Single-PanIN-seq unveils that ARID1A deficiency promotes pancreatic tumorigenesis by attenuating KRAS-induced senescence
Figures
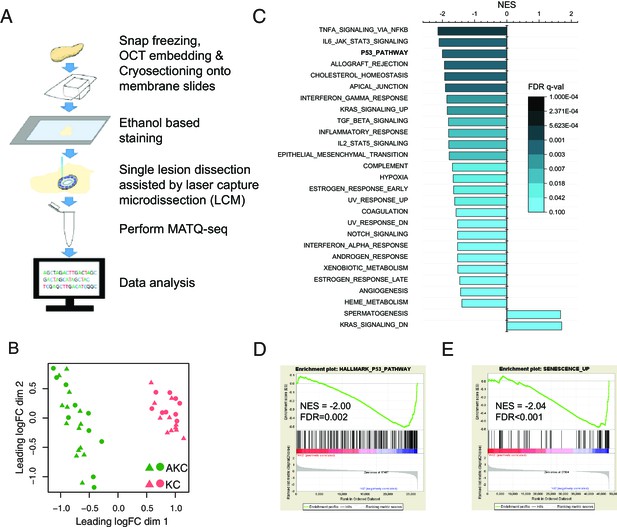
Single pancreatic intraepithelial neoplasia (PanIN) lesion RNA-seq unveils the potential player contributing to the attenuation of Kras-induced senescence in Arid1a knockout mice.
(A) Experimental scheme of transcriptome profiling of single PanIN lesions. (B) Multidimensional scaling plot showed a clear separation of the transcriptome profiles of lesions from AKC mice (Arid1afl/fl;LSL-KrasG12D/+;Ptf1aCreERT/+) and KC mice (Arid1a+/+;LSL-KrasG12D/+;Ptf1aCreERT/+). 24 lesions from 2 AKC mice and 20 lesions from 2 KC mice were used for single-lesion RNA sequencing. (C) Hallmark gene sets that are significantly enriched between lesions from AKC and KC mice. (D, E) Enrichment plots of P53_PATHWAY and SENESCENCE_UP from gene set enrichment analysis.
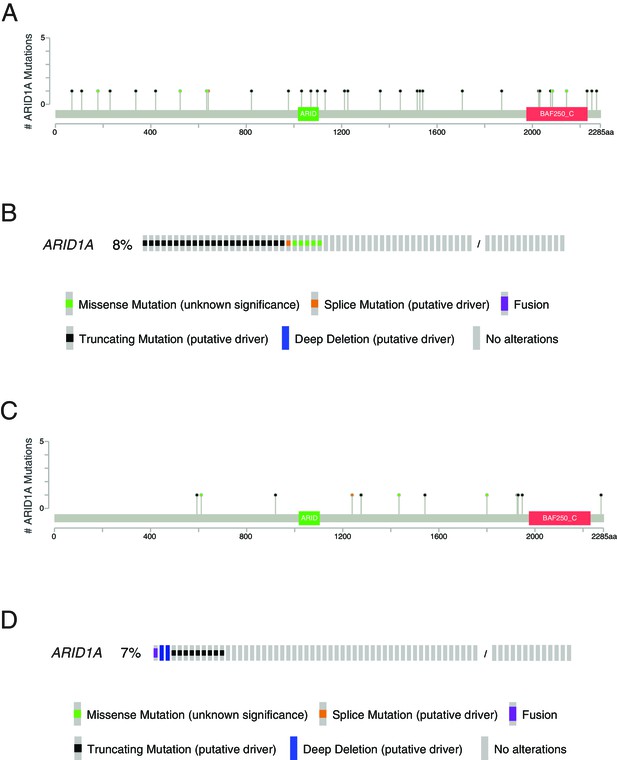
Mutations in ARID1A in ICGC-PACA-AU and TCGA pancreatic ductal adenocarcinoma (PDAC) cohorts based on cBioPortal data portal.
(A) Lollipop plot of mutations in ARID1A from ICGC-PACA-AU cohort (total sample: 383). (B) The frequency of different types of mutations in ARID1A in ICGC-PACA-AU cohort. Based on the ICGC data portal, the frequency of the mutations with high or low mutation impact is 7% for ICGC-PACA-AU cohort. (C) Lollipop plot of mutations in ARID1A from TCGA PDAC cohort (total sample: 179). (D) The frequency of different types of mutations in ARID1A in TCGA PDAC cohort.
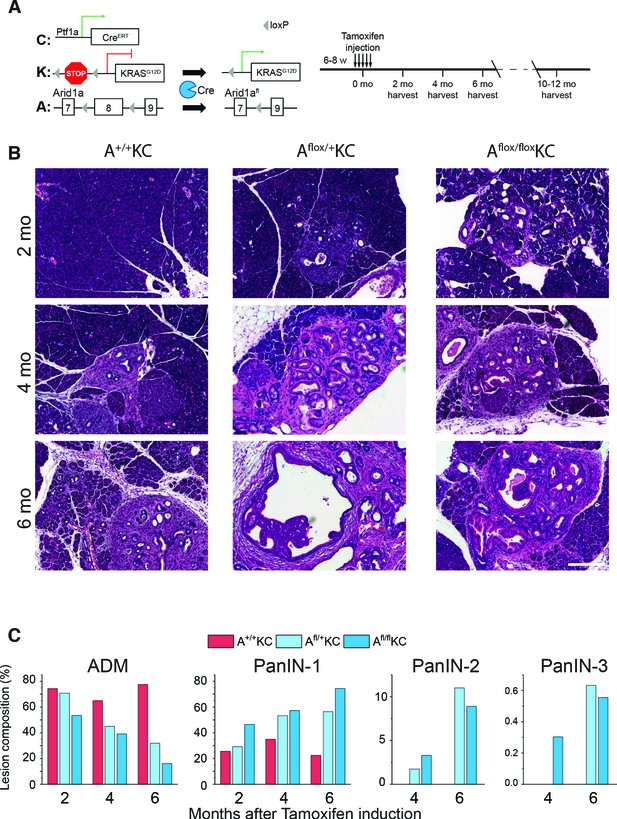
Arid1a knockout drastically accelerates pancreatic intraepithelial neoplasia (PanIN) progression.
(A) Genetic makeup and experimental scheme. (B) Representative H&E staining images of pancreata from A+/+KC, Afl/+KC, and Afl/flKC mice after administration of tamoxifen for 2, 4, and 6 months. (C) Quantification of lesions with different grades. Lesions with different grades were counted at 10 random fields under the microscope, presented as percentage. Scale bar, 200 µm.
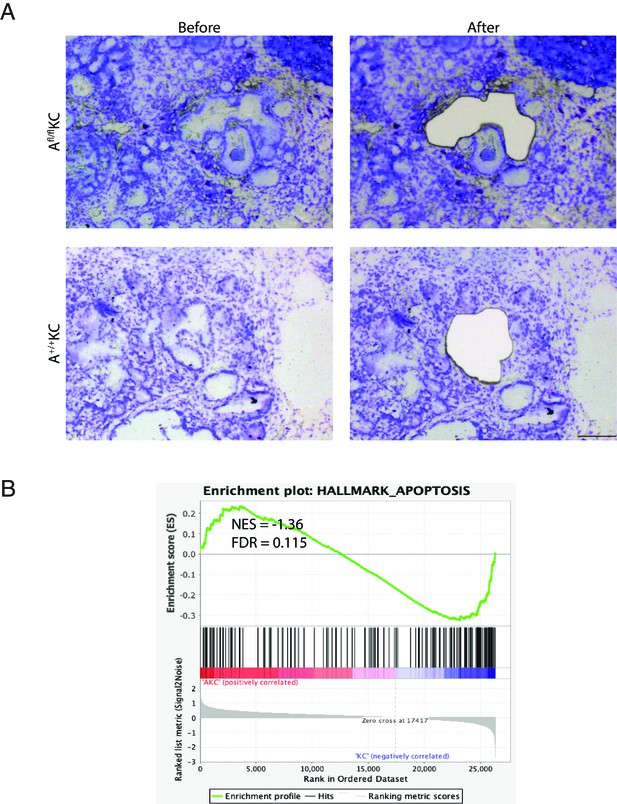
Profile of the transcriptomes of individual pancreatic intraepithelial neoplasia (PanIN) by PanIN-seq.
(A) Representative images of lesion region before and after laser capture microdissection. Scale bar, 100 µm. (B) The gene set enrichment analysis plot of apoptosis-related pathway between AKC PanIN lesions and KC PanIN lesions.

The gene set enrichment plots of 27 relevant pathways with statistical significance in Afl/flKC lesions.
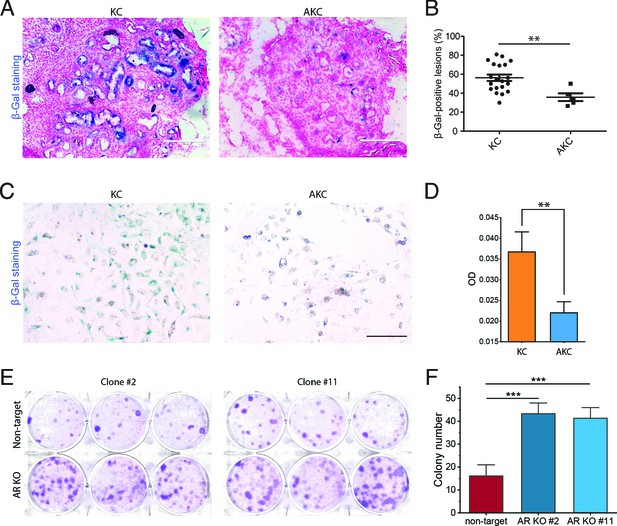
In vivo, ex vivo, and in vitro verification of attenuation of Kras-induced senescence by Arid1a deficiency.
(A) Representative images of senescence-associated beta-galactosidase (SA-β-Gal) staining of frozen pancreatic sections from KC mice and AKC mice. (B) SA-β-Gal-positive lesions were counted at five random fields under the microscope in four KC mice and one AKC mouse, presented as percentages. (C) Representative images of SA-β-Gal staining of ex vivo culture from KC and AKC mice 1 month after administration of tamoxifen. (D) Quantification of the intensity of SA-β-Gal staining at five random fields under the microscope. (E) Colony formation assay of ARID1A knockout cells and wildtype human pancreatic Nestin-expressing (HPNE) cells with KRAS induction by doxycycline (6 µg/ml) for 15 days. (F) Quantification of colony number in panel (E). The colony formation assay was performed twice. Student’s t-test: **p<0.01; ***p<0.001. Scale bars: 200 µm.
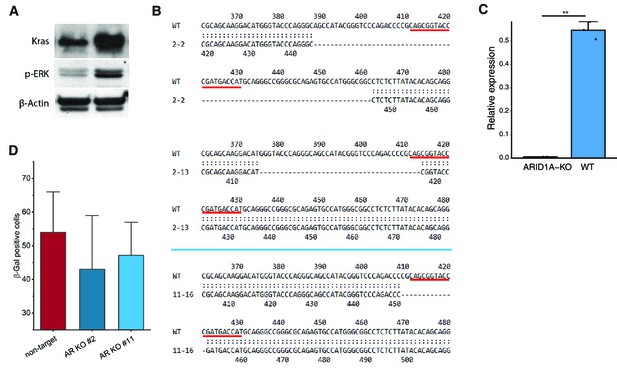
Generation of cell line with inducible KRAS overexpression and ARID1A knockout.
(A) Examination of activation of KRAS signaling by western in human pancreatic Nestin-expressing (HPNE) cells with inducible KRAS knockin (iKRAS-HPNE cells clone #4). Phosphorylation of ERK was used as the indicator for KRAS activation. (B) Confirmation of ARID1A knockout by Sanger sequencing in ARID1A-KO iKRAS-HPNE cells (clone #2 and #11). The underscored sequence is the target for guide RNA. (C) Verification of the repression of ARID1A expression in knockout cell line by qRT-PCR. **p<0.01. (D) Senescence-associatedbeta-galactosidase (SA-β-Gal) staining in HPNE cells with ARID1A knockout (clone #2 and #11) and non-target control upon KRAS induction.
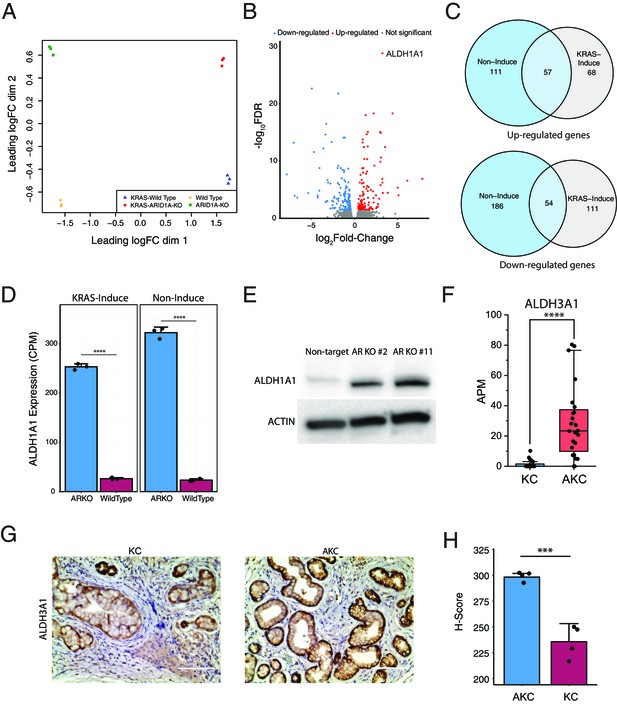
ARID1A knockout upregulates aldehyde dehydrogenase (ALDH) expression.
(A) Multidimensional scaling plot demonstrated clear separation between the transcriptome profiles of ARID1A-KO human pancreatic Nestin-expressing (HPNE) cells and wildtype cells with or without KRAS induction. RNA sequencing was performed with three biological repeats. (B) Volcano plot of differentially expressed genes between ARID1A knockout cells and wildtype cells with KRAS induction. (C) Venn diagram showing the upregulated genes (upper) and downregulated genes (bottom) that are shared between cells with (gray) or without (blue) KRAS induction. (D) ALDH1A1 mRNA levels quantified by sequencing data are significantly different between ARID1A-KO cells and wildtype cells with (left) or without (right) Kras induction. CPM: count per million reads. (E) Western blot for ALDH1A1 expression in ARID1A-KO cells and wildtype cells with KRAS induction. (F) mRNA level of Aldh3a1 in KC and AKC lesions based on pancreatic intraepithelial neoplasia (PanIN)-seq data. APM: amplicon per million reads. (G) IHC staining against ALDH3A1 in KC and AKC lesions. Scale bars: 200 µm. (H) Comparison of ALDH3A1 levels between KC and AKC lesions based on the intensity of staining in (G). H-score was calculated by counting the number of lesions with different levels of staining intensity at four random fields under the microscope. Student’s t-test: ***p<0.001; ****p<0.0001.
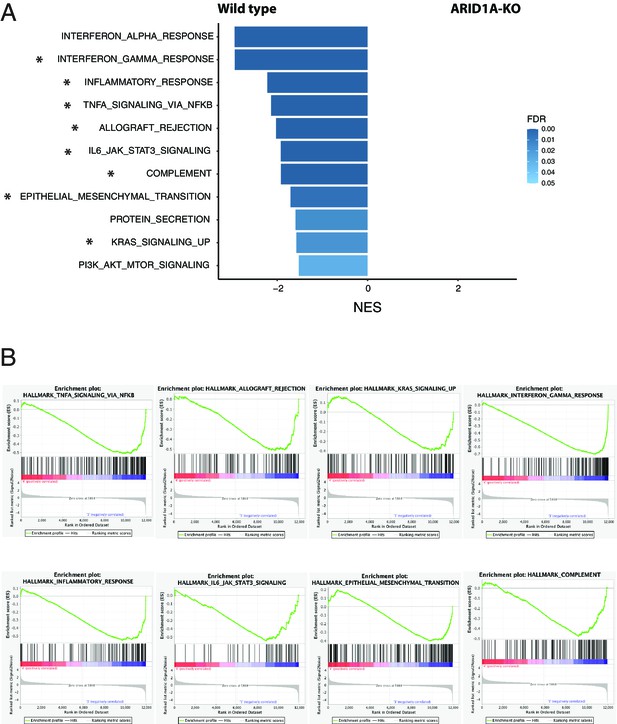
Gene set enrichment analysis on RNA-seq data.
(A) Gene set enrichment analysis between ARID1A-KO cell line and wildtype cell line under KRAS induction. (B) Gene set enrichment plots for the pathways reaching statistical significance (FDR < 0.05). The pathways that are also significantly enriched in AKC vs. KC pancreatic intraepithelial neoplasia (PanIN)-seq data are labeled with asterisks.
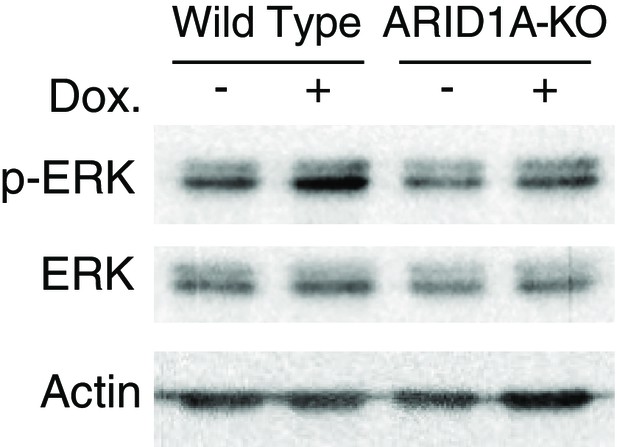
ARID1A knockout impairs phosphorylation of ERK in human pancreatic Nestin-expressing (HPNE) cells upon KRAS induction.
ARID1A-KO and wildtype HPNE cells were treated with or without 2 µg/ml doxycycline (Dox.) for 2 days. Phospho-ERK (p-ERK) was examined by western blot.
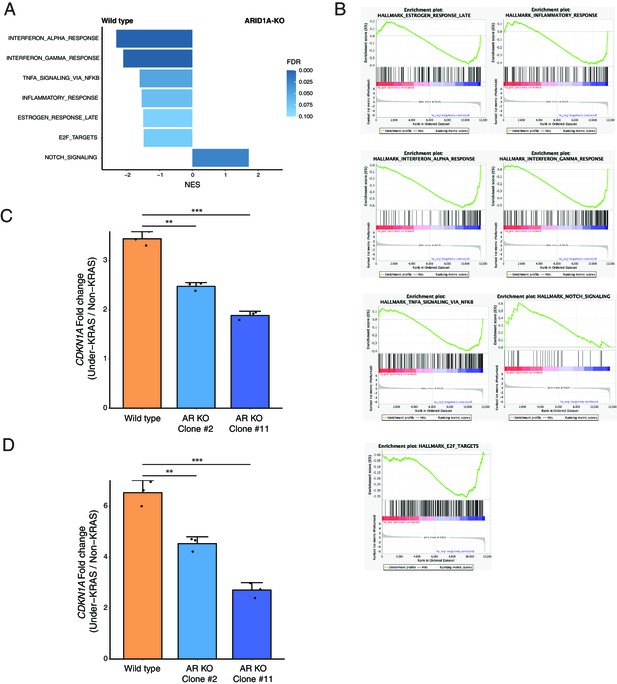
Differential response to oncogenic KRAS in ARID1A-KO and wildtype cells.
(A) Gene set enrichment analysis on the genes with differential response to oncogenic KRAS in ARID1A-KO and wildtype cells. (B) Gene set enrichment plots for the pathways with significant enrichment (FDR < 0.05). (C) The fold change of CDKN1A expression between KRAS induction and non-induction control in each cell line examined by RNA-seq. (D) The fold change of CDKN1A expression between KRAS induction and non-induction control in each cell line examined by qRT-PCR. **p<0.01; ***p<0.001.
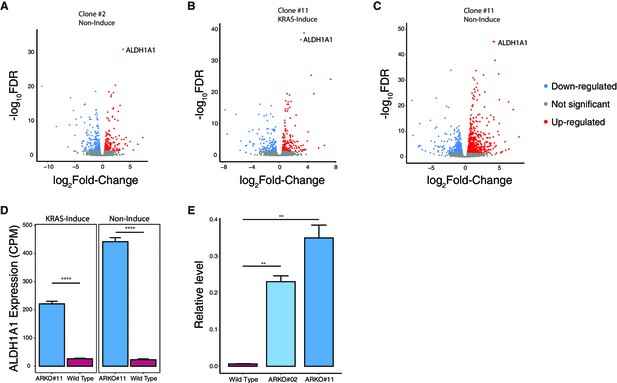
ALDH1A1 expression in ARID1A knockout human pancreatic Nestin-expressing (HPNE) cells.
(A) Volcano plot of differentially expressed genes between ARID1A knockout cells (clone #2) and wildtype cells without KRAS induction. (B, C) Volcano plot of differentially expressed genes between ARID1A knockout cells (clone #11) and wildtype cells with (B) or without (C) KRAS induction. (D) ALDH1A1 mRNA level in ARID1A knockout cells (clone #11) and wildtype cells with (left) or without (right) KRAS induction quantified by RNA-seq. CPM: count per million reads. (E) Upregulation of ALDH1A1 expression in HPNE cells with ARID1A knockout (clone #2 and #11) was confirmed by qRT-PCR.
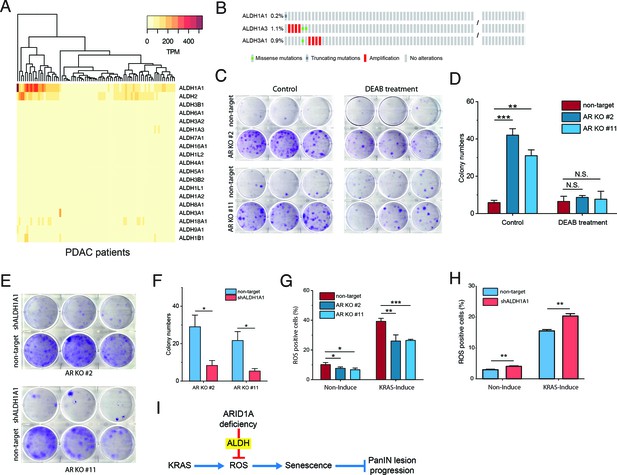
ARID1A knockout facilitates escape from KRAS-induced senescence by upregulating ALDH1A1 expression.
(A) Heatmap of the expression levels of aldehyde dehydrogenase (ALDH) family members in pancreatic ductal adenocarcinoma (PDAC) patients (Bailey et al., 2016). (B) Mutation rates of ALDH1A1, ALDH1A3, and ALDH3A1 in PDAC patients. The mutation data from these two studies (Bailey et al., 2016) and TCGA (Cancer Genome Atlas Research Network, 2017) were used for analysis. (C) Colony formation assay of ARID1A knockout cells and wildtype cells with KRAS induction, treated with and without ALDH inhibitor DEAB. (D) Quantification of colony number in panel (C). The colony formation assay was performed twice. (E) Colony formation assay of ARID1A knockout cells expressing shRNA targeting ALDH1A1 and scramble shRNA control. (F) Quantification of colony number in panel (E). The colony formation assay was performed twice. (G) Measurement of the reactive oxygen species (ROS) level using an H2DCFDA-based ROS detection assay kit. Percentage of positive cells measured by flow cytometry. (H) Measurement of the ROS levels in the ARID1A-KO cells with ALDH1A1 knockdown or scramble shRNA. (I) Working model for ARID1A-deficiency-promoted pancreatic intraepithelial neoplasia (PanIN) lesion progression via inhibition of ROS production. Student’s t-test: *p<0.05; **p<0.01; ***p<0.001.
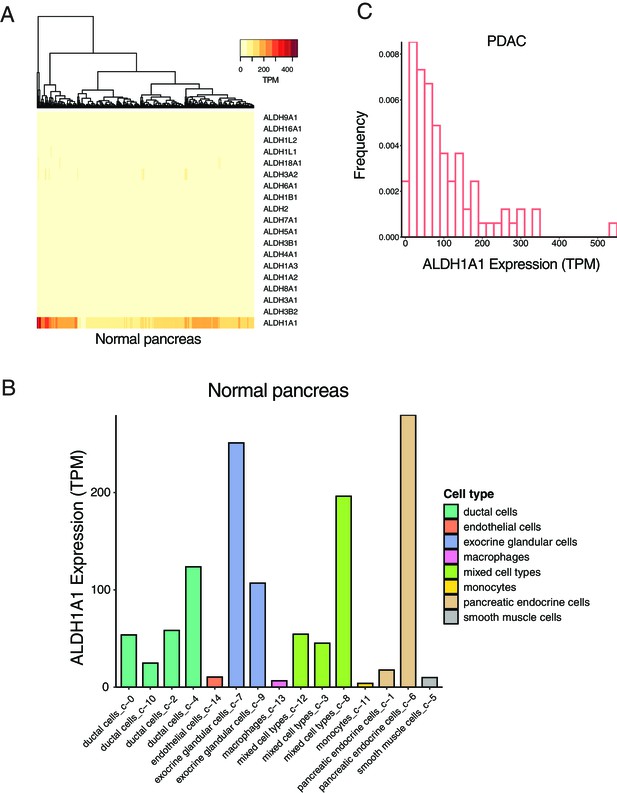
The expression of aldehyde dehydrogenase (ALDH) family members in normal pancreas and pancreatic ductal adenocarcinoma (PDAC).
(A) The expression level of different ALDH genes in normal pancreas. (B) The expression level of ALDH1A1 in different cell types of normal pancreas. (C) The distribution of the expression level of ALDH1A1 in PDAC.
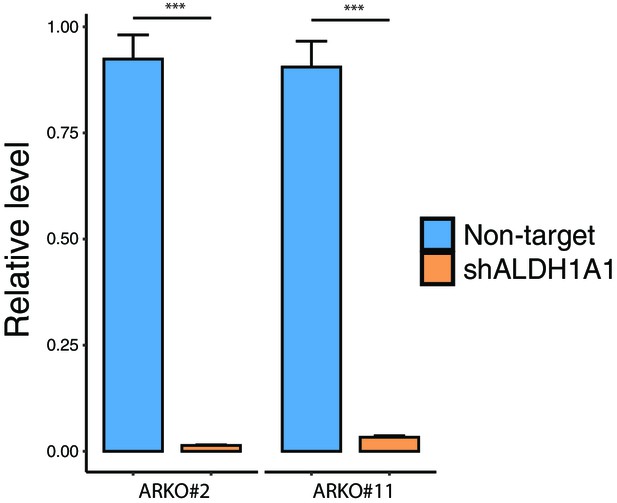
Knockdown efficiency of ALDH1A1 in human pancreatic Nestin-expressing (HPNE) cells with ARID1A knockout (clone #2 and #11) was confirmed by qRT-PCR.
Student’s t-test: **p<0.01; ***p<0.001, ****p<0.0001.
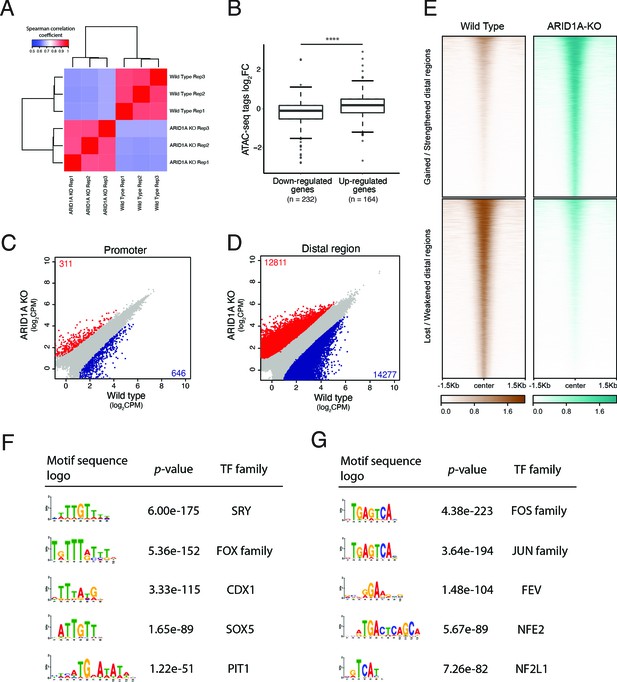
ARID1A knockout activates transcription of the ALDH1A1 gene by increasing the accessibility of its enhancer region.
(A) Spearman correlation coefficients of the read counts in peaks between ARID1A-KO human pancreatic Nestin-expressing (HPNE) cells and wildtype cells. ATAC sequencing was performed with three biological repeats. (B) The average fold change of chromatin accessibility in the promoters and enhancers of differentially expressed genes identified in RNA-seq. Mann–Whitney–Wilcoxon test: ****p<0.0001. (C, D) The scatter plot of the read counts in each peak between ARID1A-KO cells and wildtype cells for promoter (C) and distal regions (D). The peaks with significantly increased read density in ARID1A-KO cells compared with wildtype cells are colored in red. The peaks with significantly decreased read density are colored in blue. (E) Heatmap of the gained or lost distal regions between ARID1A-KO cells and wildtype cells. (F) The top five transcription factor (TF) binding motifs significantly enriched in the enhancer ATAC peaks with increased accessibility in ARID1A-KO cells. (G) The top five TF binding motifs significantly enriched in the enhancer ATAC peaks with decreased accessibility in ARID1A-KO cells.
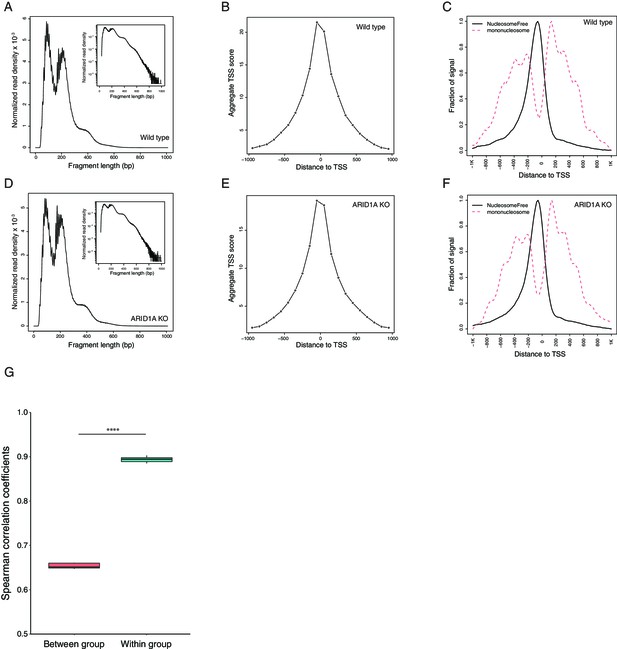
Quality control of ATAC-seq.
(A) The distribution of fragment size of wildtype cells. (B) The transcription start site (TSS) enrichment score of wildtype cells. (C) The enrichment of nucleosome-free reads at TSS compared to the reads spanning mono-nucleosome in wildtype cells. (D) The distribution of fragment size of ARID1A-KO cells. (E) The TSS enrichment score of ARID1A-KO cells. (F) The enrichment of nucleosome-free reads at TSS compared to the reads spanning mono-nucleosome in ARID1A-KO cells. (G) The Spearman correlation coefficients between the samples within the same group or across different groups.
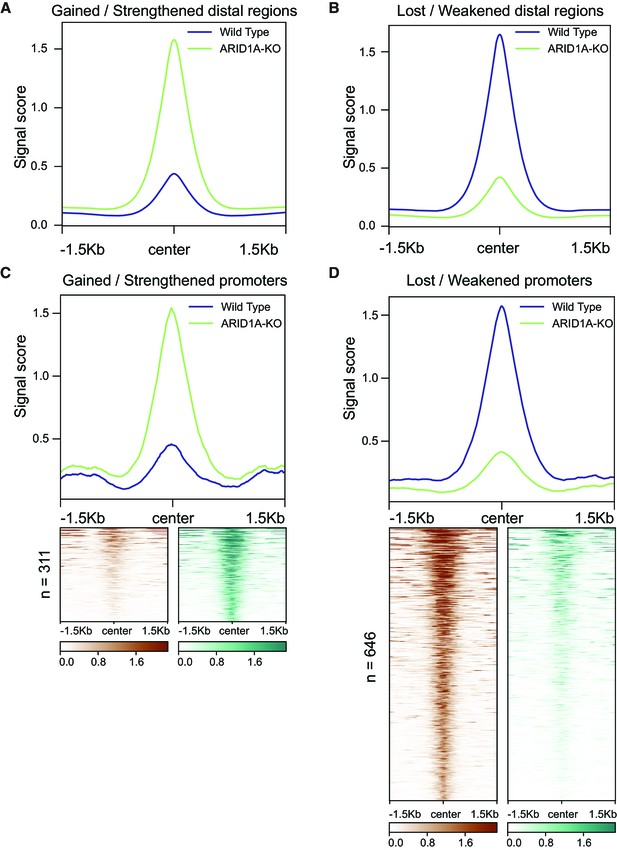
Read density profiles of differential peaks.
(A) The read density profiles of the distal regions with increased accessibility in ARID1A-KO cells compared to wildtype cells. (B) The read density profiles of the distal regions with decreased accessibility in ARID1A-KO cells compared to wildtype cells. (C) The read density profiles of the promoters with increased accessibility in ARID1A-KO cells compared to wildtype cells. (D) The read densities profiles of the promoters with decreased accessibility in ARID1A-KO cells compared to wildtype cells.
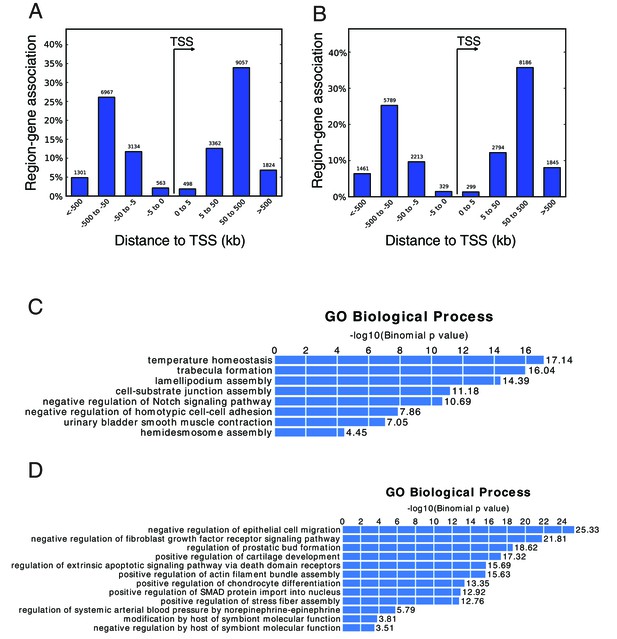
General analysis of ATAC-seq by GREAT.
(A) The genomic distribution of the peaks with significantly decreased accessibility in ARID1A-KO cells compared to wildtype cells. (B) The genomic distribution of the peaks with significantly increased accessibility in ARID1A-KO cells compared to wildtype cells. (C) GO enrichment analysis on the peaks with significantly decreased accessibility. (D) GO enrichment analysis on the peaks with significantly increased accessibility.
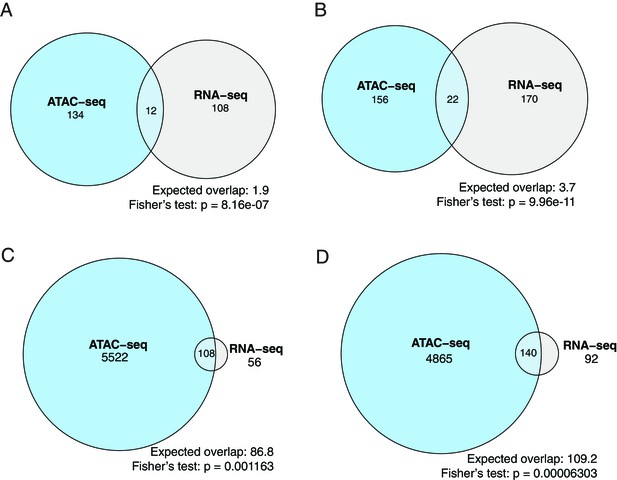
The overlap between the differentially expressed genes and differential peaks at promoter regions (A, B) and enhancer regions (C, D).
(A) The overlap between the significantly upregulated genes and the genes with significantly increased accessibility at promoter regions. (B) The overlap between the significantly downregulated genes and the genes with significantly decreased accessibility at promoter regions. (C) The overlap between the significantly upregulated genes and the genes with significantly increased accessibility in enhancer regions. (D) The overlap between the significantly downregulated genes and the genes with significantly decreased accessibility in enhancer regions.
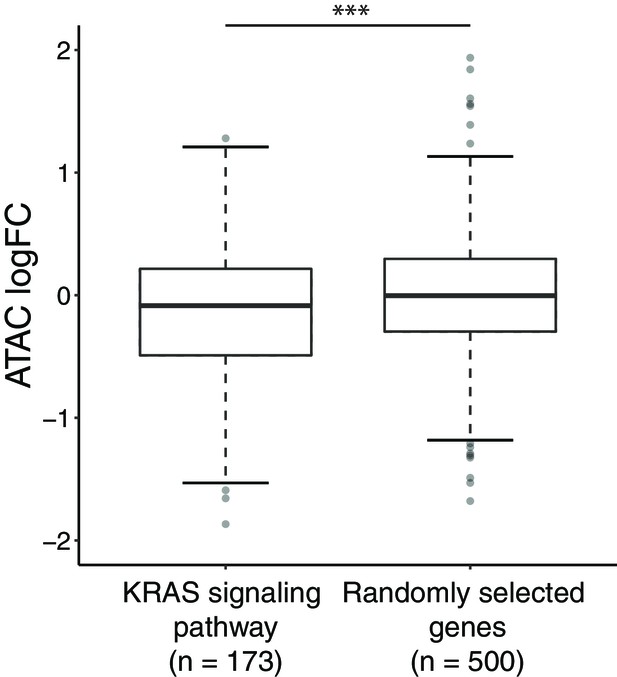
Comparison in the change of chromatin accessibility between the genes within KRAS signaling pathway and the randomly chosen genes.
For the 173 genes from KRAS_SIGNALING_UP gene set from Hallmark gene sets, we calculated the chromatin accessibility fold change of these genes between ARID1A KO and wildtype. In comparison, 500 genes without significant gene expression changes between ARID1A KO and wildtype were randomly selected.
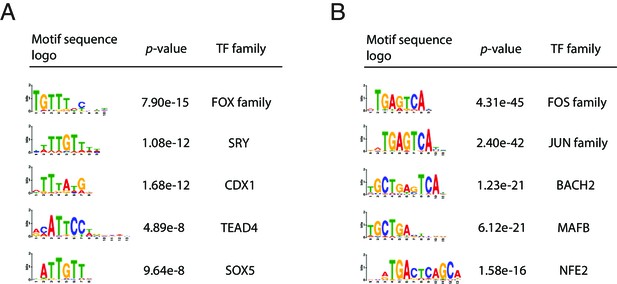
Motif analysis on the promoter regions.
(A) Motif enrichment analysis on the promoter regions with significantly increased accessibility in ARID1A-KO cells compared to wildtype cells. (B) Motif enrichment analysis on the promoter regions with significantly decreased accessibility in ARID1A-KO cells compared to wildtype cells.
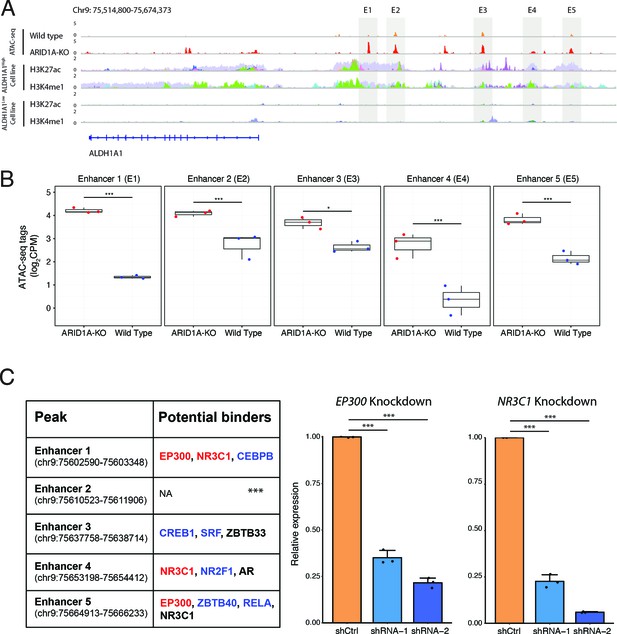
ARID1A knockout activates transcription of the ALDH1A1 gene by increasing the accessibility of its enhancer region.
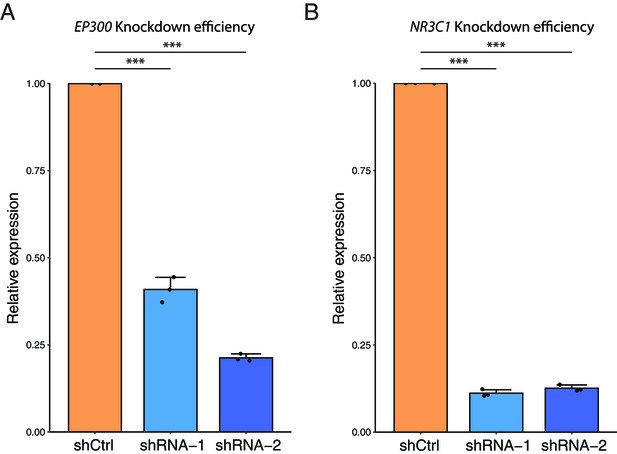
(A) The ATAC-seq tracks and H3K4me1/H3K27ac ChIP-seq tracks of the distal regions of the ALDH1A1 gene. The ChIP-seq tracks from different cell lines are labeled in different colors and overlaid. The figure with separated tracks is shown in Figure 6—figure supplement 1. The ALDH1A1high cell lines include A549, LOUCY, A673, 22RV1, VCAP, K562, and HepG2. The ALDH1A1low cell lines include HCT116, MCF7, Panc1, PC9, and DOHH2. The ChIP-seq data were obtained from the ENCODE database (ENCODE Project Consortium, 2012). (B) Read counts in the five enhancer peaks of the ALDH1A1 gene in ARID1A-KO human pancreatic Nestin-expressing (HPNE) cells and wildtype cells. p-value: *p<0.05; ***p<0.001; ****p<0.0001. (C) Transcription factors (TFs) that regulate ALDH1A1 expression by binding to its enhancer loci. Left panel: the list of TFs that can potentially bind to the enhancer regions of ALDH1A1 based on the ChIP-seq data from seven ALDH1A1high cell lines and five ALDH1A1Low cell lines. The TFs whose binding events are preferentially detected in the datasets from ALDH1A1high cell lines (Fisher’s test, p<0.05) are marked in red. The TFs that are observed in more than 50% of the datasets from ALDH1A1high cell lines but do not have enough datasets for statistical tests are marked in blue. The TFs whose binding events are observed in more than 25% but less than 50% of the datasets from ALDH1A1high cell lines are marked in black. The TFs that are not expressed in HPNE cell lines were removed. Middle panel: expression of ALDH1A1 in EP300 knockdown cells and cells with scramble shRNA. Right panel: expression of ALDH1A1 in NR3C1 knockdown cells and cells with scramble shRNA.
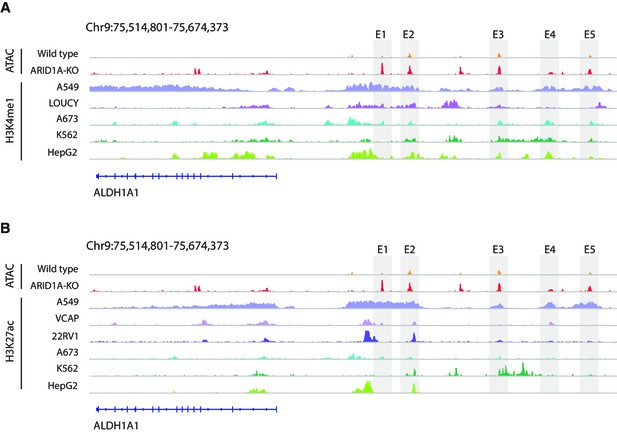
The landscape of H3K27ac and H3K4me1 at the upstream of ALDH1A1 gene in ALDH1A1high cell lines.
(A) The landscape of H3K4me1 at the upstream of ALDH1A1 gene in five ALDH1A1high cell lines. (B) The landscape of H3K27ac at the upstream of ALDH1A1 gene in six ALDH1A1high cell lines.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Ptf1aCreERT | Jackson Lab | No: 019378 | |
| Genetic reagent (M. musculus) | Lox-Stop-Lox-KrasG12D | Jackson Lab | No: 008179 | |
| Genetic reagent (M. musculus) | Arid1aflox | PMID:18448678 | Dr. Zhong Wang lab (University of Michigan) | |
| Cell line (Homo sapiens) | hTERT-HPNE | Dr. Jennifer Baily lab (UT Health) | RRID:CVCL_C466 | |
| Recombinant DNA reagent | pInducer20-KRASG12D | Dr. Haoqiang Ying lab (MD Anderson Cancer Center) | Inducible expression of KRASG12D | |
| Recombinant DNA reagent | pL-CRISPR.EFS.tRFP | Addgene | RRID:Addgene_57819 | Pol III-based sgRNA expression backbone |
| Recombinant DNA reagent | pGIPZ | Open Biosystems | Pol III-based shRNA expression backbone | |
| Sequence-based reagent | sgARID1A | This paper | sgRNA | CAGCGGTACCCGATGACCAT |
| Sequence-based reagent | shALDH1A1 | This paper | shRNA | GGAGTGTTTACCAAAGACATT |
| Sequence-based reagent | shEP300-1 | This paper | shRNA | CGGCAAACAGTTGTGCACA |
| Sequence-based reagent | shEP300-2 | This paper | shRNA | AGCTACTGAAGATAGATTA |
| Sequence-based reagent | shNR3C1-1 | This paper | shRNA | CCAACGGTGGCAATGTGAA |
| Sequence-based reagent | shNR3C1-2 | This paper | shRNA | AGCTGTAAAGTTTTCTTCA |
| Commercial assay or kit | ROS detection assay kit | BioVision | Cat# K936-250 | |
| Commercial assay or kit | Senescence β-Galactosidase Staining Kit | Cell Signaling Technology | Cat# 9860 | |
| Chemical compound, drug | ALDEFLUOR diethylaminobenzaldehyde (DEAB) reagent, 1.5 mM in 95% ethanol | Stemcell Technologies Inc | Cat# 01705 | |
| Antibody | (Rabbit polyclonal) anti-ALDH1A1 | Abcam | Abcam Cat# ab23375, RRID:AB_2224009 | WB(1:400) |
| Antibody | (Rabbit polyclonal) anti-Aldh3a1 | Abcam | Abcam Cat# ab76976, RRID:AB_1523110 | IHC(1:100) |
| Antibody | (Rabbit polyclonal) anti-Ras | Abcam | Abcam Cat# ab180772, RRID:AB_2884935 | WB(1:500) |
| Antibody | (Mouse monoclonal) anti-β-Actin | Sigma-Aldrich | Sigma-Aldrich Cat# A1978, RRID:AB_476692 | WB(1:4000) |
| Antibody | (Rabbit monoclonal) anti- Phospho-p44/42 MAPK (Erk1/2) | Cell Signaling Technology | Cell Signaling Technology Cat# 4370, RRID:AB_2315112 | WB(1:1000) |
Additional files
-
Supplementary file 1
Gene set enrichment analysis on PanIN-seq.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp1-v2.xlsx
-
Supplementary file 2
Differential gene expression analysis on PanIN-seq.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp2-v2.xlsx
-
Supplementary file 3
Differential gene expression analysis on RNA-seq.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp3-v2.xlsx
-
Supplementary file 4
Interaction test on the RNA-seq data.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp4-v2.xlsx
-
Supplementary file 5
Quality control of ATAC-seq data.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp5-v2.xlsx
-
Supplementary file 6
ATAC peaks in the regulatory regions of ALDH1A1.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp6-v2.xlsx
-
Supplementary file 7
Motif enrichment analysis on differential ATAC peaks.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp7-v2.xlsx
-
Supplementary file 8
Expression (RPKM) of ALDH1A1 in other human cell lines.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp8-v2.xlsx
-
Supplementary file 9
Binding of TFs at the regulatory region of ALDH1A1 in human cell lines.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp9-v2.xlsx
-
Supplementary file 10
List of ENCODE datasets used.
- https://cdn.elifesciences.org/articles/64204/elife-64204-supp10-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64204/elife-64204-transrepform1-v2.docx