3D in situ imaging of the female reproductive tract reveals molecular signatures of fertilizing spermatozoa in mice
Figures
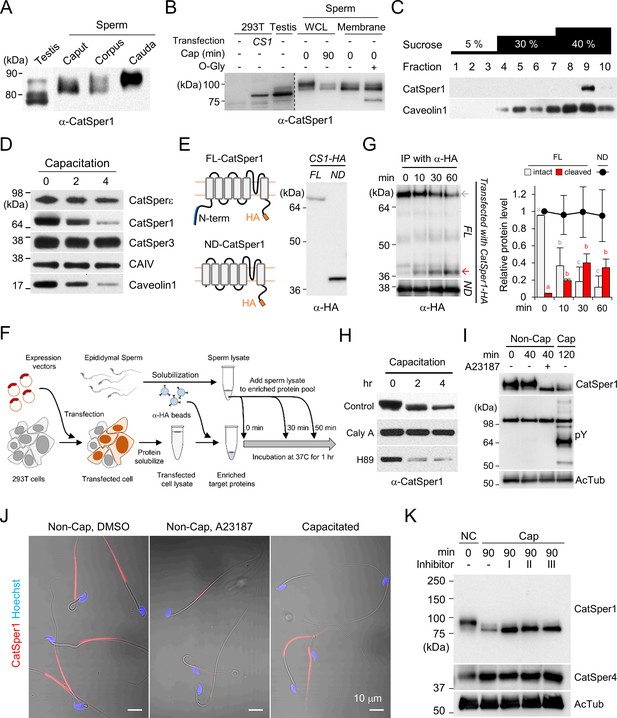
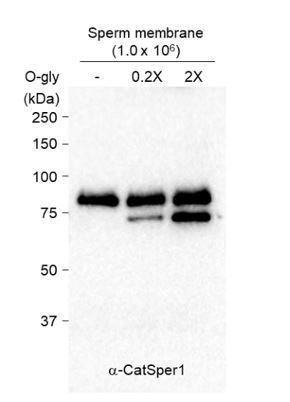
CatSper1 is specifically processed during in vitro capacitation.
(A–B) CatSper1 undergoes post-translational modification during spermiogenesis and epididymal maturation. (A) A gradual decrease in electrophoretic mobility of CatSper1 is observed by western blot analysis. (B) Mouse CatSper1 is an O-glycosylated protein. Apparent molecular weights of CatSper1 proteins were analyzed by immunoblotting recombinant CatSper1 expressed in 293 T cells (CS1) and native CatSper1 from whole cell lysate (WCL) of testis and sperm (non-capacitated, 0 min; capacitated, 90 min) compared with those of CatSper in sperm membrane fraction treated with or without O-glycosidase (O-Gly). The dotted line indicates different exposure time of the same membrane. (C) CatSper resides in lipid rafts subdomains of the plasma membrane in mature sperm. Solubilized sperm proteins were fractionized by discontinuous sucrose density gradient (5, 30, and 40%) centrifugation. (D) CatSper1 is degraded during the late stage of capacitation. Protein expression levels of CatSper1 and caveolin-1, but not CatSper3, CatSperε, or carbonic anhydrase 4 (CAIV) are altered by in vitro capacitation. (E–G) CatSper1 is cleaved within the N-terminal domain (NTD). (E) A cartoon of full-length (FL, top) and N-terminal truncated (ND, bottom) recombinant CatSper1 protein expressed in the study (left). Both proteins are tagged with HA at their respective C-termini (orange). The CatSper1 antibody used in this study is raised against the 1–150 aa region of CatSper1 (blue, Ren et al., 2001). Detection of recombinant FL-CatSper1 and ND-CatSper1 expressed in 293 T cells (right). (F) A cartoon of the experimental scheme to test NTD truncation of CatSper1. FL-CatSper1 and ND-CatSper1 expressed in 293 T cells were solubilized and pulled-down using agarose resin conjugated with HA antibody. The enriched recombinant proteins were incubated with solubilized sperm lysates at 37°C for 0, 10, 30, and 60 min and subjected to immunoblot. (G) FL-CatSper1 is cleaved at NTD by solubilized sperm lysate. FL-CatSper1 (gray arrow) decreases while truncated form (red arrow) increases by incubation with solubilized sperm lysates (top). ND-CatSper1 proteins remain largely unchanged under the same conditions (bottom). The right panel shows quantification of the protein levels by measuring the band intensity of target proteins at each time point, normalized by total FL-CatSper1 at 0 min points (n = 3; intact, gray bars; cleaved, red bars) or ND-CatSper1 (n = 4; black dots). The sum of intact and cleaved FL-CatSper1 levels was used for total FL-CatSper1. Statistical analyses were performed between relative levels at each time point within each protein. Different letters indicate the significant difference. Data is represented as mean ± SEM. See also Figure 1—source data 1. Immunoblotting was performed with the HA antibody (E and G). (H) Capacitation-associated CatSper1 degradation is regulated by phosphorylation. CatSper1 degradation is accelerated by PKA inhibition. A PKA inhibitor, H89 (50 μM), enhances capacitation-associated CatSper1 degradation. A protein phophatase1 inhibitor, calyculin A (Caly A, 0.1 μM), prevents the CatSper1 degradation during sperm capacitation in vitro. (I–J) Ca2+ influx accelerates CatSper1 degradation. (I) CatSper activation during capacitation and Ca2+ ionophore treatment (A23187, 10 μM) facilitates the CatSper1 cleavage. (J) Representative immunofluorescence images of CatSper1 in the spermatozoa incubated under the conditions used in (I). The extent of CatSper1 degradation is heterogeneous in the capacitated sperm cells (bottom) compared with A23187-treated uncapacitated sperm cells (middle). (K) Capacitation-associated CatSper1 degradation is blocked by calpain inhibitors (I, II, and III). 20 μM of each calpain inhibitor was treated with sperm during capacitation. Blots shown here are representative of three independent experiments.
-
Figure 1—source data 1
Relative amount of recombinant CatSper1 during in vitro proteolysis.
Raw data points for the quantification of protein levels for Figure 1G.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig1-data1-v3.xlsx
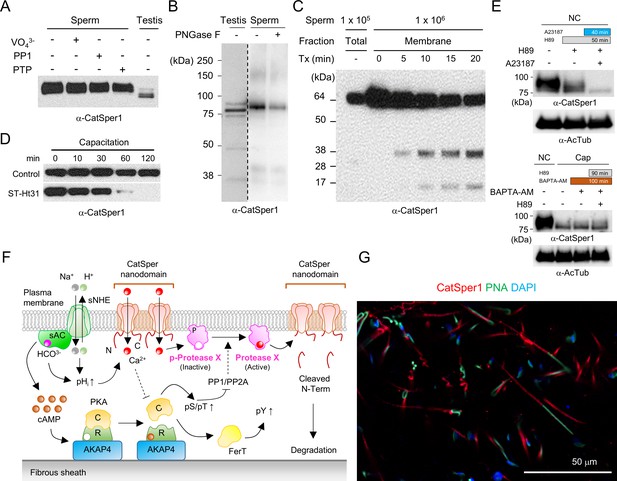
Molecular mechanism for CatSper1 down-regulation during sperm capacitation.
(A) CatSper1 is not a phosphoprotein. Treatment of sonication-solubilized cauda sperm (1 × 106 cells) with protein phosphatase 1 (PP1, 0.1 unit), protein tyrosine phosphatase (PTP1B, 5 units), and PTP inhibitor (VO43-, 1 mM) does not change the molecular weight of CatSper1. (B) N-glycosidase, PNGase F, does not change the electrophoretic mobility of CatSper1 from sperm membrane fraction. (C) CatSper complex in the flagellar membrane is compartmentalized and protected from a protease. The membrane fraction from cauda sperm cells were incubated with 0.1% Triton X-100 (Tx) in PBS at 4°C for 0, 5, 10, 15, and 20 min. The addition of Triton X-100 results in cleavage within the CatSper1 N-terminus. N-terminal truncations of CatSper1 were not observed in sperm directly lysed in the sampling buffer. (D) Inhibition of PKA-anchoring to AKAP by st-HT31 peptide (10 μM) accelerates CatSper1 degradation in capacitating sperm. (E) CatSper1 processing involves two distinct signaling pathways. Treatment with Ca2+ ionophore A23187 (10 μM, blue) accelerates CatSper1 processing in the presence of PKA inhibitor, H89 (50 μM, gray) in non-capacitating spermatozoa (top, NC). Chelating intracellular Ca2+ by loading sperm cells with BAPTA-AM (10 μM, brown) does not affect CatSper1 processing regardless of PKA inhibition under capacitating conditions (bottom, Cap). (F) A model proposes the molecular mechanism of full-length CatSper1 downregulation in capacitated sperm cells. Increase in intracellular pH activates the CatSper channel in sperm undergoing capacitation CatSper1-degrading protease X is activated by the CatSper-mediated rise in intracellular Ca2+ and cleaves the -terminal domain of CatSper1. We speculate that the activity of the Ca2+-dependent protease X can be further modulated by PKA phosphorylation cascades (i.e. dephosphorylation by PP1/PP2A); Ca2+ suppresses PKA activation (dotted lines). The cleaved N-terminal fragment from CatSper1 is subject to degradation. The CatSper channel containing the N-terminal truncated CatSper1 might become inactive. (G) A representative immunofluorescence image of conglomerated sperm in the uterus. The uterus was collected after 8 hr coitus, subjected to clearing, and immunostained. Sperm cells with intact or processed Catsper1 are visible in the uterus. Blots shown here are representative of three independent experiments.
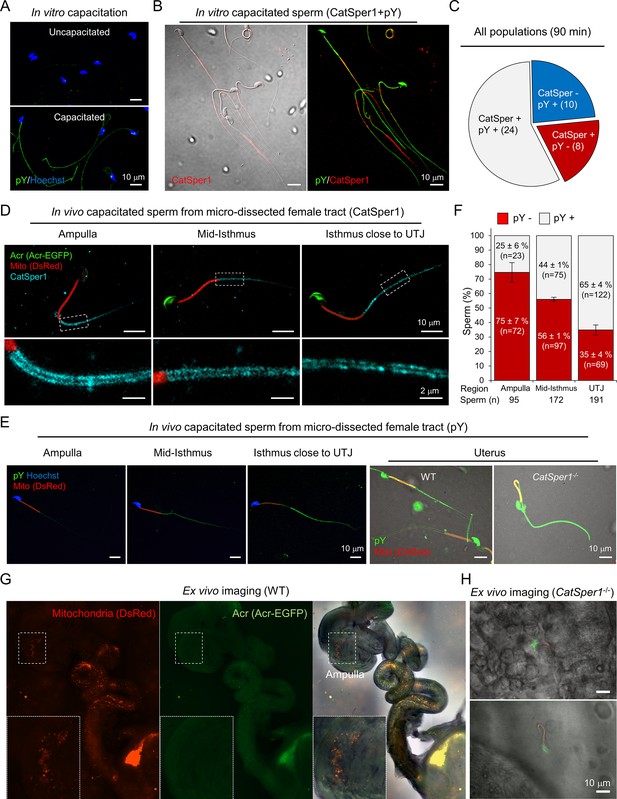
Sperm cells become heterogeneous functionally and molecularly along the female tract.
(A) Immunodetection of pY from in vitro capacitated sperm. (B) Sperm cells that maintain intact CatSper1 during in vitro capacitation exhibit reduced pY development. A representative image of CatSper1 (red) is merged with the corresponding DIC image (left) or pY image (right). Sperm were capacitated in vitro for 90 min (A–B). (C) A representative pie chart showing expression patterns of CatSper1 and pY in individual sperm cells of a single population of in vitro capacitated sperm. Sperm number in each group are indicated in parentheses. (D–F) Sperm cells capacitated in vivo show distinct molecular characteristics along the female tract. The degrees of CatSper1 processing (D) and development of tyrosine phosphorylation (pY) (E–F) during in vivo capacitation were analyzed by immunostaining of the sperm cells at different regions of microdissected female tracts 8 hr post-coitus. The indicated regions are magnified to show distributions of CatSper1 in sperm cells. Sperm cells that arrived at the ampulla are acrosome reacted (Acr-EGFP-negative) and CatSper1 intact (D) and lack pY development (E–F). Gradual increase of pY is observed in the oviductal sperm located closer to UTJ (E–F). Sperm cells that fail to pass UTJ and reside in the uterus show heterogeneous patterns of pY. Catsper1-/- sperm recovered from the uterus of a mated female show robust elevation of pY. Red, DsRed in mitochondria (Mito). (F) Sperm cells from each region of the female tract were counted and classified for pY positive (gray) or pY negative (red) from ampulla (n = 8), mid-isthmus and UTJ, respectively (n = 9) from 5 females at 8 hr post-coitus. Data is represented as mean ± SEM. See also Figure 2—source data 1. (G–H) WT sperm cells, but not Catsper1-/- sperm cells, which arrive at the ampulla are acrosome reacted. Ex vivo imaging of female tracts mated with WT (G) and Catsper1-/- (H) males (8 hr post-coitus). (G) WT sperm cells are acrosome-reacted at the ampulla (EGFP-negative, inset). (H) A few Catsper1-/- sperm cells observed at ampulla have intact acrosome (EGFP-positive). Red, DsRed in mitochondria; green, EGFP tagged to Acr (Acr-EGFP); Merged, fluorescent images merged with the corresponding DIC image. Fluorescence micrographs shown here are representative of at least three independent experiments.Su9-DsRed2/Acr-EGFP WT and Su9-DsRed2/Acr-EGFP Catsper1-/- mice (Chung et al., 2014) were used for mating (D, E, G, and H).
-
Figure 2—source data 1
Classification and quantification of pY from in vivo capacitated sperm.
Raw data points of quantification of pY signals in the sperm from different regions of the reproductive tracts from five females at 8 hr post-coitus for Figure 2F.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig2-data1-v3.xlsx
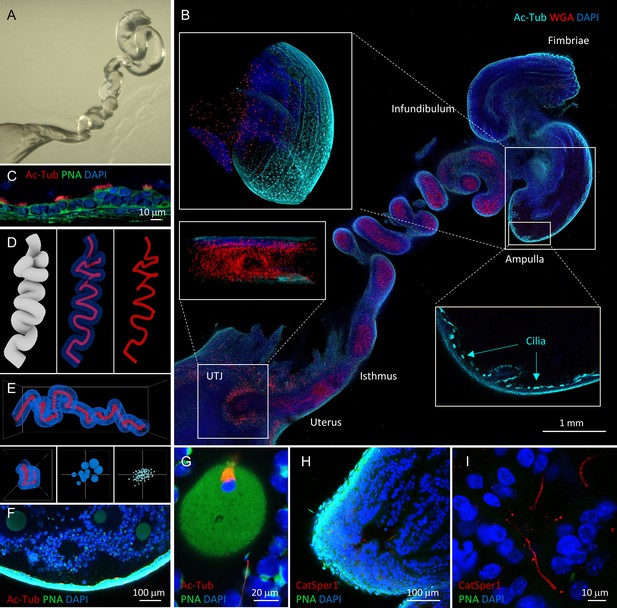
Tissue clearing preserves morphology of female reproductive tract and enables molecular imaging and post-processing of gametes in situ.
(A) Refractive index-matched cleared mouse female reproductive tract by CLARITY-based tissue clearing. (B) Optical imaging of the cleared female reproductive tract stained by WGA (red), Ac-Tub antibody (cyan) and DAPI (blue), 100×. Insets show cilia stained by Ac-Tub antibody in 2D (bottom right), a 3D (top left) projection of the ampulla, and a UTJ cross-section (bottom left). (C) Details of the ampullar epithelium stained by PNA (green), Ac-Tub antibody (red) and DAPI (blue), 400×. (D) 3D digital image reconstruction of the oviduct representing different 3D images rendered for oviductal surface (left) and central lumen of oviduct with (middle) or without (right) oviductal volume information. (E) Morphometric and fluorescent signal quantification analysis of the oviduct showing the morphometric meshwork representation of the 3D volumetric data from the oviduct imaging (top), the corresponding side view (bottom left) and the non-numerical visual representations of the basic volumetric (bottom middle) and fluorescent (bottom right) properties. (F) A fluorescent image showing a closer look of the cleared ampulla with oocytes (oocyte magnified in the panel G on the right-most side), 100×. (G) An oocyte with the meiotic spindle; a sperm cell is approaching the zona pellucida directly inside the ampulla, PNA (green), anti-AcTub antibody (red) and DAPI (blue), 630×. (H) A tile-scanned confocal image of the epithelium of the cleared ampulla (8 hr post-coitus) stained by anti-CatSper1 antibody (red), PNA (green), and DAPI (blue), 100×. (I) Details of the sperm stained directly inside the ampulla by anti-CatSper1 antibody (red). Two linear CatSper domains are clearly recognizable by confocal imaging. Cell nuclei are stained with DAPI (blue); acrosomes are stained with PNA (green). Images shown here are representative of at least three independent experiments. See also Figure 3—videos 3–7.

Multicolor 3D fluorescence in situ imaging of cleared mouse reproductive organs.
(A–C) Confocal images of cleared female reproductive organs. X-CLARITY processed ovary (A); actin stained by phalloidin (green), WGA (red), and DAPI (blue). Developing oocytes show intense WGA signal. Oviduct (B) and UTJ (C, 3D rendered) from PACT-PRESTO processed female reproductive tract; stained with WGA (green) and DAPI (blue). Mucus in the lumen of the female tract is preserved (WGA, green). (D–I) Images of X-CLARITY-cleared male reproductive organs. Whole testis before (left) and after (right) clearing (D), a seminiferous tubule from the cleared testis (E), 3D surface presentations of a seminiferous tubule (F) and sperm inside the tubule (G) of SWITCH-cleared testis from Su9-DsRed2/Acr-EGFP transgenic male. (H–I) Fluorescence images of an X-CLARITY cleared epididymis from a Su9-DsRed2/Acr-EGFP male. A 3D rendered image (H) and a confocal image (I). EGFP, green; Red, DsRed. Images shown here are representative of three independent experiments. See also Figure 3—videos 1–7.
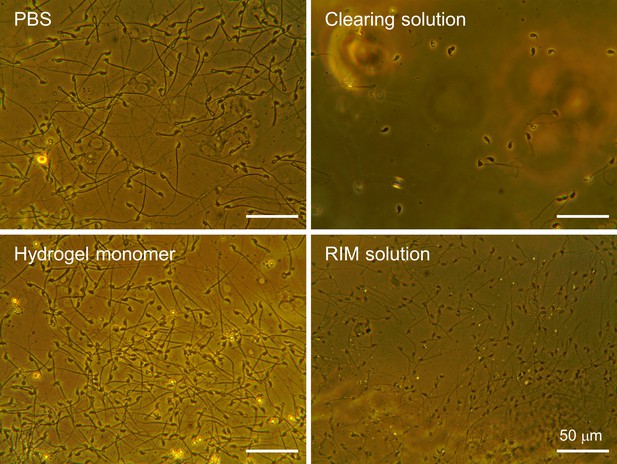
The effect of various components used in tissue clearing procedure on sperm cells.
Uterine sperm from whole-body fixed females (3 hr post-coitus) by cardiac perfusion of hydrogel monomer were adhered to glass slides and exposed for 5 days to PBS, clearing solution, hydrogel monomer, and RIM solution. Without post-fixation and polymerization, sperm cells released from the fixed female tract were lysed and detached by clearing solution containing 4% SDS, due to incomplete fixation in the lumen and no supportive matrix to fix the cell location.
3D volume imaging of a whole ovary; related to Figure 3.
(A) A movie of stitched and Z-stacked confocal slices of X-CLARITY cleared and stained ovary. (B) A 3D rendered movie from (A). Blue, DAPI; Green, Actin; Red, WGA.
3D Volume imaging of the testis and epididymis; related to Figure 3.
(A) A 3D rendered movie of stitched and Z-stacked confocal slices of the testis. (B) A movie of Z-stacked confocal images of the epididymis. X-CLARITY cleared a Su9-DsRed2/Acr-EGFP male. DAPI, blue; DsRed, red; EGFP, green.
3D rotational movie of whole female reproductive tract.
3D reconstruction of the cleared female reproductive tract corresponding to the Figure 3 showing voluminous uterus continuing into the oviduct and ampulla. DAPI, blue; WGA, red; Ac-Tub, cyan.
z-stack movie of the ampulla.
z-stack flight through video showing the ampulla with oocytes. DAPI, blue; WGA, green.
z-stack movie of the oviduct.
(A) 3D reconstruction of the oviductal isthmus. (B) z-stack flight-through projection. DAPI, blue; WGA, green.
3D rotational movie of the UTJ.
Reconstruction of the UTJ showing 3D projections. DAPI, blue; WGA, green.
Moments after fertilization.
3D reconstruction of the oocyte and sperm in the cleared ampulla. (A) A flight-through z-stack video showing the oocyte with meiotic spindle and sperm stained by AcTub signal approaching the zona-pellucida. (B) The corresponding 3D projection video to (A). DAPI, blue; PNA, green; AcTub, red.

In situ molecular imaging of sperm reveals the changes in acrosomal status and CatSper1 fluorescent patterns during capacitation along the female reproductive tract.
(A) Representative fluorescent confocal microscope images of acrosome and CatSper1 fluorescent patterns from three different regions along the cleared female reproductive tract (Ampulla, top; Middle isthmus, middle; Proximal isthmus, bottom) with different magnifications of the corresponding areas. (B) A cartoon image of the female reproductive tract showing the approximate boundaries between the regions of interest (left) used as grouping variable in the subsequent quantification (right). The total number of CatSper1-intact sperm (red columns) or acrosome-intact sperm (blue columns) counted from four independent experiments (n = 4) is shown (bottom). Circles indicate the proportion of sperm cells in each examined site from an independent experiment. Statistical analyses were carried out with one-way analysis of variance (ANOVA) with Tukey post hoc test. Means with different letters indicate significant difference (p<0.05) in pairwise comparison between the different regions of female tract. Data is represented to mean ± SEM. See also Figure 4—source data 1 and Figure 4—video 1.
-
Figure 4—source data 1
Sperm counts for intact CatSper1 and acrosome from in situ imaging.
Raw data points of sperm count for quantification of CatSper1 integrity and acrosome state for Figure 4B.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig4-data1-v3.xlsx
Sperm in the different regions of the oviduct.
(A) Sperm in the ampulla. 3D reconstruction of the representative image of the sperm cells from ampulla. Individual CatSper1 domains are recognizable in some phases of the image rotation. (B) Sperm in the mid-isthmus. 3D reconstruction of the cleared female reproductive tract with sperm inside isthmus shows CatSper1 from the tail and PNA signal from acrosome. (C) Sperm in the isthmus close to UTJ. 3D reconstruction of the cleared female reproductive tract with sperm close to UTJ region show PNA signal from acrosome, but not CatSper1. DAPI, blue; PNA, green; CatSper1, red.
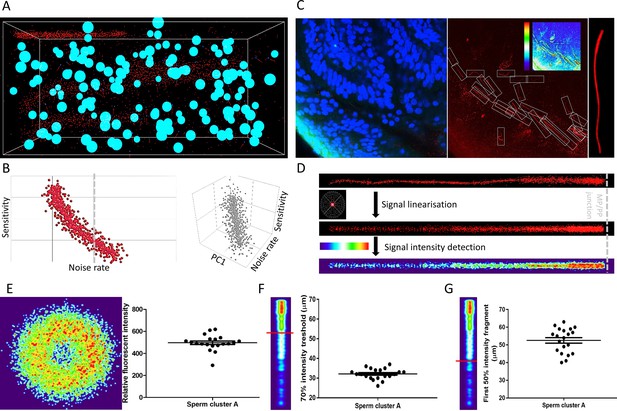
ANN automatically detects fluorescent patterns from 3D volume images of a cleared female reproductive tract, enabling isolation and statistical comparisons of sperm cells.
(A) A 3D training environment for ANN emulating sperm cells, somatic nuclei, and noise. (B) Examples of ANN training statistics showing the trend of correlation between the noise rate in the training environment and the sensitivity of ANN (left), and a three-dimensional correlation trend between noise rate in the training environment, sensitivity and principal component (PC1) consisting of sperm and nuclei abundancies (right). Nsampling environment = 1000, individual data points plotted. (C) A microscopic focal plane image of the sperm cluster inside the cleared female reproductive tract used for evaluating the ANN performance in real sample (left), its superposition in the CatSper1 channel with the individual sperm tails in detection frames with the inset analytical heatmap (middle) and the magnification of one of the analytical frames with a CatSper1-positive sperm tail (right). (D) Representation of the fluorescent signal in the sperm tail after normalizing individual voxels to signal from the corresponding sperm nucleus (top), after applying linearization and overlay algorithms (middle), and heatmap representation of the relative fluorescent intensities among multiple sperm tail (bottom). (E) Analysis of the relative intensities of the fluorescent signals from sperm located inside the mid-isthmus cleared female reproductive oviduct. The left panel represents the intensity of CatSper1 fluorescent signal in the cross-section of one sperm tail from 20 individual sperm under analysis (middle isthmus). The right panel show the distribution of relative fluorescent intensity of 20 sperm. (F) Analysis of the continuity of the fluorescent signal along the individual sperm tails; the first fragment of the 70% signal intensity decreases from the midpiece/principal piece interface. (G) The first fragment of the 50% signal intensity decreases from the midpiece/principal piece interface. Graphs in E-G represent Mean ± SEM. with individual measurements are plotted as dots (N = 20). See also Figure 5—source data 1 and Figure 5—video 1.
-
Figure 5—source data 1
ANN development source materials.
Custom scripts and 3D objects for developing training environments, ANN input feeding and training, and constructing numerical objects coordinates arrays for supervised learning, and ANN performance evaluation.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig5-data1-v3.zip

Generation of training environments for ANN development.
Examples of 12 environments with different abundancies (voxel occupancies) of sperm (left), nuclei (middle) and noise (right). The combinations of individual environments with different abundancies of their individual components (sperm, nuclei, noise) were used for ANN training (N = 104). See also Figure 5—source data 1.

A workflow diagram for ANN and performance evaluation.
(A) A workflow diagram showing the individual steps of ANN processing: 1. Primary data from fluorescent microscopy volume imaging, 2. Isolating of individual 3D environment components (sperm, nuclei, noise), 3. Generation of 3D training environments composed from the different abundancies of the individual components with exactly defined topologies of individual components; 4. Creating the virtual z-stacks (virtual ‘focal planes’) from these environments, 5. Processing the virtual z-stacks as inputs for supervised ANN training with the set of sperm positive and negative frames as output, 6. Evaluation of the ANNs performance using the ability to detect individual sperm cells as the target performance indicator during iteration process, 7. A subsequent evaluation of the individual generated ANN performance and application of the selected ANN on the real sample. (B) Examples of ANN training statistics showing trends of correlation between the number of sperm cells in the training environment and the sensitivity of ANN (left) and between the number of nuclei in the training environment and the sensitivity of ANN (right). Nsampling environment = 1000, individual data points plotted. (C) Dot plots showing the sensitivity and specificity of the sperm detection by individually generated ANNs. Graph represents Mean ± SEM. with individual measurements are plotted as dots (N = 20). See also Figure 5—source data 1.
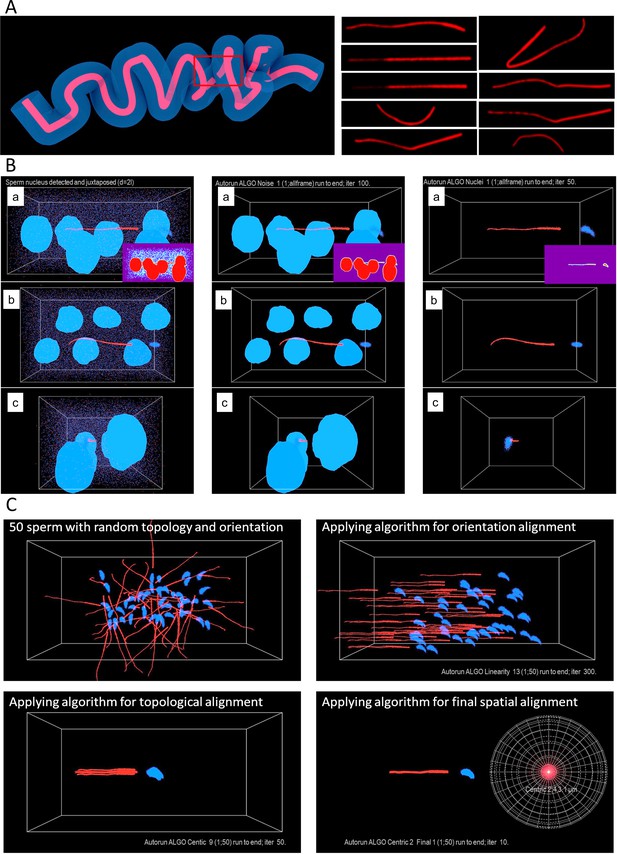
Image post-processing of sperm detected inside the cleared female reproductive tract.
(A) Localizing 3D volumetric data from a region of the oviduct (left) and examples of the processed CatSper1 sperm tail signal from the location (right). (B) Digital removal process of the signals from somatic cell nuclei and the noise by reverse ANN training environment generation. The sperm in the analytic frame with signal from somatic cell nuclei and noise in three orientation (left); the corresponding de-noised frames (middle); the final frames with sperm only after removal of somatic cell nuclei (right). (C) Demonstration of the post-processing procedure of the sperm alignment to generate composite CatSper1 intensity heatmaps. The circular map shows the minimization of the signal dispersion of the transversal tail projection from multiple aligned sperm tails. See also Figure 5—source data 1.
A flight-through movie of analytical space without noise, related to Figure 5A.
3D projection of the ANN training environment showing its two components (sperm, somatic nuclei) without noise.
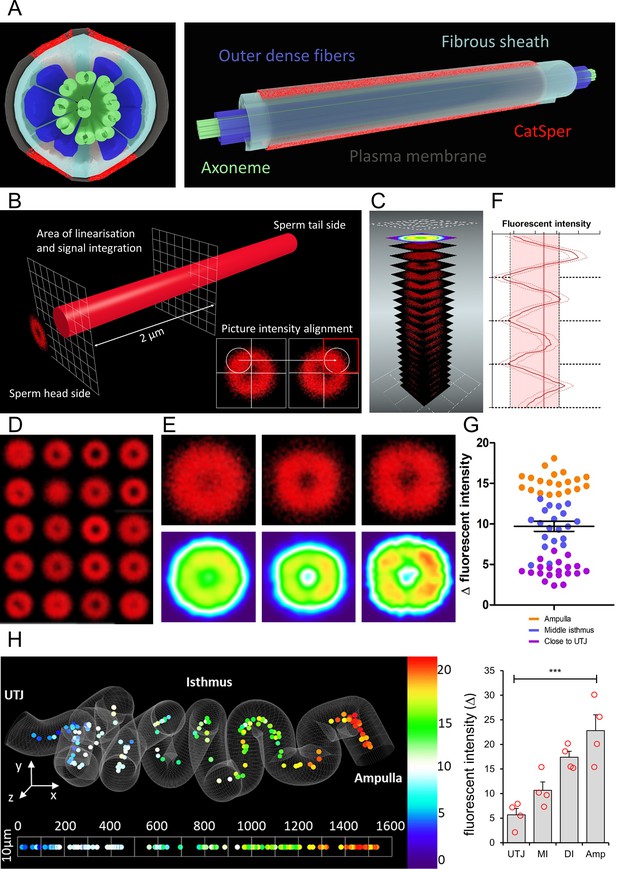
ANN assessment of quadrilateral CatSper nanodomains and Δ fluorescent intensity in sperm population along the cleared female tract immunolabeled for CatSper1 conforms to findings by other approaches used in this study.
(A) 3D perspective schematic views of quadrilateral CatSper nanodomains. A cross-section (left). A side view (right). (B) A schematic diagram describing the image processing procedure. (C) An illustration of generating a heatmap from the pre-processed micrographs. (D) 20 processed micrographs of the CatSper1 signal from the sperm cluster from middle isthmus of the cleared oviduct. (E) Processed micrographs (top) and their corresponding heatmaps (bottom) from 20 spermatozoa from the oviduct close to UTJ (left), middle isthmus (middle), and ampulla (right). (F) An example of fluorescent intensity analysis of processed images showing the four peaks corresponding to four CatSper1 quadrilateral domains and calculated averaged Δ value (transversal red line). (G) Analysis of the fluorescent intensity differences (Δ values; red area in panel F) among three sperm populations from ampulla, middle isthmus, and isthmus close to UTJ. Graph represents Mean ± SEM. with individual measurements are plotted as dots (N = 60). (H) A topological heatmap showing the integrity of the quadrilateral CatSper domain organization represented by Δ values of the fluorescent intensity along the morphometrical space of the cleared oviduct (left, Nsperm = 152) with the corresponding inferential statistical analysis of the differences of the signal intensities (Δ values, right) among four sperm populations (Amp – Ampulla, DI – Distal Isthmus close to ampulla, MI – middle isthmus, UTJ – utero-tubal junction). Bars denote Mean ± SEM. The baseline indicates homogenous angular distribution of the CatSper fluorescent signal with no quadrilateral distributions. Statistical significance was calculated using KW-ANOVA, (***p<0.001), Nanimals = 4. See also Figure 6—source datas 1 and 2.
-
Figure 6—source data 1
Quantification of Δ values of CatSper1 fluorescent intensity.
Raw data points for quantification of the integrity of the quadrilateral CatSper domain organization represented by Δ values of the fluorescent intensity along the cleared oviduct for Figure 6H.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig6-data1-v3.xlsx
-
Figure 6—source data 2
3D in situ analytical tools for CatSper quadrilateral structure.
Custom scripts for analyzing CatSper quadrilateral structure.
- https://cdn.elifesciences.org/articles/62043/elife-62043-fig6-data2-v3.zip
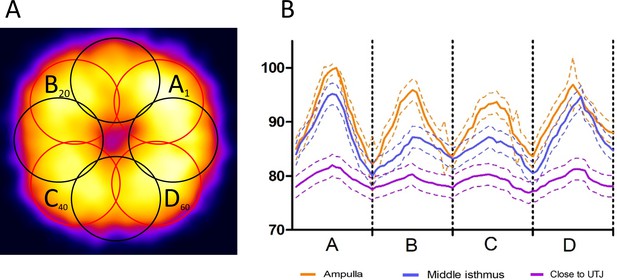
Analysis of the transversal quadrilateral CatSper1 domains organization in the tail of sperm inside the cleared female reproductive tract.
(A) Demonstration of the individual round analytical areas within the transversal projection of the CatSper1 tail signal intensity heatmap. Individual circles indicate the ROI areas of the signal intensity integration and calculation (Nshowed = 8; Ntotal = 80) with the starting point in the ROI with the highest intensity (top right corner). (B) Comparison of the transversal quadrilateral CatSper1 domains organization quality among sperm populations from three regions of the cleared oviduct. Solid lines represent Mean and dotted lines denote ± SEM. N = 60. See also Figure 6—source data 2.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Catsper1 | GenBank | Gene ID: 225865 | |
| Gene (Mus musculus) | Catsper1 | GenBank | Gene ID: 225865 | |
| Strain, strain background (Escherichia coli) | 10-b | New England BioLabs | Cat# C3019H | |
| Strain, strain background Mus musculus | B6.129S4-Catsper1tm1Clph/J | Laboratory of David E. Clapham (Ren et al., 2001) | RRID:IMSR_JAX:0 18311 | |
| Strain, strain background Mus musculus | B6D2-Tg(CAG/Su9 -DsRed2,Acr3-EGFP)RBGS002Osb/OsbRbrc | Hasuwa et al., 2010 | RRID:IMSR_RBRC03743 | |
| Strain, strain background Mus musculus | C57BL/6 | Charles River Laboratories | Crl:029 | |
| Strain, strain background Mus musculus | B6D2F1 | Charles River Laboratories | Crl:099 | |
| Strain, strain background Mus musculus | CD1 | Charles River Laboratories | Crl:CD1(ICR) RRID:IMSR_CRL:022 | |
| Strain, strain background (Escherichia coli) | b-10 | New England BioLabs | CMC0016 | |
| Cell line (Homo sapiens) | 293T | ATCC | Cat# CRL-3216; RRID:CVCL_0063 | |
| Biological sample (Mus musculus) | Testes | Chung lab, Yale School of Medicine | N/A | |
| Biological sample (Mus musculus) | Spermatozoa | Chung lab, Yale School of Medicine | N/A | |
| Antibody | Rabbit polyclonal anti-mCatSper1 | Laboratory of David E. Clapham (Ren et al., 2001) | RRID:AB_2314097 | Western blot, (2 μg/mL); Immunocytochemistry, (10 μg/mL); In situ imaging, (7 μg/mL) |
| Antibody | Rabbit polyclonal anti-mCatSper3 | Laboratory of David E. Clapham (Qi et al., 2007) | N/A | (2 μg/mL) |
| Antibody | Rabbit polyclonal anti-mCatSperε | Laboratory of Jean-Ju Chung (Chung et al., 2017) | N/A | (1.6 μg/mL) |
| Antibody | Rabbit Polyclonal CA-IV antibody | SantaCruz | Cat# sc-25598 | (1:500) |
| Antibody | Mouse monoclonal anti-caveolin1 (clone 2297) | BD Biosciences | Cat# 610406 RRID:AB_397788 | (1:500) |
| Antibody | Mouse monoclonal anti-phosphotyrosine (clone 4G10) | EMD milipore | Cat# 05–321; RRID:AB_309678 | Western blot, (1:1,000); Immunocytochemistry, (1:1000) |
| Antibody | Mouse monoclonal anti-acetylated tubulin (clone 6-11b-1) | EMD milipore | Cat# 7451; RRID:AB_609894 | Western blot, (1:20,000); In situ imaging, (1:100) |
| Antibody | Rabbit monoclonal anti-beta actin (clone 13E5) | Cell signaling technology | Cat# 4970 RRID:AB_2223172 | (1:100) |
| Antibody | Rabbit monoclonal anti-HA (clone C29F4) | Cell signaling technology | Cat# 3724 RRID:AB_1549585 | (1:2,000) |
| Antibody | Goat polyclonal anti-mouse IgG-HRP | Jackson ImmunoResearch | Cat# 115-035-003; RRID:AB_10015289 | (1:10,000) |
| Antibody | Goat polyclonal anti-rabbit IgG-HRP | Jackson ImmunoResearch | Cat# 111-035-144; RRID:AB_2307391 | (1:10,000) |
| Antibody | Goat polyclonal anti-rabbit IgG-Alexa 568 | Invitrogen | Cat# A-11036 RRID:AB_10563566 | Immunocytochemistry, (1:1000); In situ imaging, (1:500) |
| Antibody | Goat polyclonal anti-rabbit IgG-Alexa 647 | Invitrogen | Cat# A-21245 RRID:AB_2535813 | Immunocytochemistry, (1:1000); In situ imaging, (1:500) |
| Antibody | Goat polyclonal anti-mouse IgG-Alexa 488 | Invitrogen | Cat# A-11029 RRID:AB_138404 | Immunocytochemistry, (1:1000); In situ imaging, (1:500) |
| Antibody | Goat polyclonal anti-mouse IgG-Alexa 647 | Invitrogen | Cat# A21236 RRID:AB_2535805 | Immunocytochemistry, (1:1000); In situ imaging, (1:500) |
| Antibody | Mouse monoclonal anti-HA agarose (clone HA-7) | EMD milipore | Cat# A2095 AB_257974 | (10 uL) |
| Antibody | Anti-HA magnetic beads (clone 2–2.2.14) | Thermo scietific | Cat# 88836 RRID:AB_2749815 | (5 uL) |
| Recombinant DNA reagent | pcDNA3-mCatsper1 | Laboratory of David E. Clapham | N/A | |
| Recombinant DNA reagent | pcDNA3.1(-)-Flag-Catsper1-FL-HA | This paper | N/A | Encoding Mouse CatSper1, 1–686 aa |
| Recombinant DNA reagent | pcDNA3.1(-)-Flag-Catsper1-ND-HA | This paper | N/A | Encoding Mouse CatSper1, 345–686 aa |
| Commercial assay or kit | O-glycosidase | New England BioLabs | Cat# P0733S | (80,000 Unit) |
| Commercial assay or kit | PNGase F | Sigma-Aldrich | Cat# P9120 | (2 nM Unit) |
| Peptide, recombinant protein | Protein phosphatase 1 (PP1) | New England BioLabs | Cat# P0754 | (0.1 Unit) |
| Peptide, recombinant protein | Protein tyrosine phosphatase 1B (PTP1B) | Abcam | Cat# Ab42574 | (5 Unit) |
| Chemical compound, drug | Sodium orthovanadate | New England BioLabs | Cat# P0758S | (1 mM) |
| Chemical compound, drug | H89 | Calbiochem | Cat# 371963 | (50 μM) |
| Chemical compound, drug | ST-Ht31 | Promega | Cat# V8211 | (10 μM) |
| Chemical compound, drug | Calyculin A | Calbiochem | Cat# 208851 | (100 nM) |
| Chemical compound, drug | A23187 | Sigma-Aldrich | Cat# C7522 | (10 μM) |
| Chemical compound, drug | Calpain inhibitor I | Calbiochem | Cat# 208719 | (20 μM) |
| Chemical compound, drug | Calpain inhibitor II | Enzo life science | Cat# BML-PI100 | (20 μM) |
| Chemical compound, drug | Calpain inhibitor III | Enzo life science | Cat# BML-PI130 | (20 μM) |
| Chemical compound, drug | PNA-Alexa 568 | Invitrogen | Cat# L32458 | (1:500) |
| Chemical compound, drug | WGA-Alexa 555 | Invitrogen | Cat# W32464 | (1:500) |
| Chemical compound, drug | WGA-Alexa 647 | Invitrogen | Cat# W32466 | (1:500) |
| Chemical compound, drug | Azo-initiator | Wako | Cat# VA-044 | |
| Chemical compound, drug | 32% Paraformaldehyde | Electron Microscopy Sciences | Cat# 15714 s | |
| Chemical compound, drug | 50% Glutaraldehyde | Electron Microscopy Sciences | Cat# 16310 | |
| Chemical compound, drug | Electrophoretic Tissue Clearing Solution | Logos Biosystems | Cat# C13001 | |
| Chemical compound, drug | N-methyl-d-glucamine | Sigma-Aldrich | Cat# M2004 | |
| Chemical compound, drug | Diatrizoic acid | Sigma-Aldrich | Cat# D9268 | |
| Chemical compound, drug | 60% Iodixanol | Sigma-Aldrich | Cat# D1556 | |
| Chemical compound, drug | DAPI | Thermo scietific | Cat# D1306 | 1:1000 |
| Chemical compound, drug | Hoechst | Thermo scietific | Cat# 62249 | 1 μg/mL |
| Chemical compound, drug | Acrylamid | BioRad | Cat# 1610140 | |
| Software, algorithm | ImageJ | National Institue of Health | Public domain: https://imagej.nih.gov/ij/ | |
| Software, algorithm | Zen software | Carl Zeiss AG | ZEISS ZEN lite www.zeiss.com | |
| Software, algorithm | Imaris | BitPlane; Oxford instruments | Imaris 9.6 | |
| Software, algorithm | MATLAB | MathWorks | MATLAB 9.3 (R2017b) | |
| Software, algorithm | Blender | Blender Foundation, community | Blender 2.79 www.blender.org | |
| Software, algorithm | GraphPad | GraphPad Software | GraphPad Prism 5 | |
| Software, algorithm | ANNs development source materials | This paper | Source archive attached to Figure 5 | Set of ANN development tools |
| Software, algorithm | CatSper quadrilateral structure 3D in situ analytical tools | This paper | Source archive attached to Figure 6 | Set of 3D CatSper analytical tools |