Flagellar energetics from high-resolution imaging of beating patterns in tethered mouse sperm
Figures
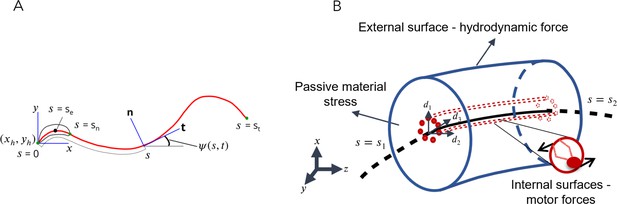
Schematic representations for the Soft, Internally driven Kirchhoff rod model.
(A) Geometric variables defined along the centerline. (B) An arbitrary control volume used for deriving the equations of the model: the volume consists of the passive flagellar material; hydrodynamic forces act on the external surface while axonemal motors act on the internal surfaces. The passive material adjacent to the cross-sectional faces at either end exerts stresses on those faces.
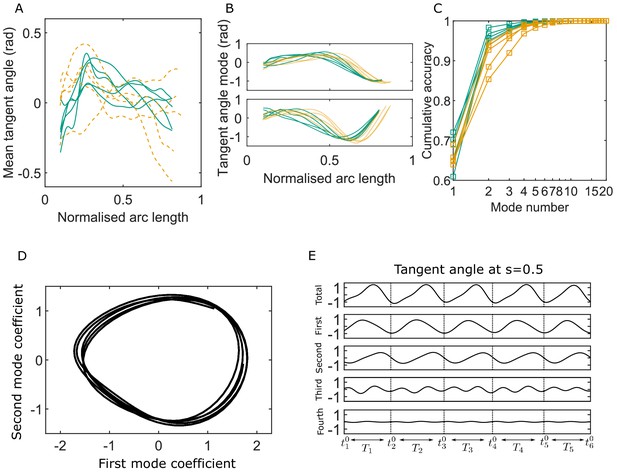
Key variables of the Chebyshev polynomial-based proper orthogonal decomposition (C-POD) of experimental tangent-angle data.
(A) Time-averaged tangent-angle profiles for five wildtype (WT) (continuous curves) and five knockout (KO) (dashed curves) samples. (B) First (top) and second (bottom) C-POD shape modes for WT (continuous curves) and KO (dashed curves) samples; the colors are as in (A). (C) Cumulative accuracy of the C-POD representation for WT and KO samples; the colors are as in (A). A representation using the first four modes captures 95% or more of the observed centerline shapes for all samples. (D) Five shape cycles for a single WT sample in the parameter space defined by the time-dependent coefficients of the first two C-POD shape modes. The zero-crossing of the second modal coefficient marks the start of a new cycle. (E) Contributions of the first four modes to the tangent angle at the midpoint of the sperm body in the five tangent-angle cycles in (D): the horizontal line in the top plot is the time-averaged tangent angle for this WT sample. The starting time of the th cycle is denoted as , and its duration (i.e., cycle time) is .
-
Figure 2—source data 1
Numerical data for Figure 2A, B.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig2-data1-v2.mat
-
Figure 2—source data 2
Numerical data for Figure 2C.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig2-data2-v2.mat
-
Figure 2—source data 3
Numerical data for Figure 2D.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig2-data3-v2.mat
-
Figure 2—source data 4
Numerical data for Figure 2E.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig2-data4-v2.mat
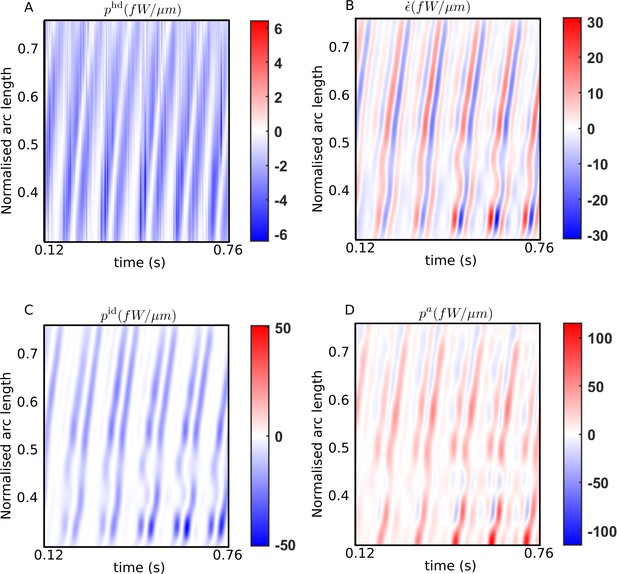
Spatiotemporal distributions of the rates of (A) hydrodynamic dissipation, (B) elastic storage, and (C) internal dissipation, and (D) the active power along the flagellum of a wildtype sperm over several beat cycles: red indicates positive rates, while blue indicates negative rates.
The data in (C) and (D) have been obtained using the scaling value of 103 Pa s μm4 for the internal friction coefficient.
-
Figure 3—source data 1
Numerical data for all kymographs.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig3-data1-v2.mat
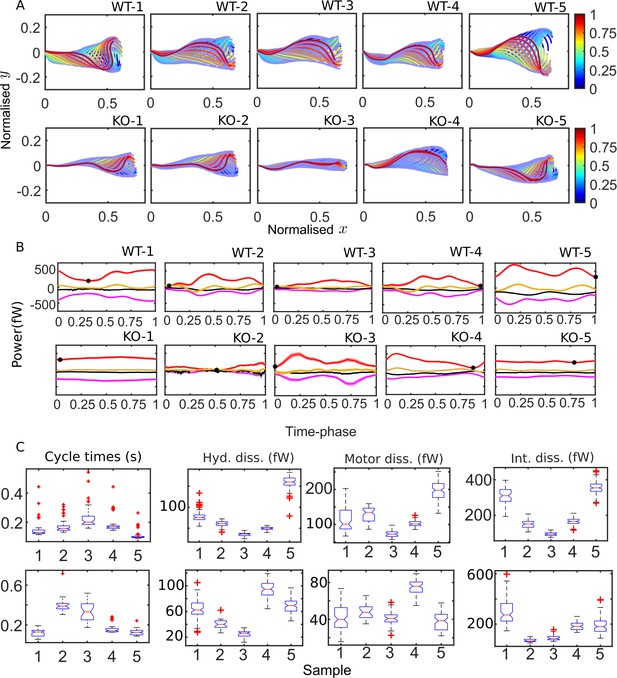
Mean cycles of beat patterns and energetics.
(A) Each colored curve shows the mean shape at a particular phase of the mean cycle for the five wildtype (WT) (top row) and knockout (KO) (bottom row) samples. The color bands around each curve indicate the standard error in the mean component. (B) Mean cycles for the magnitudes of the net elastic storage (yellow), hydrodynamic dissipation (black), internal frictional dissipation (magenta), and active power (red) in WT (top panel) and KO (bottom panel) sperm samples corresponding to those in (A). Bands show standard errors in means. (C) Statistical distributions of cycle times and dissipation rates in each of the WT (top panel) and KO (bottom panel) samples. The box-plots present the median (red line), the first and third quartile (bottom and top box edges), and minimum and maximum (lower and upper whiskers) values for 40–60 cycles. Outliers that are more than 1.5 times the interquartile range away from the top or bottom of the box are indicated by red crosses. The notch extremes correspond to , where q1, q2, and q3 are the first, second (median), and third quartiles, respectively, and is the number of observations (McGill et al., 1978).
-
Figure 4—source data 1
Numerical data for Figure 4A.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig4-data1-v2.mat
-
Figure 4—source data 2
Numerical data for Figure 4B.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig4-data2-v2.mat
-
Figure 4—source data 3
Numerical data for Figure 4C (cycle times).
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig4-data3-v2.mat
-
Figure 4—source data 4
Numerical data for Figure 4C (hyd. dissn., motor dissn., and int. dissn.).
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig4-data4-v2.mat
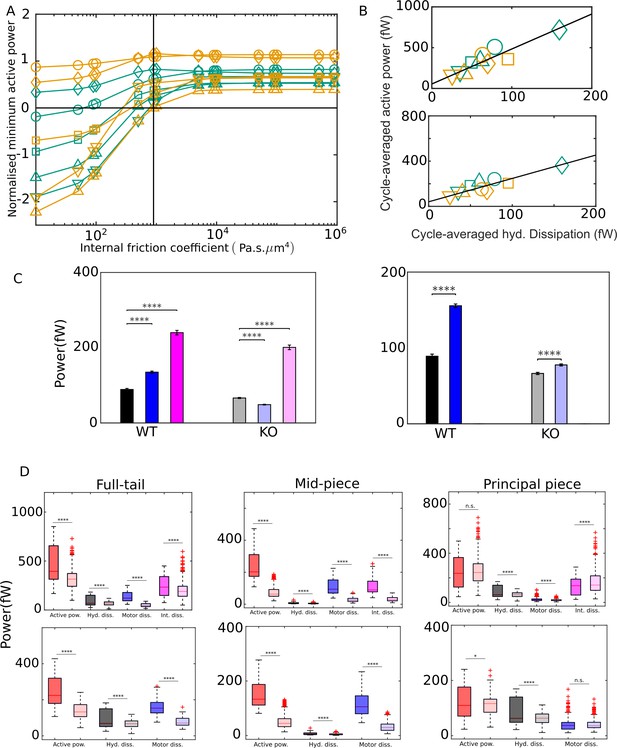
External versus internal dissipation in flagella.
(A) Effect of the value of internal friction coefficient, , on the minimum value of the net active power required to overcome dissipation for wildtype (WT) (blue) and knockout (KO) (red) samples. At each , and for each sample, the minimum in the mean cycle of the net active power, , is normalized by the time average of the motor input, , over all cycles. The vertical line is the scaling value of 103 Pa s μm4. (B) Correlation of time averages of the hydrodynamic dissipation and net active power for Pa s μm4 (top) and 0 (bottom). The lines are linear fits through data for both species. (C) Comparison of the external hydrodynamic dissipation (black) with the motor (blue) and passive internal frictional (magenta) dissipations obtained with Pa s μm4 (left) and 0 (right). The bars represent the averages of the cycle-means of dissipations pooled from all the five sperm samples in each genotype; the error bars represent 1 standard deviation in each direction in the set of pooled cycle-means. (D) Statistical distributions of the cycle-means of powers from the WT (dark color boxes) and KO (light color boxes) samples pooled together over the entire tail (left), mid-piece (middle), and principal piece (right). The top and bottom panels are for Pa s μm4 and 0, respectively. In (C) and (D), unpaired two-tailed t-tests are used to compare population means; **** refers to a significance level of , *** , ** , *. Differences are not significant (n.s.) when .
-
Figure 5—source data 1
Numerical data for Figure 5A.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig5-data1-v2.mat
-
Figure 5—source data 2
Numerical data for Figure 5B.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig5-data2-v2.mat
-
Figure 5—source data 3
Numerical data for Figure 5C.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig5-data3-v2.mat
-
Figure 5—source data 4
Numerical data for Figure 5D.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig5-data4-v2.mat
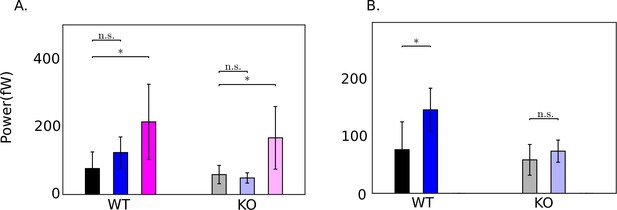
Comparison of population means of time-averaged dissipations obtained with the five wildtype and Crisp2 knockout mice sperm samples obtained with (A) =103 Pa.s.m4 and (B) = 0 Pa.s.m.
Error bars shown correspond to 1 standard deviation in the set of samples. Unpaired two-tailed t-tests are used to compare population means; ** , *. Differences are not significant (n.s.) when .
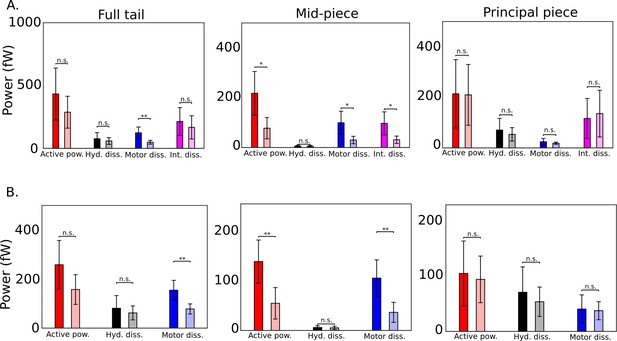
Comparison across genotypes of population means of time-averaged powers obtained with the five wildtype and Crisp2 knockout mice sperm samples o =103 Pa.s.m4 is shown in A and = 0 Pa.s.m4 in B.
Error bars shown correspond to 1 standard deviation in the set of samples. Unpaired two-tailed t-tests are used to compare population means; **, *. Differences are not significant (n.s.) when .
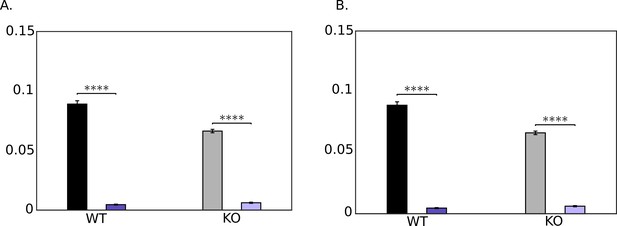
Comparison of pooled averages of the cycle-means of hydrodynamic dissipation in the tail (black) and dissipation at head due to hydrodynamic and tethering resistances (purple): , *. =103 Pa.s.m4 is shown in A and = 0 Pa.s.m4 in B.
Differences are not significant (n.s.) when .
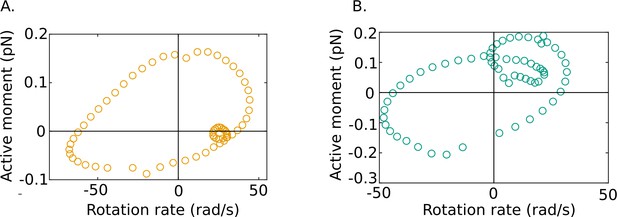
Out-of-phase mean beat cycles of active moment density and angular rotation rate at in (A) wildtype (WT)-1 and (B) knockout (KO)-1 samples.
-
Figure 6—source data 1
Numerical data for Figure 6.
- https://cdn.elifesciences.org/articles/62524/elife-62524-fig6-data1-v2.mat
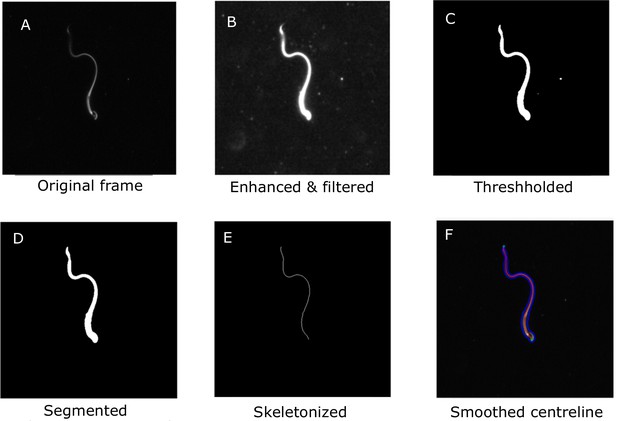
Main steps in the image-processing algorithm shown for a single frame.
A. The original frame B. Enhanced and filtered C. Thresholded frame D. Segmented E. Skeletonized frame F. Smoothed centreline.
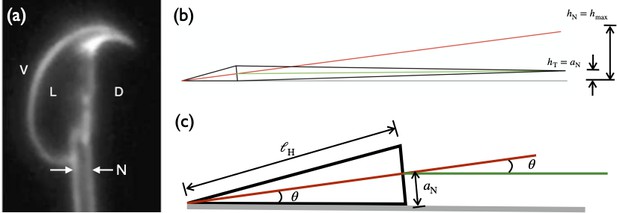
Orientation of the sperm cell with respect to the glass slide.
(a) Image from Woolley, 2003 showing the left side (L) of a mouse sperm head facing the viewer with the ventral (V) and dorsal (D) sides of the head indicated. The concave side of the hook is towards the dorsal side. Also shown is the neck (N) at the proximal end of the flagellum. (b) Schematic showing the mouse sperm body as viewed from its dorsal side when the left side of its head is against the wall. The intrinsic angle made by the head with the flagellum at the neck enables planar beating when the left side of the head is against the wall. The red and green lines indicate the axes of the head and the tail. In this orientation, the flagellum beats in a plane parallel to the wall and its centerline is at a distance equal to the neck radius ,, from the wall. If there had been no angle at the neck, the tip of the tail would be at a height of . (c) The angle (in radians) of the head axis to the wall, .
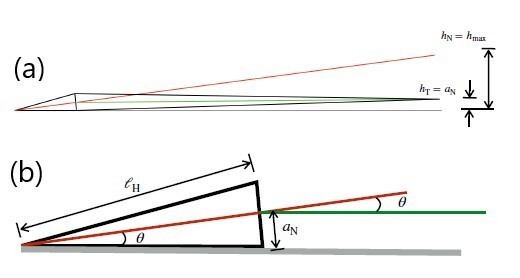
(a) Schematic showing the mouse sperm body as viewed from its dorsal side when the left side of its head is against the wall. The intrinsic angle made by the head with the flagellum at the neck enables planar beating when the left side of the head is against the wall. The red and green lines indicate the axes of the head and the tail. In this orientation, the flagellum beats in a plane parallel to the wall and its centreline is at a distance equal to the neck radius ,an, from the wall. If there had been no angle at the neck, the tip of the tail would be at a height of hT = hmax. (b) The angle (in radians) of the head axis to the wall, θ ≈ an/ ℓh.
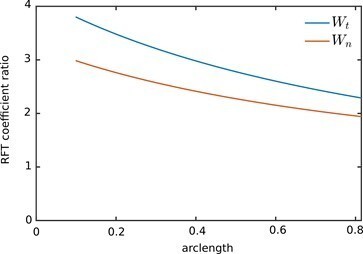
Ratio of the Wall-RFT coefficients in revised manuscript to the bulk coefficients in the original submission.
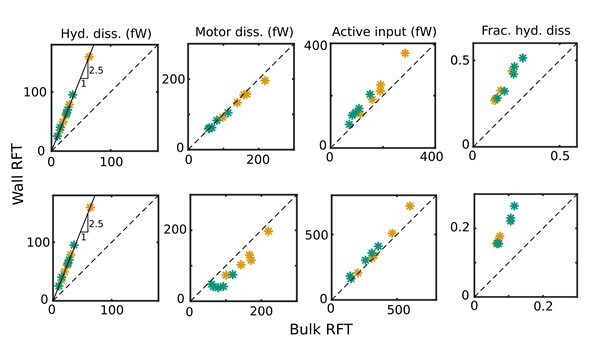
Comparison of revised values of the cycle-means of (i) hydrodynamic dissipation, (ii) motor dissipation, (iii) active power input, and (iv) the proportion of hydrodynamic dissipation out of the total dissipation against their values in the original manuscript for WT (green symbols) and KO (yellow symbols) samples, with ηn = 0 (top panel) and 103 Pa s µm4(bottom panel): the values in the original submission are along the horizontal axes and the revised values are along the vertical axes.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Crisp2 | Lim et al., 2019 | - | - |
| Strain, strain background (Mus musculus) | C57BL/6N | Lim et al., 2019 | PMID:30759213 | Mice produced through the Australian Phenomics Network |
| Biological sample (Mus musculus) | Sperm | Lim et al., 2019 | - | Collected from the cauda epididymis and vas deferens using the backflushing method |
| Chemical compound, drug | TYH medium with 0.3 mg/ml BSA | Lim et al., 2019 | - | Buffer media for sperm |
| Software, algorithm | MATLAB, MATLAB Image Processing Toolbox, Fiji | SCR 001622,SCR 002285 | Code (Nandagiri, 2021) and original videos (Nandagiri et al., 2020) available for public access |
One-way ANOVA data for establishing that sample means of cycle variables are significantly different from population means in the wildtype (WT) and knockout (KO) genotypes; denotes the -test statistic and denotes probability of the null hypothesis.
| Genotype | Flagellar region | Power | Treatments DOF | Error DOF | ||
|---|---|---|---|---|---|---|
| WT | Full | Cycle time | 4 | 306 | 50.31 | |
| WT | Full | Input power | 4 | 306 | 833.19 | |
| WT | Full | Hyd. dissn. | 4 | 306 | 1441.37 | |
| WT | Full | Internal dissn. | 4 | 306 | 730.68 | |
| WT | Full tail | Motor dissn. | 4 | 306 | 246.17 | |
| WT | Mid-piece | Input power | 4 | 306 | 483.02 | |
| WT | Mid-piece | Hyd. dissn. | 4 | 306 | 186.86 | |
| WT | Mid-piece | Internal dissn. | 4 | 306 | 436.92 | |
| WT | Mid-piece | Motor dissn. | 4 | 306 | 421.04 | |
| WT | Principal piece | Input power | 4 | 306 | 786.93 | |
| WT | Principal piece | Hyd. disspn. | 4 | 306 | 1716.31 | |
| WT | Principal piece | Internal dissn. | 4 | 306 | 520.19 | |
| WT | Principal piece | Motor dissn. | 4 | 306 | 140.96 | |
| KO | Full tail | Cycle time | 4 | 270 | 235.45 | |
| KO | Full tail | Input power | 4 | 270 | 110.04 | |
| KO | Full tail | Hyd. dissn. | 4 | 270 | 198.37 | |
| KO | Full tail | Internal dissn. | 4 | 270 | 90.02 | |
| KO | Full tail | Motor dissn. | 4 | 270 | 120.31 | |
| KO | Mid-piece | Input power | 4 | 270 | 528.12 | |
| KO | Mid-piece | Hyd. dissn. | 4 | 270 | 216.51 | |
| KO | Mid-piece | Internal dissn. | 4 | 270 | 247.53 | |
| KO | Mid-piece | Motor dissn. | 4 | 270 | 374.02 | |
| KO | Principal piece | Input power | 4 | 270 | 125.73 | |
| KO | Principal piece | Hyd. dissn. | 4 | 270 | 206.07 | |
| KO | Principal piece | Internal dissn. | 4 | 270 | 109.09 | |
| KO | Principal piece | Motor dissn. | 4 | 270 | 20.3 |
Comparison of the external hydrodynamic dissipation with the internal frictional and motor dissipations: denotes the Student's -test statistic in an unpaired, two-tailed test; is the probability of the null hypothesis.
| Genotype | Flagellar region | (Pa s μm4) | Hyd. dissn. (fW) | Int. dissn. (fW) | ||
|---|---|---|---|---|---|---|
| WT | Full tail | 103 | 89.07 | 240.05 | 22.71 | |
| WT | Mid-piece | 103 | 6.69 | 108.43 | 36.39 | |
| WT | Principal piece | 103 | 82.21 | 131.11 | 9.82 | |
| KO | Full tail | 103 | 66.39 | 200.91 | 21.44 | |
| KO | Mid-piece | 103 | 4.80 | 32.40 | 29.16 | |
| KO | Principal piece | 103 | 61.48 | 168.28 | 16.73 | |
| Genotype | Flagellar region | (Pa s μm4) | Hyd. dissn. (fW) | Motor dissn. (fW) | ||
| WT | Full tail | 103 | 89.07 | 135.06 | 11.48 | |
| WT | Mid-piece | 103 | 6.69 | 109.75 | 36.21 | |
| WT | Principal piece | 103 | 82.21 | 25.09 | 20.71 | |
| KO | Full tail | 103 | 66.39 | 48.72 | 9.74 | |
| KO | Mid-piece | 103 | 4.80 | 29.14 | 24.87 | |
| KO | Principal piece | 103 | 61.48 | 19.49 | 28.4 | |
| WT | Full tail | 0 | 89.07 | 155.58 | 11.48 | |
| WT | Mid-piece | 0 | 6.69 | 112.56 | 36.21 | |
| WT | Principal piece | 0 | 82.21 | 42.71 | 20.71 | |
| KO | Full tail | 0 | 66.39 | 77.61 | 9.74 | |
| KO | Full tail | 0 | 4.80 | 35.23 | 24.87 | |
| KO | Principal piece | 0 | 61.48 | 42.28 | 28.4 |
Comparison of wildtype (WT) and knockout (KO) samples.
The values in in columns 2 and 3 are the means of the distributions created by pooling together values from the individual cycles of all the sperm samples in each genotype.
| Quantity | WT | KO | ||
|---|---|---|---|---|
| Cycle time (s) | 0.16 | 0.18 | 2.72 | 0.01 |
| Reciprocal cycle time (Hz) | 7.19 | 7.2 | 0.03501 | 0.97 |
| Full tail hyd. dissn. (fW) | 89.07 | 66.39 | 6.91 | 1.26 × 10−11 |
| Mid-piece hyd. dissn. (fW) | 6.69 | 4.80 | 6.89 | 1.49 × 10−11 |
| Principal piece hyd. dissn. (fW) | 82.21 | 61.48 | 6.71 | 4.63 × 10−11 |
| Full tail motor dissn. (fW), Pa s μm4 | 135.06 | 48.72 | 26.89 | |
| Full tail int. dissn. (fW), Pa s μm4 | 240.05 | 200.91 | 4.55 | 6.54 × 10−6 |
| Mid-piece motor disspn. (fW), Pa s μm4 | 109.75 | 29.14 | 25.57 | |
| Mid-piece int. dissn. (fW), Pa s μm4 | 108.43 | 32.40 | 24.59 | |
| Principal piece motor dissn. (fW), Pa s μm4 | 25.08 | 19.49 | 5.62 | 3.01 × 10−8 |
| Principal piece int. dissn. (fW), Pa s μm4 | 131.11 | 168.28 | 5.03 | 3.01 × 10−8 |
| Full tail motor dissn. (fW), Pa s μm4 | 155.58 | 77.61 | 24.87 | |
| Mid-piece motor dissn. (fW), Pa s μm4 | 112.56 | 35.23 | 29.10 | |
| Principal piece motor dissn. (fW), Pa s μm4 | 42.71 | 42.28 | 0.22 | 0.82 |





