dashed area) and gut (red dashed area) tissue partially squeezed outside of the body cavity. Time is indicated in min:s, with time 0 corresponding to ~6 hr after first cleavage at 17°C, when the first lateral movements of ectoderm towards the midline begin and become visible as a slight lateral contraction of the embryo. See also Video 6. (C) Postembryonic phenotypes in control and upshifted lin-5(ev571) animals recovered as embryos shortly after the upshift (Materials and methods). pv: protruding vulva; ste: sterile; egl: egg laying defective. See source data for detailed breakdown of phenotypes in individual embryos. See also Figure 1—figure supplement 3—source data 1 for detailed breakdown of phenotypes in individual embryos.
-
Figure 1—figure supplement 3—source data 1
Postembryonic phenotypes in individual animals.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig1-figsupp3-data1-v3.xlsx
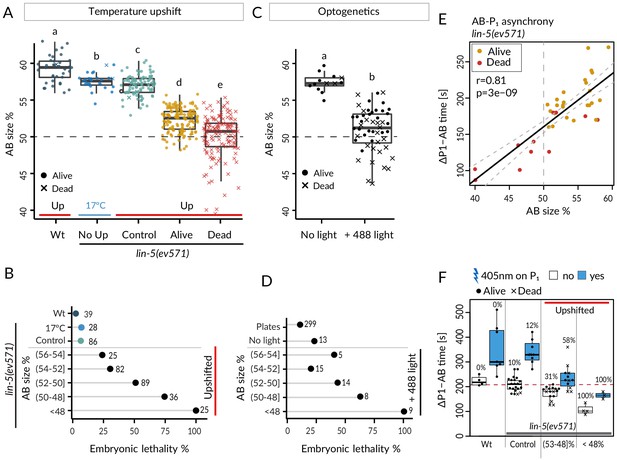
Equalized first cell division decreases embryonic viability.
(A) Boxplots of relative AB size distribution determined in the mid-plane (here and thereafter) in embryos of the indicated conditions; upshifted (Up) wild-type (Wt), lin-5(ev571) kept at 17°C (No Up), lin-5(ev571) upshifted in the early 2-cell stage (Control), as well as lin-5(ev571) upshifted during the first mitotic division and binned into embryos that later lived (Alive) or died (Dead). Here and in other figure panels, two boxplots that do not share the same letter are significantly different from each other with p<0.05; Welch's t-test. See Supplementary file 6 for exact p value and complete statistical analyses for this and all other figures. Dashed lines here and in C indicate equal AB and P1 sizes. (B) Lethality of embryos of from A, with Alive and Dead categories binned as a function of relative AB size and an indication of the number of embryos in each bin. Brackets indicate a mathematical notation for the size interval, [with an inclusion of the value next to the square bracket], (but not of that next to the round bracket), so that individual bins do not overlap. (C) Relative AB size in embryos expressing LIN-5::ePDZ::mCherry and PH::eGFP::LOV, either not subjected to 488 nm laser light (left, n = 13) or equalized through optogenetic recruitment of LIN-5 to the anterior cortex (right, n = 56). (D) Lethality of embryos from C as a function of relative AB size; binning as in B. ‘Plates’ refers to embryos expressing LIN-5::ePDZ::mCherry and PH::eGFP::LOV scored for hatching on plates, without filming. (E) Division asynchrony between AB and P1 as a function of AB size in upshifted lin-5(ev571) embryos; time was determined using anaphase onset monitored with mCherry::H2B. Shown is the subset of lin-5(ev571) control and upshifted embryos from A for which AB-P1 asynchrony was measured. Pearson’s correlation value (r) is shown here and in subsequent figure panels, together with significance of association between two variables determined via the asymptotic t approximation (cor.test function in R, p). (F) Division asynchrony between AB and P1 in embryos of indicated conditions; upshifted lin-5(ev571) embryos were split into two bins as a function of AB sizes, as shown. Percentage above each boxplot indicates proportion of embryonic lethality; blue: illumination of P1 with 405 nm laser to delay cell cycle progression and restore asynchrony between AB and P1. Non-illuminated embryos: same data as in E. Dashed line indicates mean time difference in control embryos. See also Figure 2—source data 1 and Figure 2—source code 1 for underlying analysis and data for this figure.
-
Figure 2—source code 1
Cell size manipulation and lethality analysis in lin-5(ev571) and optogenetic embryos.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig2-code1-v3.zip
-
Figure 2—source data 1
Cell size measurements and division timing in lin-5(ev571) and optogenetic embryos.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig2-data1-v3.xlsx

Aberrant blastomere arrangement at the 4-cell stage.
(A, C) Maximal Z-projection from fluorescent time-lapse recording of lin-5(ev571) embryos expressing GFP::PH and mCherry::H2B to mark the plasma membrane and chromatin, respectively. In the wild type, as well as in control lin-5(ev571) embryos kept at 17°C (shown here), AB divides before P1, resulting in a characteristic 4-cell rhomboid arrangement. Time is indicated in min:s since the beginning of the recording. (B) Normally, P2 is in contact with ABp and EMS, so that Delta signaling from P2 induces proper fate acquisition of ABp, distinguishing it from ABa, whereas Wnt/Src signaling from P2 induces polarization of EMS. (C) A minor fraction of upshifted lin-5(ev571) embryos fail to sufficiently skew the AB cell during division, leading to a T-arrangement at the 4-cell stage, whereby P2 does not contact any of the two AB daughter cells, presumably preventing Delta signaling. See also Video 7. (D) Schematic of abnormal T-arrangement at the 4-cell stage in a minor fraction of upshifted lin-5(ev571) embryos, which prevents instructive signaling between P2 and ABp. Note that upshifted lin-5(ev571) embryos with a T-arrangement were more elongated than those with a normal rhomboid arrangement (59.6 ± 2.83 and 55.0 ± 2.75 µm, p<0.0003 for Welch’s two-sample t-test, n = 10 and 80 measured for this trait, respectively). However, the eggshell length of lin-5(ev571) embryos is not different from that of wild-type embryos (54.4 ± 2.8 µm and 55 µm ± 1.7, respectively, p=0.39, n = 10 and 80).
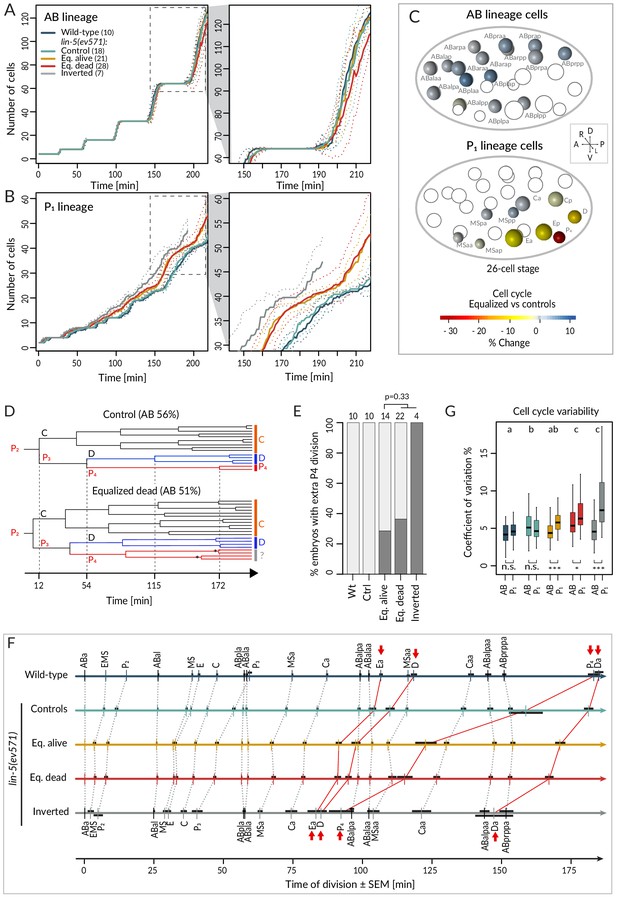
Faster cell cycle progression in P1 descendants of equalized embryos results in altered division sequence.
(A, B) Number of cells in the AB (A) and P1 (B) lineages starting from the 4-cell stage in embryos of indicated conditions. Dashed rectangular region is enlarged on the right. In this and subsequent figure panels, time 0 corresponds to ABa cleavage. Dotted lines – most visible in the enlargements on the right – indicate standard deviations. (C) Graphical depiction of differences in cell cycle timing in individual cells of the AB (top) and P1 (bottom) lineages at the 26 cell stage; colors indicate relative change of cell cycle timing in equalized lin-5(ev571) embryos compared to control lin-5(ev571) condition. In each representation, empty circles represent cells of the other lineage. (D) Partial lineage tree of a control lin-5(ev571) embryo (top) and an equalized lin-5(ev571) embryo (bottom). Normally, P4 divides only once to produce the germline precursors Z2 and Z3, which remain quiescent until hatching (top). In some equalized and inverted embryos, P4 divides more than once, suggesting a repetition of the P3 fate or a fate transformation towards a D-like state (bottom, labeled by question mark). Also, a global acceleration of the C and D lineages is apparent in the equalized embryo compared to the control. Vertical grid lines and time indicated on the x axis correspond to division of P2, P3, D, and P4 in the control embryo. (E) Frequency of embryos with one or more extra divisions in the P4 lineage in indicated conditions (dark gray), with number of analyzed embryos shown on top. Light gray corresponds to embryos with a normal phenotype here and in similar panels in other figures. Ectopic P4 lineage divisions are not significantly more frequent in dying embryos than in those that are alive (Fisher’s exact test, p=0.33). (F) Temporal division sequence in embryos in indicated conditions. Note that the faster cell cycle of P1 lineage cells in equalized and inverted embryos leads to an altered division sequence, highlighted by connecting red lines and arrows for Ea, D, P4, and Da. Dark horizontal bars indicate standard error of the mean. Not all AB cells are shown due to space constrains, but the first and last AB cell in each division round are indicated. (G) Variability in cell cycle duration in AB and P1 lineage expressed as variation coefficient to normalize for cell cycle duration in different cells. Color code as in A. Here and in Figure 4C, D, letters above groups indicate statistical comparison of overall variability among five groups using ANOVA and Tukey's honest significant difference post-hoc test for all pairwise combinations, whereas comparisons between AB and P1 lineages within groups are indicated below boxplots using Welch’s two-sample t-test; asterisks indicate significance levels at 0.05 (*) or 0.001 (***).
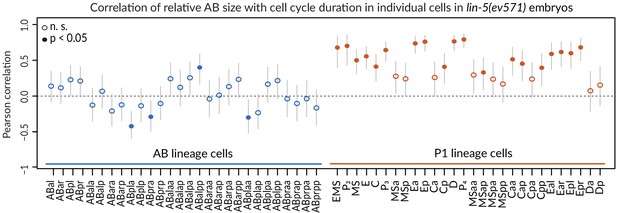
Correlation of cell cycle with AB size in lin-5(ev571) embryos.
Cell cycle duration correlates with initial size asymmetry in most P1 lineage cells (orange), but rarely in AB lineage cells (blue). Note that this analysis included all upshifted and control lin-5(ev571) embryos to obtain most reliable correlation of cell-cycle duration with initial size asymmetry. Vertical lines indicate 95% confidence interval for correlation coefficient value. Cells with significant Pearson correlation are indicated by filled discs (p<0.05, corrected for multiple testing with the Benjamini–Hochberg method), whereas empty circles indicate differences that are not statistically significant. Detailed results of statistical tests for each cell are in Supplementary file 3.
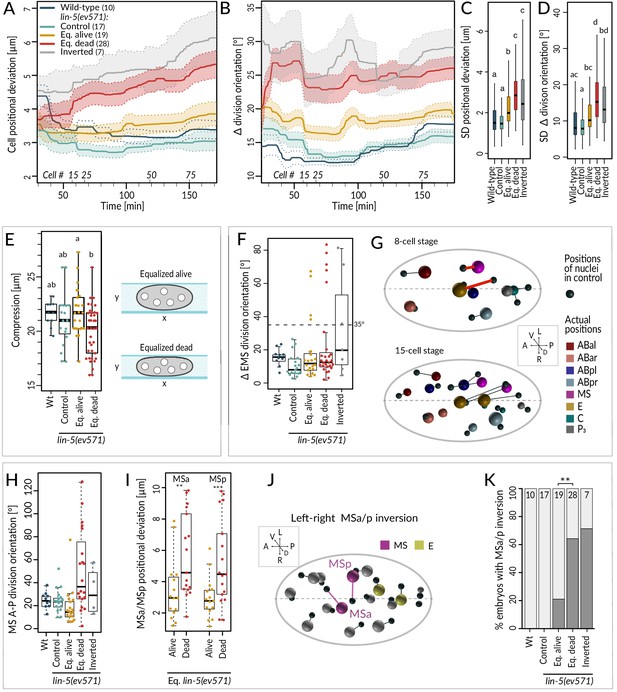
Cell division axis and cell position are frequently altered in equalized embryos.
(A, B) Mean positional deviation (A) or angular deviation (B) ± SD per cell over time in embryos of indicated conditions compared to control lin-5(ev571) reference. The mean positional deviation per cell was calculated at 1 min intervals as the sum of Euclidean distances of individual cells divided by the number of cells at that stage. Only embryos lineaged past the 15-cell stage were included in this analysis. (C, D) Variability in cell position deviation (C) and angular deviation of division orientation (D) in embryos of indicated conditions expressed as SD for cells between 25 and 180 min of development (time 0 corresponds to ABa division). (E) Compression (sample height) in embryos of indicated conditions. Comparisons between groups were performed using Tukey's honest significant difference post-hoc test. (F) Angular deviation of EMS spindle at anaphase onset in embryos of indicated conditions compared to the control lin-5(ev571) reference. Note that the four live equalized embryos exhibiting a >35o EMS skew at anaphase onset corrected spindle orientation to near normality during late anaphase. (G) Nuclear positions in the control lin-5(ev571) reference shown with black spheres, connected by lines with the position of the corresponding nuclei in an equalized lin-5(ev571) embryo exhibiting the EMS skew; sphere size is proportional to nuclear diameter. The skew in EMS division leads to mispositioning of E and MS at the 8-cell stage (highlighted by red line; top), resulting in widespread positioning defects at the 15-cell stage (bottom) and thereafter (not shown). Here and in J, dashed line indicates center of the L-R axis. The directions of the three embryonic axes are depicted here and in other panels. (H) Deviation of MS division orientation angle in embryos of indicated conditions compared to the control lin-5(ev571) reference. (I) Mispositioning of MSa and MSp in equalized lin-5(ev571) embryos measured as the distance to corresponding positions in the control lin-5(ev571) reference. (J) Positions of nuclei in equalized lin-5(ev571) embryo with L-R inversion of MSa and MSp (MSa and MSp: magenta; Ea and Ep: yellow), connected by lines to cell positions in the control lin-5(ev571) reference, indicated as small black spheres. (K) Quantification of MSa/MSp inversion phenotype in indicated conditions scored prior to MSa/MSp division; the phenotype is significantly associated with the outcome of development among equalized lin-5(ev571) embryos (Fisher’s exact test, p=0.007).
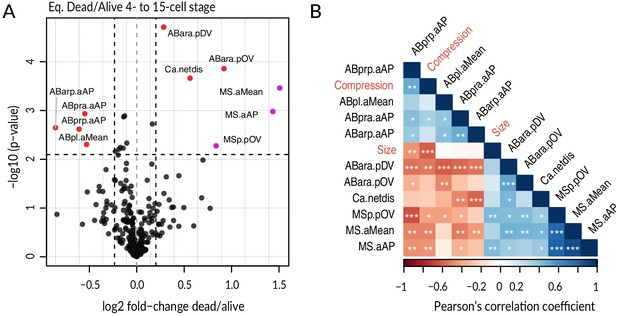
Systematic analysis of features in lineaged embryos up to 15-cell stage.
(A) Volcano plot illustrating selection of significant features (red and purple, with purple indicating MS-related features) comparing equalized dead and equalized alive lin-5(ev571) embryos up to the 15-cell stage (p-value<0.008, corresponding to a <10% false discovery rate and a change of at least ±15% between groups, indicated by dashed lines). Features are encoded as a combination of cell name and measured value in the following way: netdis: net total displacement of given cell during its lifetime; aXX and pXX: division angle and positional deviation, respectively, with XX referring to the direction, i.e., AP = anterioposterior, DV = dorsoventral, LR = left–right; overall angular (aMean) or spatial (pOV) deviation. Note that embryos with skewed EMS division were excluded from this analysis due to their broad substantial defects. See Supplementary file 4 for details. (B) Correlogram of significant features from A. Significant pairwise Pearson correlations are indicated with overlaid white asterisks (p-value *<0.05, **<0.001, ***<0.001, association test based on the t statistics for two normally distributed variables). Embryo size and compression (sample height) were added to explore possible effects of physical factors on the selected features.
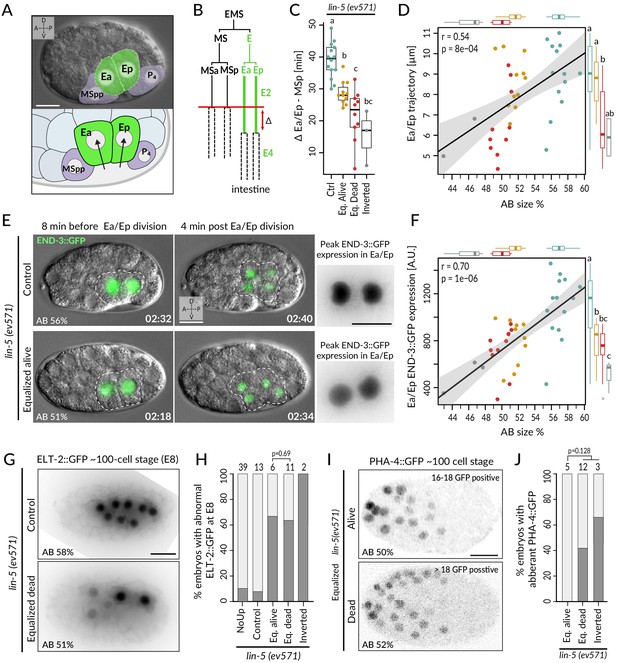
Incomplete fate acquisition in endodermal and pharyngeal lineages in equalized lin-5(ev571) embryos.
(A) Gastrulation in C. elegans begins with ingression of Ea/Ep. Actual embryo viewed by DIC with partial overlay (top) and enlarged corresponding schematic (bottom). (B) Gastrulation is accompanied by a nearly twofold lengthening of the Ea/Ep cell cycle compared to that of their MSa/MSp cousins. Ea/Ep normally divide after internalization. Red arrow indicates usual delay between MSp and Ea/Ep divisions shown in C. (C) Time delay between MSp and Ea/Ep divisions in embryos of indicated conditions. (D) Average trajectory of Ea/Ep interphase nucleus as a function of initial AB size. Here and in F: same color code as in C; moreover, marginal boxplots show overall distribution of individual groups of corresponding color, with letters indicating whether the mean differs statistically between groups (Welch's t-test, p<0.05). (E) END-3::GFP expression in Ea/Ep (dashed area) in control and equalized lin-5(ev571) embryo; Ea/Ep cells in the latter divide close to the eggshell, before completing ingression. Higher magnification views on the right show the time point with peak END-3::GFP expression. Time is indicated in h:min from the time of first cleavage. See also Video 8. (F) Quantification of END-3::GFP peak expression during the Ea/Ep cell cycle as a function of initial AB size. Statistical comparisons are indicated using the letters code above the marginal boxplot (Welch’s test, BH-corrected p-value<0.05). See also Figure 5—source data 1 and Figure 5—source code 1 for analysis for C–F. (G) Control (top) or equalized dead lin-5(ev571) (bottom) ~100-cell stage embryo expressing GFP::PH (not shown) and ELT-2::GFP in inverted black and white rendition. Note that the image of the control embryo has been rotated, explaining the white areas in the corners. (H) Quantification of embryos with abnormal ELT-2::GFP expression (defined as <8 GFP-positive cells at the ~100-cell stage) in indicated conditions. Lethality of equalized embryos is not significantly associated with ELT-2::GFP expression pattern (p=0.69, Fisher’s exact test). (I) Expression of PHA-4::GFP in inverted black and white rendition at the ~100-cell stage in an equalized alive (top) and equalized dead (bottom) lin-5(ev571) embryo. (J) Quantification of embryos with abnormal PHA-4::GFP expression at the ~100-cell stage in indicated conditions. Lethality of equalized embryos is not significantly associated with PHA-4::GFP expression pattern (p=0.128, Fisher’s exact test). See Figure 5—figure supplement 1E for detailed annotation of expression patterns.
-
Figure 5—source code 1
Analysis of Ea/Ep migration, cell cycle duration, and END-3::GFP expresion in lin-5(ev571) and control embryos.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig5-code1-v3.zip
-
Figure 5—source data 1
Ea/Ep migration, cell cycle duration, and END-3::GFP expresion in individual lin-5(ev571) and control embryos.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig5-data1-v3.xlsx
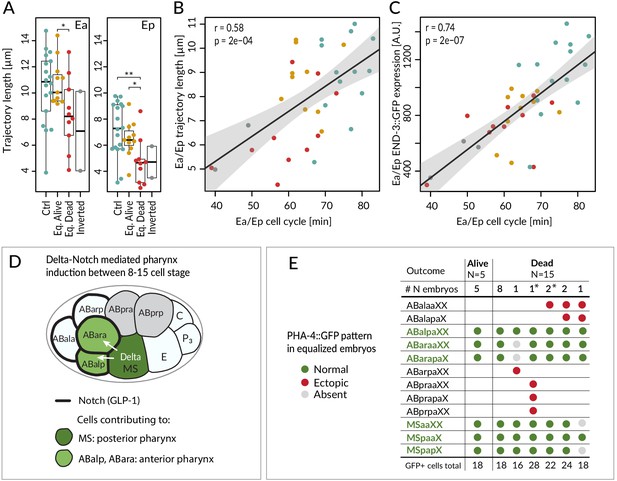
Fate acquisition in endodermal and pharyngeal lineages.
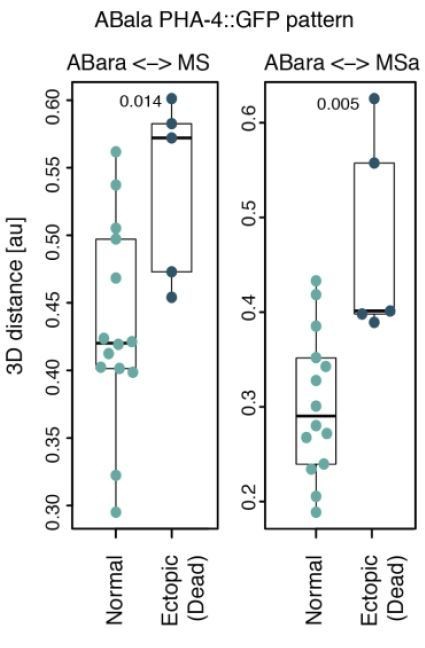
(A) Trajectory of Ea and Ep nuclei in embryos of indicated conditions compared with Welch's two-sample t-test; *<0.05, **<0.001. (B) Correlation between average trajectory and cell cycle duration in Ea/Ep. Same color code as in A here and in C. (C) Correlation between average END-3::GFP expression levels and average cell cycle duration in Ea/Ep. See also Figure 5—source data 1 and Figure 5—source code 1 for underlying data and analysis for A–C. (D) Schematic of anterior pharynx induction in ABalp and ABara descendants via Delta/Notch signaling emanating from MS/MSa/MSp. The GLP-1 Notch receptor (thick black lines) is expressed on the surface of all ABa cells, but ABala and ABarp normally have no physical contact with the signaling MS cells, such that they are not induced. (E) PHA-4::GFP distribution assessed by scoring the identity of GFP-positive cells at the ~100-cell stage in equalized lin-5(ev571) embryos. X in the cell designations (e.g., MSpaaXX) stands for a/p; therefore, XX comprises four cells: aa, ap, pa, pp. Asterisks indicate two inverted embryos with aberrant pattern that were included in this analysis.

Logistic Lasso regression machine learning analysis predicts lethality in early equalized embryos.
(A) Prediction accuracy of lethality versus survival in equalized embryos for individual logistic Lasso models generated by fivefold cross-validation (cv) over 250 repetitions in indicated stages. Note that embryos with a T-arrangement at the 4-cell stage or with a skewed EMS division at the 6-cell stage were excluded from this analysis, as were embryos with missing values for some of the cells. (B) Number of predictive features in individual Lasso models over cv 250 iteration; 14 alive and 17 dead equalized lin-5(ev571) embryos were used for training. Y-axis: number of iterations in which a model with the indicated number of features was selected. Zero features dominant at the 4-cell stage indicates that an empty model (intercept only) was selected in the majority of iterations and performed poorly overall. (C) Inclusion frequency of individual features in an optimal model at each cv iteration over 250 repetitions at indicated stages. Selected variables were chosen from 18, 72, 150, and 249 features present at 4-, 8-, 15-, and 28-cell stage, respectively. Supplementary file 7 lists all features used at each stage for Lasso analysis. (D) Receiver–operator curve (ROC) of best model selected at each stage with the minimal cross-validation error. Shown are selectivity and sensitivity as a function of changing the threshold value for assigning embryos to dead or alive category; diagonal line: random classification. We noted also that even though the models at the 8-cell stage had inferior accuracy, MS division orientation was repeatedly selected among four predictive features at that stage, highlighting the importance of the MSa and MSp inversion phenotype described in Figure 4.
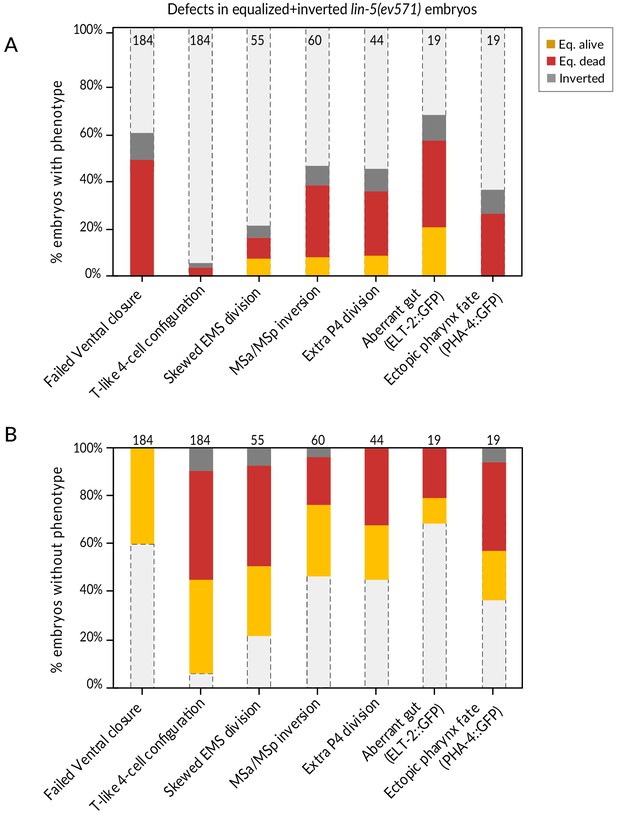
Summary of phenotypes observed in equalized and inverted lin-5(ev571) embryos.
(A, B) Fraction of equalized (alive or dead, as indicated) and inverted lin-5(ev571) embryos displaying a given phenotype (A), or not (B), with number of embryos analyzed for each trait indicated above the bars. Unequal control lin-5(ev571) embryos showed no or minimal occurrence of these phenotypes; refer to previous figures for control scoring.
-
Figure 6—figure supplement 1—source data 1
Counts of embryos with phenotypes indicated in the Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig6-figsupp1-data1-v3.xlsx
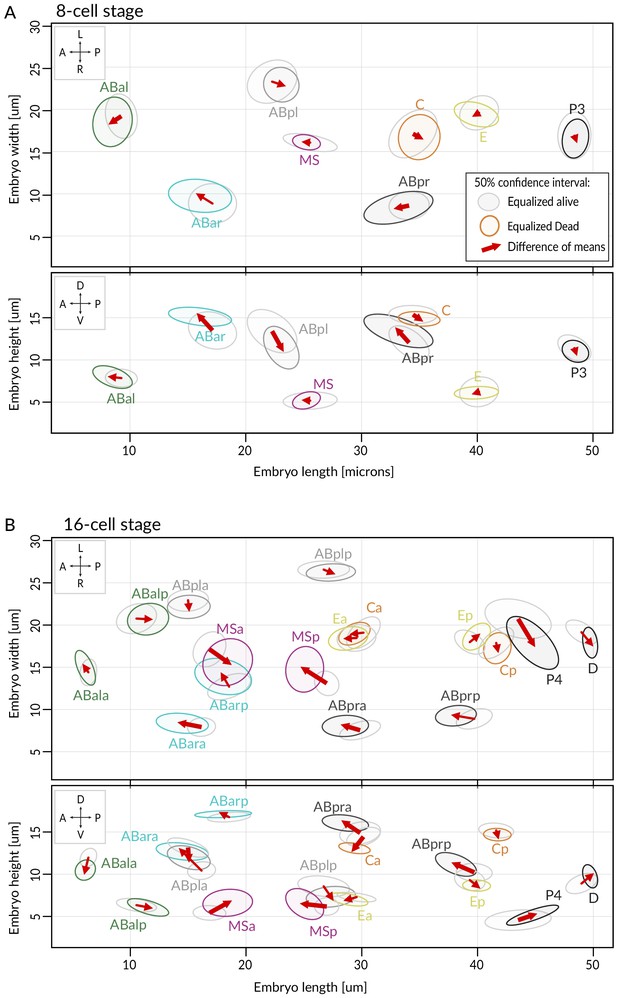
Comparison of cell positions at the 8- and 16-cell stage in equalized embryos.
(A, B) Shown are dorsal (XY, top) and lateral (XZ, bottom) views at the 8- (A) and 16-cell stage (B). Ellipses indicate 50% confidence interval for nuclear position distributions among alive (gray) and dead (colored) lin-5(ev571) embryos. Arrows show direction of change between mean cell positions of the two groups. Arrows with a bold line indicate significant difference in at least one direction (see Supplementary file 2 for statistics).
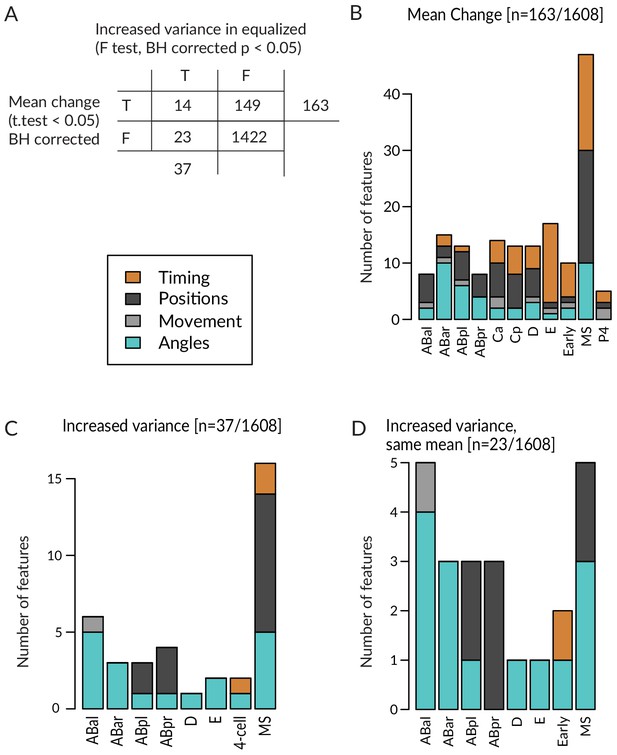
Comparison of variance and mean changes between equalized and control lin-5(ev571) embryos.
(A) Contingency table cross-tabulating features scored in the lineaging data set with a significant change in mean and/or variance among equalized embryos compared to lin-5(ev571) control embryos. Welch’s t-test and F-test with Benjamini–Hochberg (BH) correction were used; T: true; F: false. (B, C) Number of significant features from (A) according to cell lineage and feature type (colored according to the legend), with significant change of mean or variance among equalized embryos. (D) Twenty-three features exhibit an increase in variance without a change in the mean. Note that there is not a single variable with increased variance among controls. See the source data for this figure for all 1608 features, p-values, and the degree of change.
-
Figure 6—figure supplement 3—source data 1
Results of statistical tests comparing means and variance between equalized and control embryos for each variable lin-5.
- https://cdn.elifesciences.org/articles/61714/elife-61714-fig6-figsupp3-data1-v3.xlsx
Videos
Unequal division of the wild-type C. elegans zygote.
The resulting 2-cell stage of this particular embryo had an AB size of 62% of the total embryo in the mid-plane. Note vertical rocking of spindle as it moves towards the posterior pole during anaphase. Time is indicated in min:s in this and all other videos.
Unequal division of the lin-5(ev571) mutant zygote at the permissive temperature of 17°C (relative AB size 58%).
Note severely dampened anaphase spindle rocking indicating reduced cortical pulling forces in this video and in Video 3.
Transient upshift of lin-5(ev571) embryo from 17°C to 27°C from metaphase until the completion of cytokinesis results in equalized or even inverted division, as in this particular embryo, in which AB is smaller than P1 (AB size 47%).
Optogenetic recruitment of transgenic LIN-5 to the anterior cortex during mitosis counteracts posterior forces and results in equalized division (relative AB size 49%).
Montage showing DIC recording (left), LIN-5::EPDZ::mCherry (middle), and PH::EGFP::LOV (right). LIN-5::EPDZ::mCherry was recruited to membrane-bound PH::EGFP::LOV by exposing a small rectangular region at the anterior cortex with a 488 nm laser during mitosis.
Ventral closure of epidermis at the end gastrulation in control lin-5(ev571) embryo at the permissive temperature of 17°C.
Lateral cells migrate over ventral neuroblasts to enclose the embryo in a continuous epidermal layer called the hypodermis. Subsequently, differentiating muscles start to contract and the embryo gradually elongates into a tubular body shape.
Upshifted equalized lin-5(ev571) embryo failing to undergo ventral epidermis closure, resulting in the extrusion of internal tissues from the body cavity as muscles start contracting.
Embryo shown here had a relative AB size of 53%.
Equalized size of AB and P1 leads to an aberrant T-arrangement at the 4-cell stage in ~4% of equalized embryos (relative AB size is 50% in this case).
Expression of the endodermal marker END-3::GFP in control (left) and equalized (right) lin-5(ev571) embryos (relative AB size 58% and 53%, respectively).
In the equalized embryo, Ea/Ep express less END-3::GFP than they normally do, and divide prematurely, before completing ingression.
Additional files
-
Supplementary file 1
Annotation of all lineaged embryos reported in this study in Figures 3 and 4.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp1-v3.xlsx
-
Supplementary file 2
Statistical comparisons of all features (cell cycle timing, cell positions, division angles, migration) for individual cells between pairs of embryo groups (wild type, controls, equalized alive, equalized dead, inverted).
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp2-v3.statistics.xlsx
-
Supplementary file 3
Analysis of correlation between relative AB size and cell cycle duration in all lin-5(ev571) embryos from Supplementary file 1.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp3-v3.xlsx
-
Supplementary file 4
Lst of features between 4- and 15-cell stage and comparison of mean values between equalized alive and equalized dead lin-5 (ev571) embryos for the volcano plot in Figure 4—figure supplement 1A.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp4-v3.hatched.csv
-
Supplementary file 5
List of C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp5-v3.xlsx
-
Supplementary file 6
Statistical comparisons, average values, and number of observations for the figures in this study, with the exception of lineaged embryos (Figures 3 and 4), for which values are provided in other supplementary files.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp6-v3.xlsx
-
Supplementary file 7
List of features used for Lasso analysis at 4-, 8-, 15-, and 28-cell stage, including inclusion frequency and model coefficients for best predictive models.
- https://cdn.elifesciences.org/articles/61714/elife-61714-supp7-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61714/elife-61714-transrepform-v3.docx