In vitro proteasome processing of neo-splicetopes does not predict their presentation in vivo
Figures
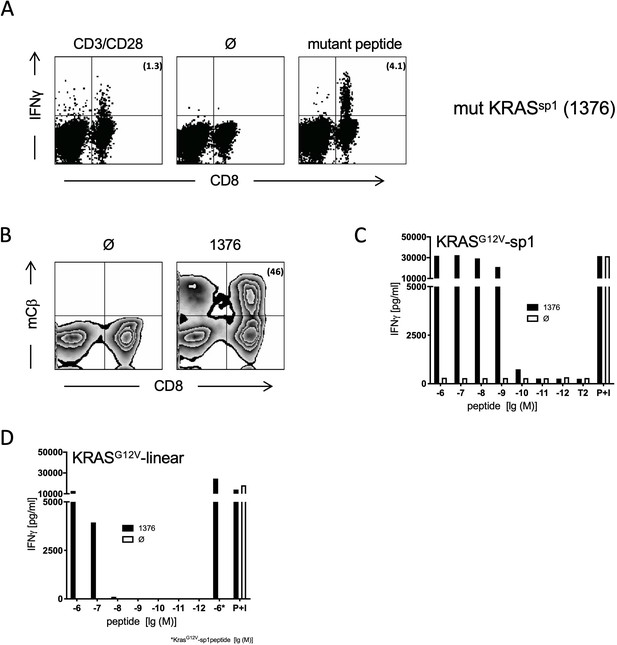
Generation and characterization of TCRs specific for spliced epitope 1 (sp1) of mutant KRASG12V.
(A) A representative example of ex vivo intracellular cytokine staining (ICS) analysis of KRAS mutant peptide immunized ABabDII mice (Li et al., 2010) 7 days after the last immunization with sp1 (KLVVGAVGV). Stimulation with CD3/CD28 beads served as positive control, co-culture without peptide (Ø) was used as negative control. Numbers in brackets represent percent IFNγ+ CD8+ T cells, respectively. Spleens of mice with IFNγ-reactive CD8+ T cells were cultured for 10 days in the presence of 10−8 M of sp1 KRAS peptide, and reactive CD8+ T cells were purified by IFNγ-capture assay for isolation of TCR α and β chains by RACE-PCR. (B) The corresponding TCR α and β chains isolated from one KRASG12V sp1 peptide immunized ABabDII mouse, respectively (1376), were cloned into retroviral vector pMP71 and reexpressed in human PBMC. Transduction efficacy was measured by staining of the mouse TCRβ constant chain on CD8+ T cells, and the number of positive CD8+ T cells is shown in brackets. (C) TCR gene transfer confers specificity for mutant spliced KRASG12V peptide KLVVGAVGV (sp1). IFNγ production of KRASG12V splice-specific 1376 TCR-transduced T cells upon co-culture with sp1-peptide-loaded T2 cells (1376 [solid bars]). As negative control, T2 cells were not peptide loaded. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (p + I) were added to the co-culture. All target cells were also co-cultured with non-transduced T cells (Ø, open bars). (D) TCR gene transfer confers cross-reactivity for mutant linear KRASG12V peptide KLVVVGAVGV. IFNγ production of KRASG12V splice-specific 1376 TCR-transduced T cells upon co-culture with KRASG12V linear peptide-loaded T2 cells (1376 [solid bars]). As negative control, T2 cells were not peptide loaded. For maximal stimulation, PMA and ionomycin (p + I) were added to the co-culture. All target cells were also co-cultured with non-transduced T cells (Ø, open bars). Experiments were done at least in duplicate.
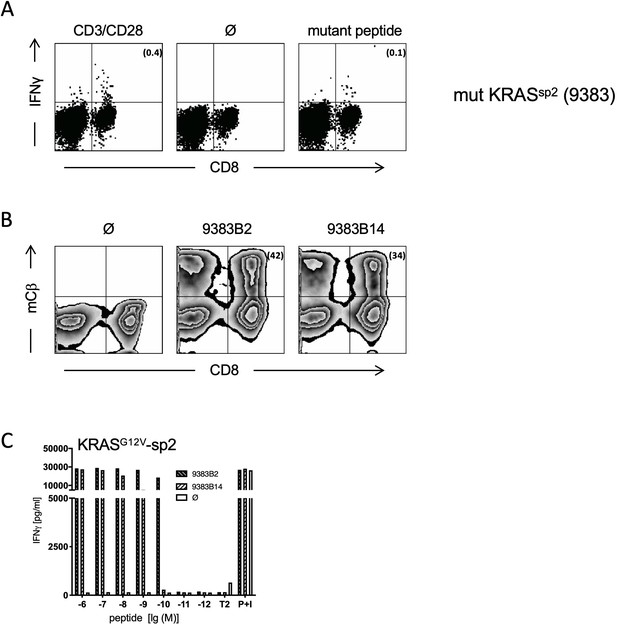
Generation and characterization of TCRs specific for spliced epitope 2 (sp2) of mutant KRASG12V.
(A) A representative example of ex vivo ICS analysis of KRAS mutant peptide KLVVVAVGV immunized ABabDII mice (Textoris-Taube et al., 2019) 7 days after the last immunization with sp2 (KLVVVAVGV). Stimulation with CD3/CD28 beads served as positive control, and stimulation without peptide (Ø) was used as negative control. Numbers in brackets represent percent IFNγ+ CD8+ T cells, respectively. Spleens of mice with IFNγ-reactive CD8+ T cells were cultured for 10 days in the presence of 10−8 M of sp2 KRAS peptide, and mutation-specific CD8+ T cells were purified by IFNγ-capture assay for isolation of TCR α and β chains by RACE-PCR. (B) The corresponding TCR α and β chains isolated from one KRASG12V sp2 peptide immunized ABabDII mouse (9383B2/B14, respectively) were cloned into retroviral vector pMP71 and reexpressed in human PBMC. Transduction efficacy was measured by staining of the mouse TCRβ constant chain on CD8+ T cells, and the number of positive CD8+ T cells is shown in brackets. (C) TCR gene transfer confers specificity for mutant spliced KRASG12V peptides KLVVVAVGV (sp2). IFNγ production of KRASG12V TCR-transduced T cells upon co-culture with sp2-peptide-loaded T2 cells (9383B2 [hatched bars] and 9383B14 [open hatched bars]). As negative control, T2 cells were not loaded. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (p + I) were added to the co-culture. All target cells were also co-cultured with non-transduced T cells (Ø, open bars). Experiments were done at least in duplicate.
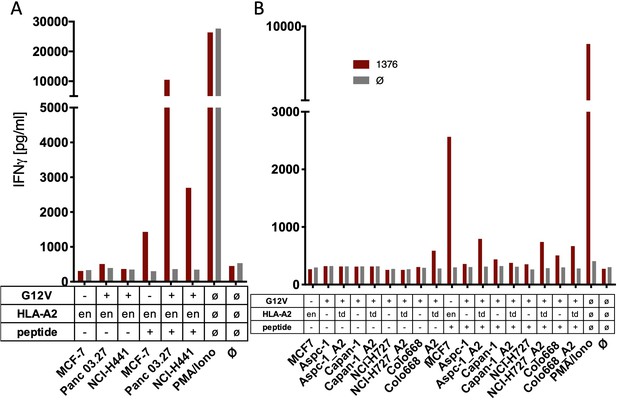
The spliced KRASG12V epitope one is not recognized by spliced epitope 1 (sp1)-TCR-redirected T cells.
(A) For analysis of natural processing and recognition of KRASG12V epitopes, cell lines naturally expressing HLA-A2:01 and harboring the KRASG12V mutation and HLA-A2:01+ KRASwt cell line MCF7 were co-cultured with KRASG12V TCR1376-redirected T cells. (B) HLA-A2:01-negative cell lines were transiently transduced with an HLA-A02:01 expressing retroviral construct (td) and co-cultured as in (A). IFNγ production of transduced T cells is shown (red bars). As positive control, peptide-loaded cells (+) were used, respectively. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (PMA/Iono) were added, and all target cells were also co-cultured with non-transduced T cells (gray bars; Ø); en: endogenous expression of HLA-A2:01. Representative measurements are shown, and experiments were done at least in duplicate.
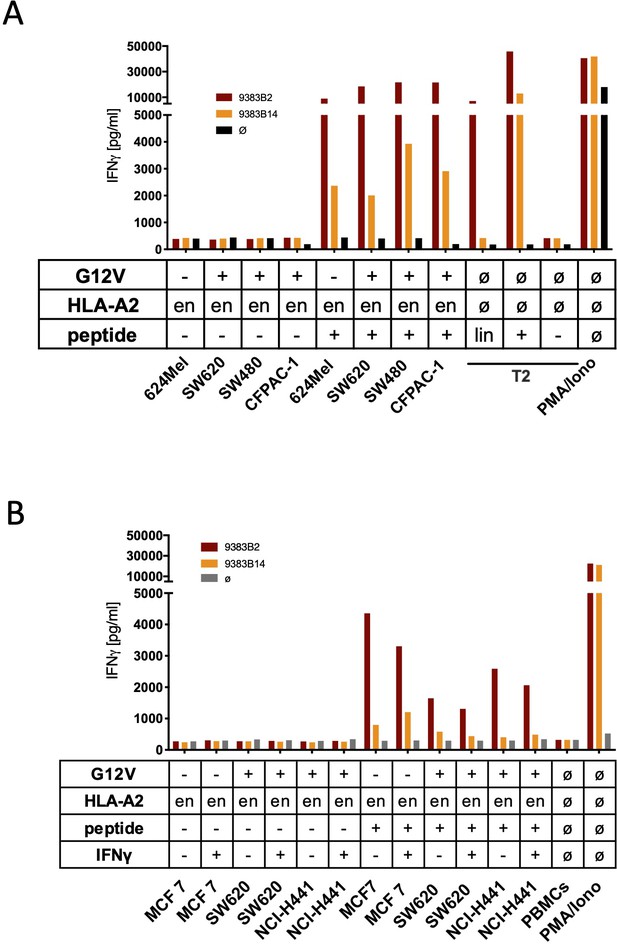
The spliced KRASG12V epitope 1 (sp1) is not recognized by sp2-TCR-redirected T cells.
(A) For analysis of natural processing and recognition of KRASG12V epitopes, cell lines harboring the KRASG12V mutation and KRASwt cell line MCF7 were co-cultured with KRASG12V splice-specific TCR9383B2 and TCR9383B14- redirected T cells, respectively. IFNγ production of transduced T cells is shown (red bars: TCR9383B2; orange bars: TCR9383B14). (B) Pre-treatment of cell lines harboring KRASG12V mutation with recombinant IFNγ for 48 hr neither induces recognition by KRASG12V TCR9383B2 nor KRASG12V TCR9383B14-redirected T cells. IFNγ production of transduced T cells is shown (red bars: TCR9383B2; orange bars: TCR9383B14, respectively). As positive control, peptide-loaded cells (+) were used. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (p + I) were added, and all target cells were also co-cultured with non-transduced T cells (black/gray bars; Ø); en: endogenous expression of HLA-A2:01. Representative measurements are shown, and experiments were done at least in duplicate.
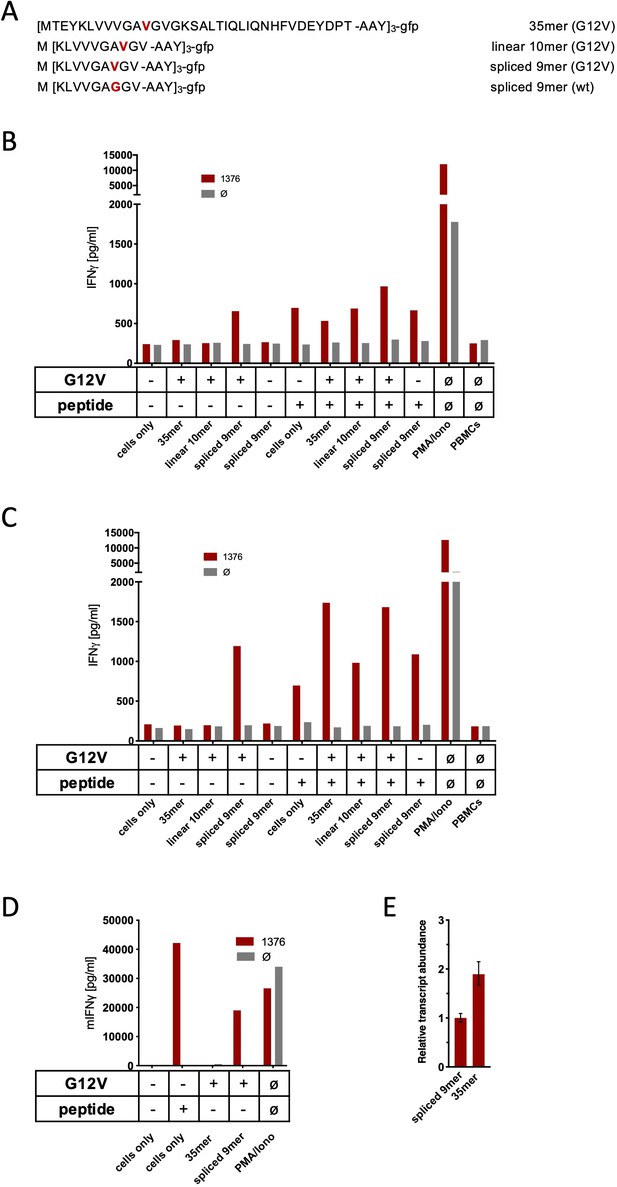
Co-culture of KRASG12V splice-specific TCR (TCR1376) with human and mouse cells expressing KRASG12V cDNA or triple epitopes.
(A) Schematic representation of KRASG12V/wt triple epitopes used for recombinant overexpression in MCF7, 624Mel, and NIH-HHD cells. TCR1376 was retrovirally transduced into human PBMCs or TCR1xCD45.1xRag1-/- mouse splenocytes, respectively, and 104 transduced cells were co-cultured 1:1 with target cells (B: MCF7; C: 624Mel; D: NIH-HHD). Target cells were loaded with 10−6 M spliced peptide or transduced with either KRASG12V triple minigene 35mer or KRASG12V triple epitope spliced nonamer. KRASwt triple epitope spliced nonamer and KRASG12V triple epitope linear decamer were used as control. IFNγ production of transduced T cells is shown (red bars). For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (PMA/Iono) were added, and all target cells were also co-cultured with non-transduced T cells (gray bars; Ø). Representative measurements are shown, and experiments were done at least in duplicate. (E) Relative amounts of KRASG12V triple minigene 35mer and KRASG12V triple epitope spliced nonamer were determined by qPCR on transduced NIH-HHD cells. KRASG12V triple epitope spliced nonamer expression is arbitrarily set to 1.
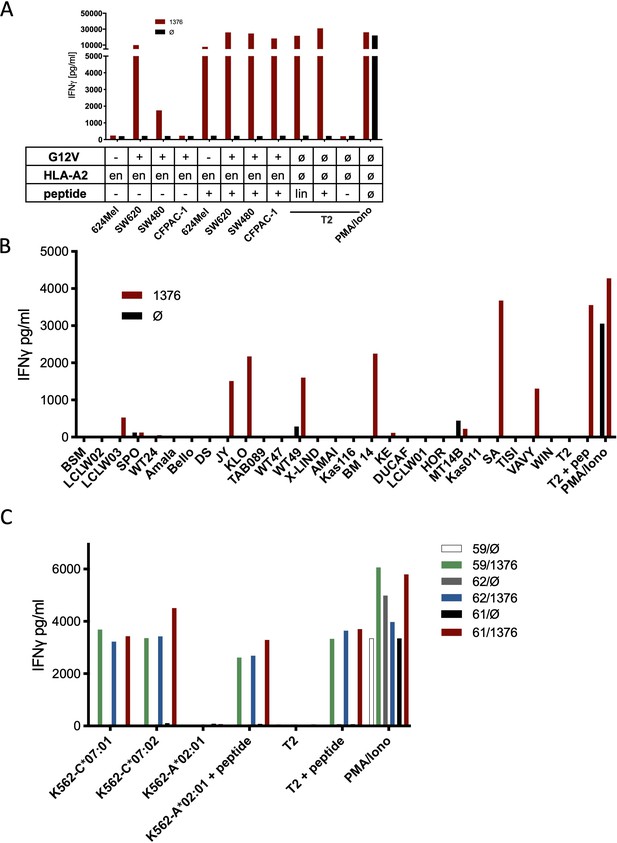
Co-culture of KRASG12V splice peptide-specific TCR (TCR1376) cross-reacts with HLA-C07 allele.
(A) Cell lines SW480, SW620, and CFPAC-1 naturally expressing HLA-A2:01 and harboring the KRASG12V mutation and HLA-A2:01+ KRASwt cell line 624Mel were co-cultured with KRASG12V TCR1376-redirected T cells. As positive control peptide loaded tumor or T2 cells were used, respectively. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (PMA/Iono) were added, and all target cells were also co-cultured with non-transduced T cells (black bars); en: endogenous expression of HLA-A2:01. IFNγ as readout in the supernatant was determined by ELISA. (B) TCR1376-transduced T cells were co-cultured with lymphoblastoid B cell lines (B-LCLs) expressing different HLA allotypes (Supplementary file 1). One representative example out of three different T cell donors is given. (C) K562 transduced with HLA-C07:01, HLA-C07:02, and HLA-A02:01 molecules were co-cultured with TCR1376-transduced T cells. Representative measurements are shown, and experiments were done at least in duplicate.
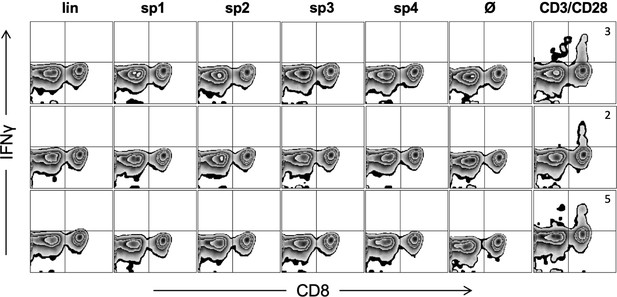
Triple KRASG12V1-35 minigene immunization does not generate cytotoxic T lymphocyte (CTL) response against predicted linear or spliced HLA-A02:01 epitopes.
Representative examples of ex vivo ICS analysis of KRAS mutant peptide immunized ABabDII mice 7 days after the last immunization, and linear as well as spliced epitopes 1, 2, and 4 were used. Stimulation with CD3/CD28 beads served as positive control, and stimulation without peptide (Ø) was used as negative control. Numbers in brackets represent percent IFNγ+ CD8+ T cells, respectively. lin: KLVVVGAVGV; sp1: spliced epitope 1, KLVVGAVGV; sp2: spliced epitope 2, KLVVVAVGV; sp3: spliced epitope 3, YLVVVGAVGV; sp4: spliced epitope 4, KLVVVGVGV.
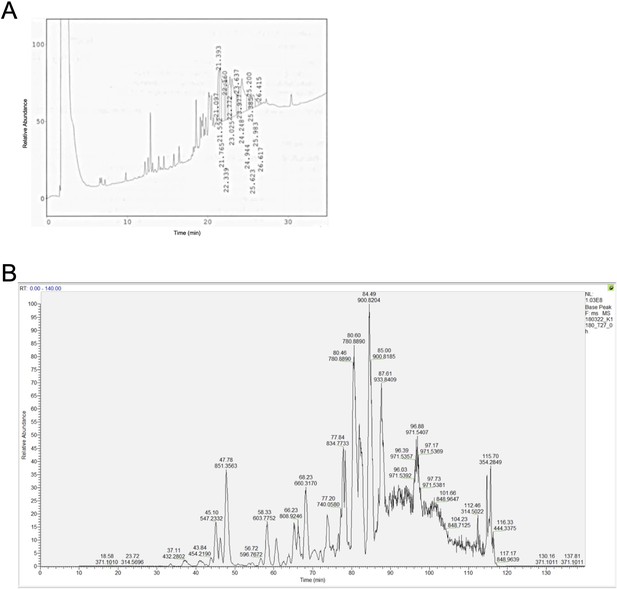
Base peak chromatogram of the synthetic polypeptides (A) KRASG12V2-35 and (B) KRASG12V2-32.
The polypeptides were highly insoluble and did not allow any substantial further purification.
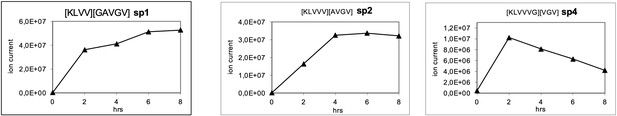
Generation of the predicted putative KRASG12V-derived nonamer neo-splicetopes sp1, sp2, and sp4 from the synthetic polypeptide substrate KRASG12V2-14 in a kinetic proteasome-catalyzed peptide splicing experiment.
The synthetic KRASG12V2-14 polypeptide at a concentration of 30 µM was processed in the presence of 6 µg 20S proteasome for various time points. To identify spliced peptides, a fasta data file was generated with ProtAG and loaded onto PD2.1. The kinetics were analyzed with LC Quan 2.7. The predicted decamer KRASG12V4-8/10-14 (sp3, see Figure 3—figure supplement 2) was not detected.
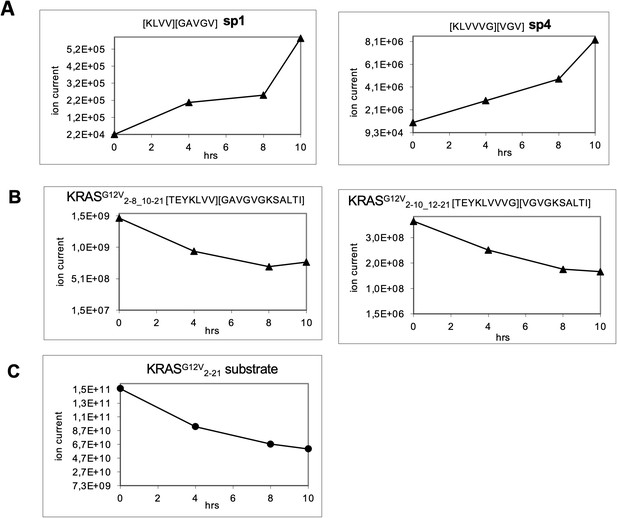
Generation of the predicted putative KRASG12V-derived nonamer neo-splicetopes sp1 and sp4 from the synthetic polypeptide substrate KRASG12V2-21 in a kinetic proteasome-catalyzed peptide splicing experiment.
The synthetic KRASG12V2-14 polypeptide at a concentration of 60 µM was processed in the presence of 8 µg 20S erythrocyte proteasome for various time points. To identify spliced peptides, a fasta data file was generated with ProtAG and loaded onto PD2.1. The kinetics were analyzed with LC Quan 2.7. All kinetics are derived from the same experiment. (A) The generation of KRASG12V5-8/10-14 (sp1) and KRASG12V5-10/12-14 (sp4) is shown. (B) Degradation kinetics of the faulty polypeptides KRASG12V2-8_10-21 and KRASG12V2-10_12-21 reflecting in sequence the splicing reaction for KRASG12V5-8/10-14 and KRASG12V5-10/12-14, respectively. (C) Degradation kinetics of the synthetic KRASG12V2-21 polypeptide substrate. The decamer KRASG12V4-8/10-14 (sp3, see Figure 3—figure supplement 2) was not detected.
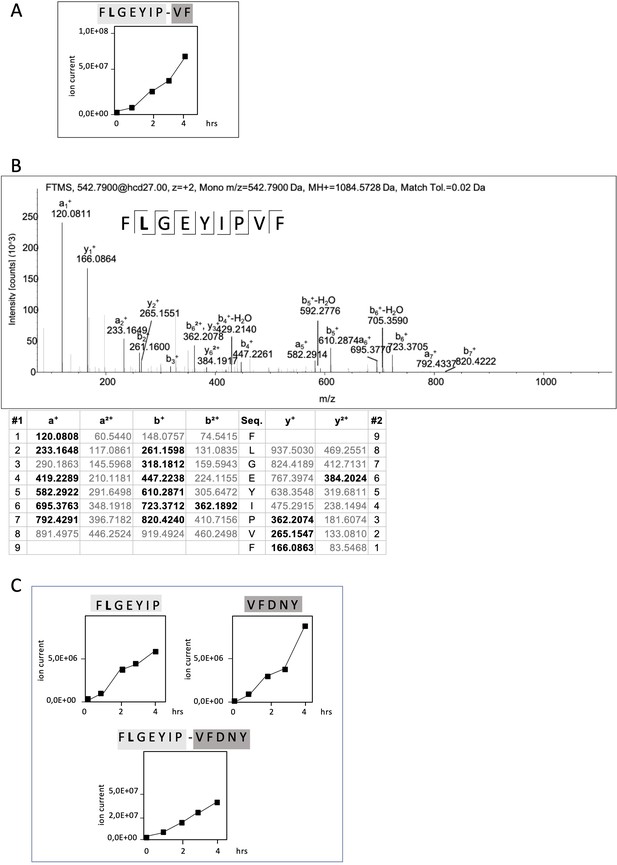
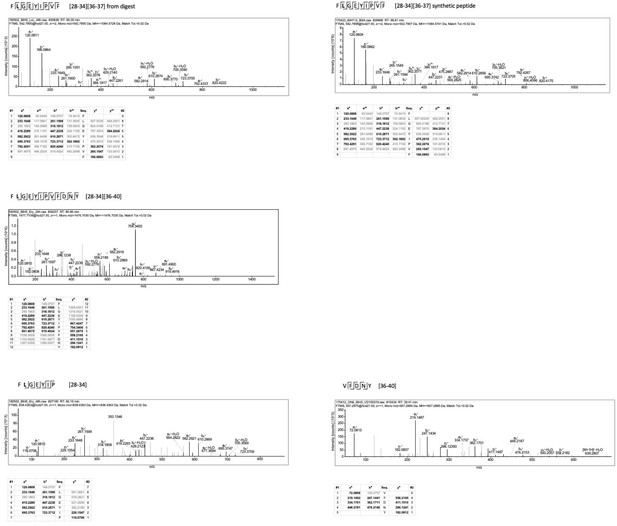
Non-spliced and spliced peptides generated from RAC2P29L20–44 in kinetic proteasome-catalyzed peptide splicing reactions.
The candidate RAC2P29L neo-splicetope is generated via a C-terminally extended precursor peptide. (A) Kinetics of the generation of the 9mer candidate RAC2P29L28-34/36-37 neo-splicetope and the non-spliced RAC2P29L28-36 neoepitope. Note that generation of the non-spliced RAC2P29L28-36 peptide is significantly more efficient than the generation of the spliced RAC2P29L28-34/36-37. (B) MS/MS spectra of the candidate RAC2P29L28-34/36-37 neo-splicetope. (C) Kinetics of the generation of the non-spliced acceptor FLGEYIP and donor VFDNY peptides and the generation of the C-terminally extended spliced precursor peptide RAC2P29L 28-34/36-40 FLGEYIP-VFDNY. The MS/MS spectra for the identified RAC2P29L-derived peptides are shown in Figure 4—figure supplement 1.
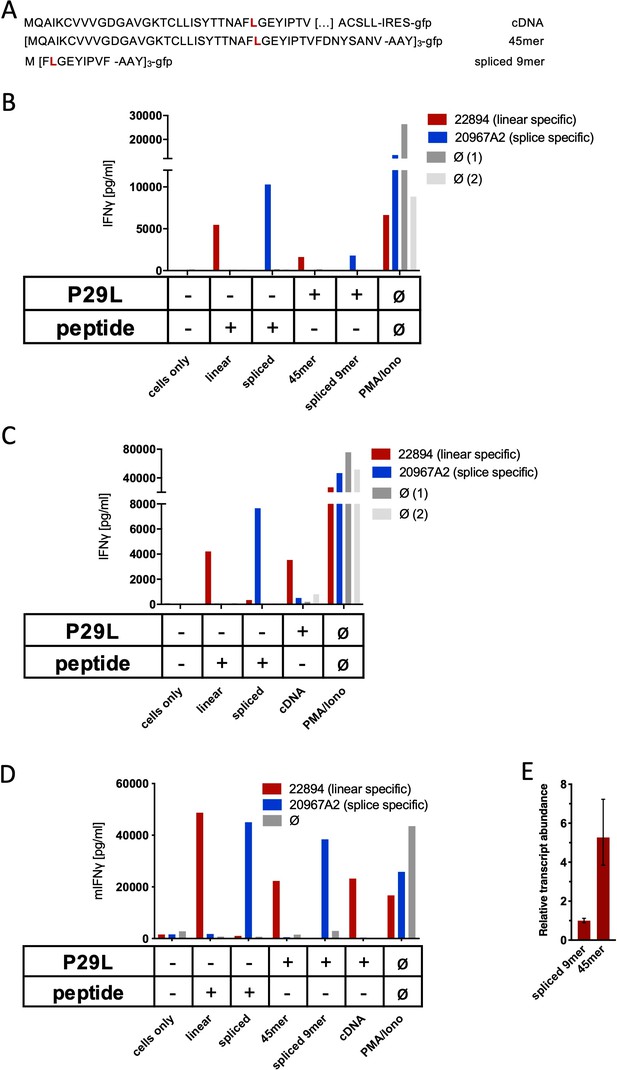
Co-culture of RAC2P29L linear-specific TCR (TCR22894) and Rac2P29L splice-specific TCR (TCR20967A2) with cells expressing Rac2P29L triple epitopes and cDNA.
(A) Schematic representation of RAC2P29L cDNA and triple epitopes used for recombinant overexpression in Mel21a and NIH-HHD cells. (B, C) TCRs were retrovirally transduced into human PBMCs and 104 transduced cells were co-cultured 1:1 with Mel21a target cells. (D) TCRs were retrovirally transduced into TCR1xCD45.1xRag1-/- mouse splenocytes, and 104 transduced cells were co-cultured 1:1 with NIH-HHD target cells. Respective human and mouse target cells were loaded with 10−6 M spliced or non-spliced RACP29L peptide, or transduced with either Rac2P29L triple epitope 45mer, Rac2P29L triple epitope nonamer, or Rac2P29L cDNA. Upon co-culture with recombinant TCR+ T cells, IFNγ release was measured. For maximal stimulation, phorbol myristate acetate (PMA) and ionomycin (PMA/Iono) were added, and all target cells were also co-cultured with non-transduced T cells (gray bars; Ø). Representative measurements are shown, and experiments were done at least in duplicate. (E) Relative amounts of Rac2P29L triple epitope 45mer and Rac2P29L triple epitope nonamer were determined by qPCR on transduced NIH-HHD cells. Rac2P29L triple epitope nonamer expression is arbitrarily set to 1.
Additional files
-
Supplementary file 1
HLA-ABC haplotypes of lymphoblastoid B cell lines (BLCLs) and tumor cell lines SW480 and SW620.
- https://cdn.elifesciences.org/articles/62019/elife-62019-supp1-v3.docx
-
Supplementary file 2
Faulty peptides identified within the KRASG12V polypeptide substrates.
- https://cdn.elifesciences.org/articles/62019/elife-62019-supp2-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62019/elife-62019-transrepform-v3.pdf