Disparate bone anabolic cues activate bone formation by regulating the rapid lysosomal degradation of sclerostin protein
Figures
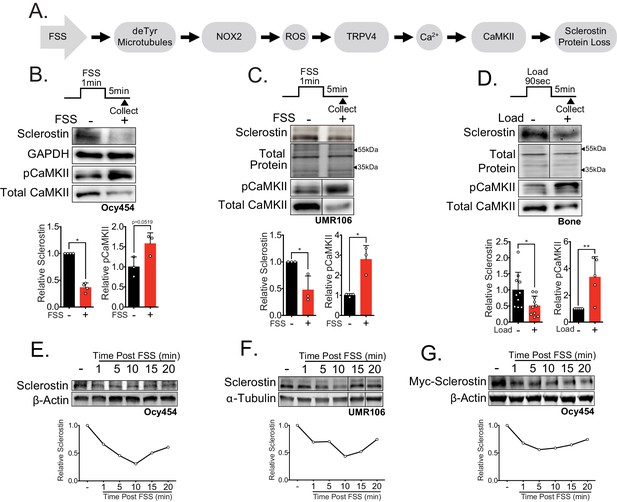
Sclerostin protein is rapidly degraded after mechanical stimulus in vitro and in vivo.
(A) FSS causes the rapid loss of sclerostin protein through a number of molecular mediators. (B) Ocy454 cells (n = 3–4) or (C) UMR106 cells (n = 3) were exposed to 1 min of FSS at 4 dynes/cm2 and lysed 5 min post-flow. Western blots were probed for sclerostin, GAPDH, pCaMKII, and total CaMKII. (D) Sixteen week old female C57Bl/6 mice were ulnar loaded (1800 με, 90 s, 2 Hz), cortical osteocyte-enriched lysates isolated 5 min post-load, and western blots probed for sclerostin (n = 10 mice), pCaMKII, and total CaMKII (n = 5 mice). Sclerostin abundance relative to the loading control or pCaMKII relative to total CaMKII was quantified. (E) Ocy454 cells with endogenous sclerostin (n = 2), (F) UMR106 cells with endogenous sclerostin (n = 4), or (G) Ocy454 cells transfected with Myc-tagged sclerostin (n = 1) were subjected to 5 min of FSS at 4 dynes/cm2 and lysed at the indicated times post-flow. Western blots were probed for sclerostin and β-actin. A representative time course is shown for each. Sclerostin abundance relative to the loading control was quantified. For each antibody, western blots are from a single gel and exposure; a vertical black line indicates removal of irrelevant lanes. Graphs depict mean ± SD. *p<0.05, **p<0.01 by unpaired two-tailed t-tests (B–D).
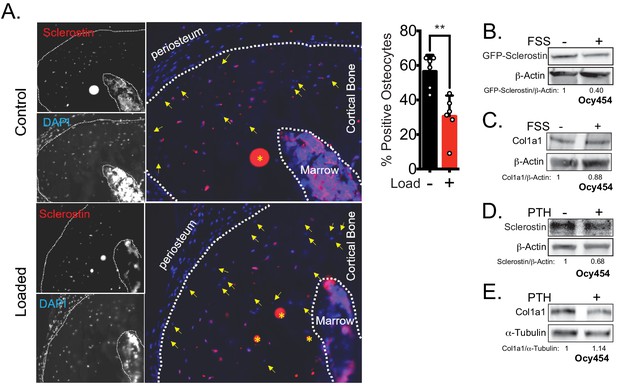
Rapid loss of sclerostin protein occurs in osteocytes in vivo and in vitro.
(A) Seventeen week old male mice were subjected to a single bout of ulnar loading (1800 με), and ulnae were fixed in warm formalin 5 min after loading. Cryosections were stained for sclerostin to identify sclerostin-positive osteocytes, which were counted and presented as a proportion of total osteocytes in a selected ROI (n = 6). Yellow arrows indicate sclerostin-negative osteocytes, defined by the presence of DAPI staining but without detectable sclerostin. Yellow asterisks indicate non-specific staining. (B) Ocy454 cells transfected with GFP-sclerostin were subjected to 5 min of FSS at 4 dynes/cm2 and lysed immediately post-flow. Western blots were probed for sclerostin and β-actin. (C) Ocy454 cells were subjected to 5 min of FSS at 4 dynes/cm2 and lysed immediately post-flow. Western blots were probed for pro-collagen type 1α1 and β-actin. (D, E) Ocy454 cells were treated with PTH (10 nM) for 30 min and lysed. Western blots were probed for sclerostin, β-actin, pro-collagen type 1α1, and α-tubulin.
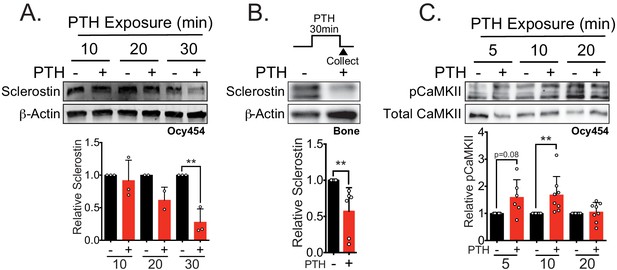
Sclerostin protein is rapidly degraded after PTH exposure in vitro and ex vivo.
(A) Ocy454 cells transfected with GFP-sclerostin were treated with vehicle or PTH (1–34) (10 nM) for the indicated time and were lysed. Western blots were probed for sclerostin and β-actin (n = 2–3). (B) Dissected tibiae flushed of marrow were treated with vehicle or PTH (1–34) (10 nM) for 30 min ex vivo and homogenized. Western blots were probed for sclerostin and β-actin (n = 6 mice). (C) Ocy454 cells were treated with vehicle or PTH (1–34) (10 nM) for the indicated time and were lysed. Western blots were probed for pCaMKII and total CaMKII (n = 6–8). Graphs depict mean ± SD. *p<0.05, **p<0.01 by two-way ANOVA with Holm–Sidak post hoc correction (A, C) or unpaired two-tailed t-test (B).

Acute treatment with PTH does not affect Sost mRNA hours later.
Ocy454 cells were treated with PTH (10 nM) for 30 min and then collected 5.5 hr after treatment to examine Sost mRNA levels as normalized to Gapdh, Rpl13, and Hprt by RT-qPCR (n=6).
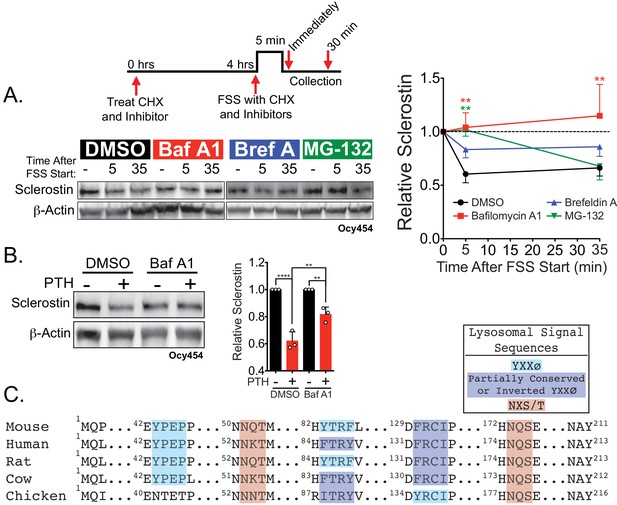
Sclerostin is rapidly degraded by the lysosome following bone anabolic stimuli.
(A) Ocy454 cells transfected with GFP-sclerostin were treated with cycloheximide (150 µg/mL) to prevent new protein synthesis and either DMSO (0.1%), bafilomycin A1 (100 nM) to inhibit lysosomal degradation, brefeldin A (2 μm) to inhibit secretion, or MG-132 (10 μm) to inhibit the proteasome 4 hr prior to FSS. Cells were subjected to 5 min of FSS at 4 dynes/cm2 and lysed immediately after the end of FSS or 30 min after the conclusion of FSS. Western blots were probed for sclerostin and β-actin. Time courses show mean ± SEM (n = 3–6 independent experiments/group). (B) Ocy454 cells transfected with GFP-sclerostin were pre-treated with DMSO (0.1%) or bafilomycin A1 (100 nM) to inhibit lysosomal degradation for 30 min prior to the addition of vehicle or PTH (1–34) (10 nM) for an additional 30 min (n = 3). Sclerostin abundance relative to the loading control was quantified. Graph depicts mean ± SD. *p<0.05, **p<0.01, ****p<0.0001 by two-way ANOVA with Holm–Sidak post hoc correction. (C) Amino acid sequences for sclerostin from mouse, human, rat, cow, and chicken were aligned using NCBI COBALT. Abbreviated sequences are shown and are annotated for putative lysosomal signal sequences. Full sequences are presented in Figure 3—figure supplement 1.
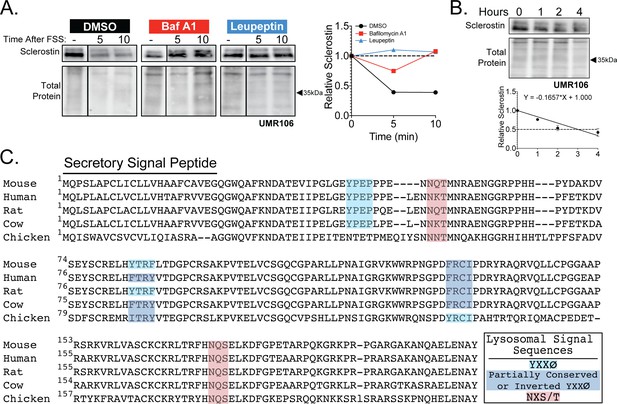
Sclerostin is rapidly degraded by the lysosome.
(A) UMR106 cells were treated with cycloheximide (150 µg/mL) and either DMSO (0.1%), bafilomycin A1 (100 nM) for 6 hr, or leupeptin (200 μM) for 4 hr prior to FSS. Cells were subjected to 1 min of FSS at 4 dynes/cm2 and were lysed 5 or 10 min after the conclusion of FSS. Western blots were probed for sclerostin. Sclerostin abundance relative to total protein was quantified. For each antibody, blots are from a single gel and exposure; a vertical black line indicates removal of irrelevant lanes. (B) UMR106 cells were treated with cycloheximide (150 µg/mL), and lysates were collected at 0, 1, 2, and 4 hr after treatment in the absence of stimuli. Whole-cell lysates were western blotted for sclerostin abundance (n = 2). Sclerostin abundance relative to the total protein was quantified. Graph shows mean ± SD and best-fit linear regression constrained through y = 1. (C) Full amino acid sequences for sclerostin from mouse, human, rat, cow, and chicken were aligned using NCBI COBALT. Putative lysosomal signal sequences are annotated. The secretory signal peptide is annotated for amino acids 1–23.
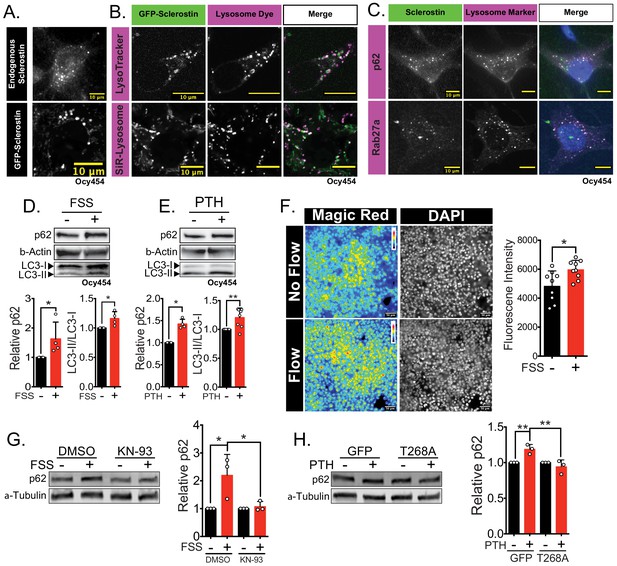
Sclerostin co-localizes with lysosomal markers, and both FSS and PTH induce lysosome activation through CaMKII.
(A) Endogenous sclerostin (top) and GFP-tagged sclerostin (bottom) form discrete puncta in Ocy454 cells. (B) Ocy454 cells were transfected with GFP-sclerostin, and lysosomes were visualized with Lysotracker (1 mM, 1 hr) or siR-Lysosome (1 μM, 4 hr). Scale bar represents 10 μm. (C) Ocy454 cells were stained for endogenous sclerostin and either p62/sequestosome-1 or Rab27a to evaluate co-localization with these lysosome-associated proteins. (D) Ocy454 cells were exposed to 1 min of FSS at 4 dynes/cm2, lysed immediately post-flow, and western blotted for p62/sequestosome-1, β-actin, and LC3 (n = 4). (E) Ocy454 cells were treated with PTH (1–34) (10 nM) for 5 min, lysed, and western blotted for p62/sequestosome-1 and β-actin (n = 4) and LC3 (n = 8). (F) UMR106 cells were subjected to FSS for 5 min, then Magic Red Cathepsin B was applied for 10 min, fixed, and imaged to assess lysosome activity (n = 9). (G) Ocy454 cells were treated with DMSO or KN-93 (10 μM) to inhibit CaMKII for 1 hr prior to FSS at 4 dynes/cm2 for 5 min before lysing immediately after FSS. Western blots were probed for p62/sequestosome-1 and β-actin (n = 3). (H) Ocy454 cells were transfected with a plasmid expressing either GFP or dominant negative CaMKII T286A prior to treatment with PTH (1–34) (10 nM) for 30 min. Western blots were probed for p62/sequestosome-1 and β-actin (n = 3). Graphs depict mean ± SD. *p<0.05, **p<0.01 by unpaired two-tailed t-test (D–F) or by two-way ANOVA with Holm–Sidak post hoc correction (G, H).
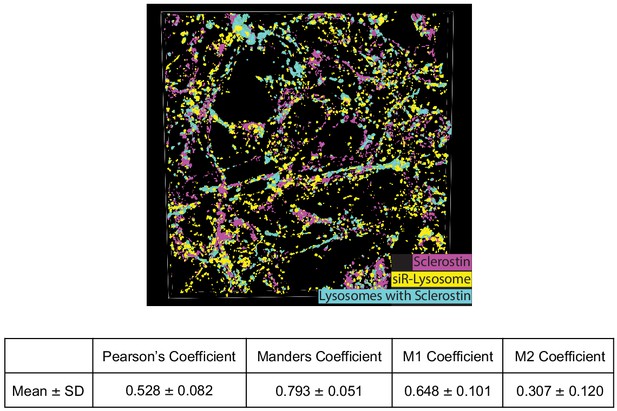
Co-localization of sclerostin with lysosomes.
GFP-sclerostin-transfected and siR-lysosome-stained Ocy454 cells were imaged in Z-stacks with 0.5 μm steps and reconstructed into 3D images. Nikon general analysis was programmed to identify lysosomes containing sclerostin. GFP-sclerostin is pseudocolored in pink, siR-lysosome in yellow, and lysosomes containing sclerostin in cyan. Z-stack images from GFP-sclerostin- and siR-lysosome-labeled cells were analyzed for co-localization coefficients. M1 represents the overlap of sclerostin with siR-lysosome; M2 represents the overlap of siR-lysosome with sclerostin (n = 3).
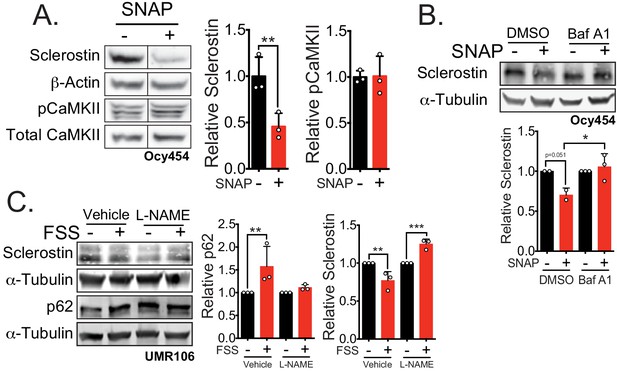
Nitric oxide, known mechanical load effector in bone and a molecular controller of crinophagy, contributes to sclerostin degradation.
(A) Ocy454 cells transfected with GFP-sclerostin were treated with vehicle (water) or 10 μM SNAP, a nitric oxide donor, and lysed after 5 min. Western blots were probed for sclerostin, α-tubulin, pCaMKII, and total CaMKII (n = 3). For each antibody, blots are from a single gel and exposure; a vertical black line indicates removal of irrelevant lanes. (B) Ocy454 cells transfected with myc-tagged sclerostin were treated with DMSO or bafilomycin A1 (100 nM) to inhibit lysosomal degradation, for 30 min, then treated with SNAP, a nitric oxide donor, for 5 min and lysed. Western blots were probed for sclerostin and α-tubulin (n = 2–3). (C) UMR106 cells were treated with vehicle or L-NAME (1 mM) to inhibit nitric oxide synthases (NOSs) for 1 hr and then exposed to 1 or 5 min of FSS. Lysates from cells exposed to 1 min of FSS were probed for p62/sequestosome-1 and α-tubulin abundance and lysates from cells exposed to 5 min of sclerostin were probed for sclerostin and α-tubulin abundance (n = 3). Graphs depict mean ± SD. *p<0.05, **p<0.01, ***p<0.001 by unpaired two-tailed t-test (A) or two-way ANOVA with Holm–Sidak post hoc test (B, C).
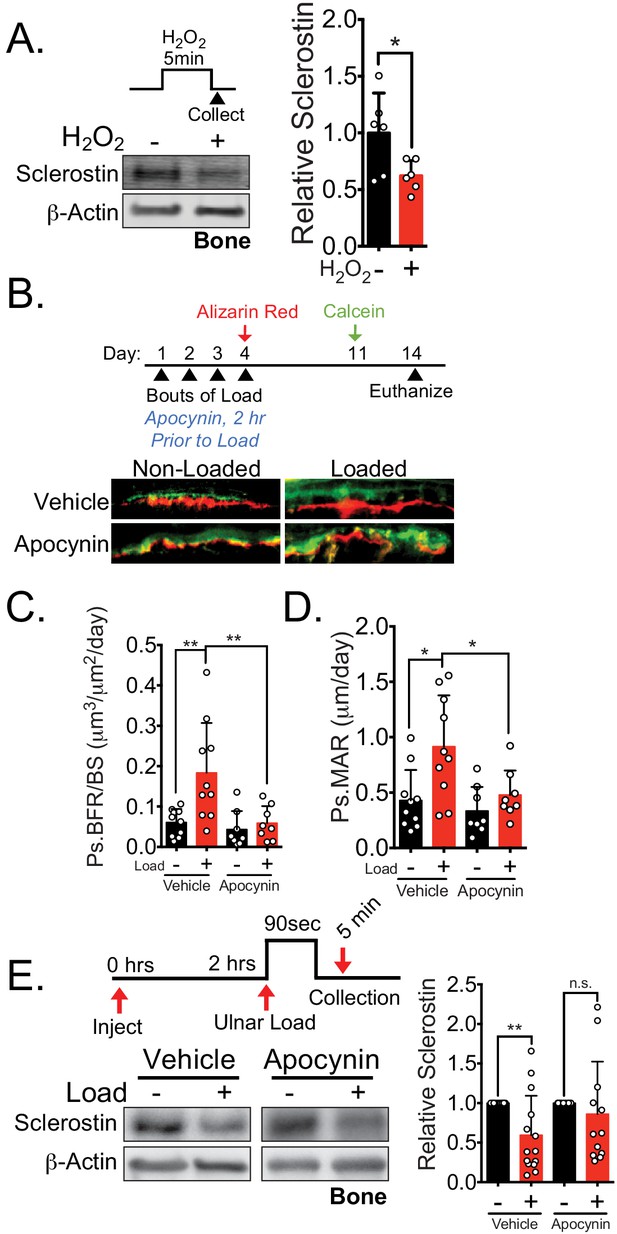
NOX2-dependent ROS are necessary for load-induced sclerostin degradation and bone formation in vivo.
(A) Dissected ulnae and radii flushed of marrow were treated with hydrogen peroxide (100 μM) as a source of ROS for 5 min before homogenization. Western blots were probed with sclerostin and β-actin (n = 6 mice). (B) Thirteen week old male C57Bl/6 mice treated with vehicle (saline, n = 10 mice) or apocynin (3 mg/kg, n = 8 mice) to inhibit NOX2 were forearm loaded (1800 με, 90 s, 2 Hz) and labeled with calcein and alizarin red at the indicated times for dynamic histomorphometry. Representative periosteal double labeling is shown. (C) Periosteal bone formation rate (Ps.BFR) and (D) periosteal mineral apposition rate (Ps.MAR) were calculated. (E) Fourteen to 17 week old male and female C57Bl/6 mice treated with vehicle (saline + 4% DMSO, i.p., n = 14) or apocynin (3 mg/kg in saline, i.p., n = 12 mice) to inhibit NOX2 were treated 2 hr prior to ulnar loading (2000 με, 90 s, 2 Hz). Non-loaded and loaded limbs were isolated 5 min post-load, and western blots were probed for sclerostin and β-actin. Vehicle data is duplicated in Figure 7D as all animals were run and processed together. Graphs depict mean ± SD. *p<0.05, **p<0.01 by unpaired two-tailed t-test (A), two-way ANOVA with Holm–Sidak post hoc correction (C, D), or Kruskal–Wallis with Dunn’s post hoc correction (E).
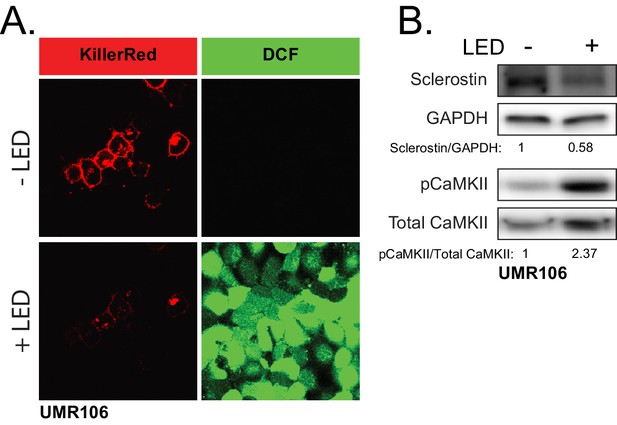
ROS is sufficient to drive CaMKII activation and loss of sclerostin protein.
(A) UMR106 cells transfected with KillerRed imaged before and after stimulation with LED light. DCF was used to track ROS production. (B) UMR106 cells transfected with KillerRed were stimulated with LED light for 5 min and lysed 5 min after. Westerns were probed for sclerostin, GAPDH, pCaMKII, and total CaMKII.
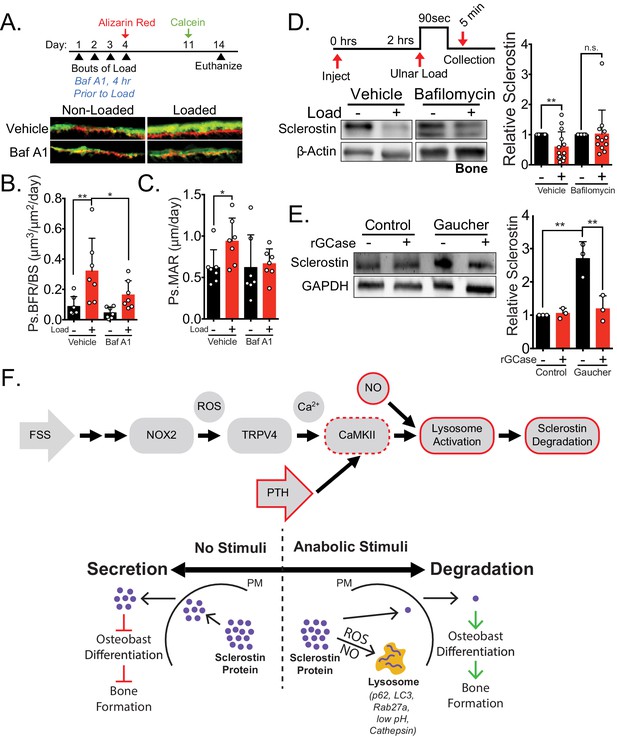
Lysosomal function is necessary for load-induced sclerostin degradation and subsequent bone formation and is implicated in human disease.
(A) Fifteen week old male C57Bl/6 mice treated with vehicle (saline + 4% DMSO, n = 7 mice) or bafilomycin A1 (1 mg/kg, n = 7 mice) to inhibit lysosomal degradation were forearm loaded (2000 με, 90 s, 2 Hz) and labeled with calcein and alizarin red at the indicated times for dynamic histomorphometry. Representative periosteal double labeling are shown. (B) Periosteal bone formation rate (Ps.BFR) and (C) periosteal mineral apposition rate (Ps.MAR) were calculated. (D) Fourteen to 17 week old male and female C57Bl/6 mice treated with vehicle (saline + 4% DMSO, i.p., n = 14) or bafilomycin A1 (1 mg/kg in saline + 4% DMSO, i.p., n = 12 mice) to inhibit lysosomal degradation were treated 2 hr prior to ulnar loading (2000 με, 90 s, 2 Hz). Non-loaded and loaded limbs were isolated 5 min post-load, and western blots were probed for sclerostin and β-actin. Vehicle data is duplicated in Figure 6E as all animals were run and processed together. (E) Human iPSC-derived osteoblasts from either control (non-diseased) or Gaucher disease patients were treated with vehicle or recombinant glucocerebrosidase (rGCase, 0.24 U/mL) for 5 days, then lysed for western blotting. Western blots were probed for sclerostin and GAPDH (n = 3 independent patient-derived iPSC lines/group). Graphs depict mean ± SD. *p<0.05, **p<0.01 by two-way ANOVA with Holm–Sidak post hoc correction (B, C, and E) or Kruskal–Wallis with Dunn’s post hoc correction (D). (F) FSS causes the rapid degradation of sclerostin protein by the lysosome through a number of molecular mediators. PTH, converging with this FSS mechano-transduction pathway at CaMKII, also mediates the lysosomal degradation of sclerostin protein. Icons outlined red are molecular mechanisms controlling sclerostin abundance that have been described within this manuscript that integrate into our previously described mechano-transduction cascade. Osteoanabolic stimuli, working through reactive oxygen (ROS) and reactive nitrogen species (RNS), direct sclerostin to the lysosome for degradation. This results in reduced sclerostin to allow for bone formation. PM: plasma membrane; ROS: reactive oxygen species; NO: nitric oxide.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Sost | National Center for Biotechnology Information | AAK13455 | |
| Gene (Homo sapiens) | Sost | National Center for Biotechnology Information | AAK16158.1 | |
| Gene (Rattus norvegicus) | Sost | National Center for Biotechnology Information | EDM06161.1 | |
| Gene (Bos Taurus) | Sost | National Center for Biotechnology Information | NP_001159986.1 | |
| Strain, strain background (Mus musculus, male and female) | C57Bl/6 | Jackson Laboratories | 000664 | RRID:IMSR_JAX:000664 |
| Cell line (Mus musculus) | Ocy454 | P. Divieti-Pajevic, Boston University | RRID:CVCL_UW31 | |
| Cell line (Rattus norvegicus) | UMR106 | ATCC | CRL-1661 | RRID:CVCL_3617 |
| Transfected construct (Mus musculus) | CaMKII T286A | Addgene | #29430 | |
| Transfected construct (Homo sapiens) | GFP-tagged sclerostin | Origene | #RG217648 | |
| Transfected construct (Mus musculus) | myc-tagged sclerostin | Origene | #MR222588 | |
| Transfected construct (Mus musculus) | KillerRed | Evrogen | FP966 | |
| Biological sample (Homo sapiens) | WT and GD iPSCs | PMID:23071332 | ||
| Antibody | Goat Polyclonal Anti-sclerostin | R and D Systems | AF1589 RRID:AB_2270997 | (1:250–500) |
| Antibody | Mouse Monoclonal Anti-GAPDH | Millipore | MAB374 RRID:AB_2107445 | (1:2500) |
| Antibody | Mouse Monoclonal Anti-βActin | Sigma | A1978 RRID:AB_476692 | (1:5000) |
| Antibody | Mouse Monoclonal Anti-αTubulin | Sigma | T9026 RRID:AB_477593 | (1:2000) |
| Antibody | Rabbit Polyclonal Anti-Col1a1 | Sigma | ABT257 RRID:AB_2890134 | (1:1000) |
| Antibody | Rabbit Monoclonal Anti-pCaMKII | Cell Signalling Technology | 12716S RRID:AB_2713889 | (1:1000) |
| Antibody | Rabbit Polyclonal Anti-Total CaMKII | Cell Signalling Technology | 3362S RRID:AB_2067938 | (1:1000) |
| Antibody | Rabbit Monoclonal Anti-p62/Sequestosome-1 | Cell Signalling Technology | 23214 RRID:AB_2798858 | (1:250–500) |
| Antibody | Rabbit Polyclonal Anti-LC3B | Cell Signalling Technology | 2775 RRID:AB_915950 | (1:500) |
| Antibody | Rabbit Monoclonal Anti-Rab27A | Cell Signalling Technology | 69295 RRID:AB_2799759 | (1:250) |
| Antibody | HRP Anti-Rabbit | Cell Signalling Technology | 7074 RRID:AB_2099233 | (1:1000–5000) |
| Antibody | HRP Anti-Mouse | Cell Signalling Technology | 7076 RRID:AB_330924 | (1:1000–5000) |
| Antibody | HRP Anti-Goat | Thermo Fisher Scientific | A27014 RRID:AB_2536079 | (1:1000) |
| Antibody | Donkey anti-Goat IgG Alexa Fluor 546 | Thermo Fisher Scientific | A-11056 RRID:AB_142628 | (1:100) |
| Antibody | Chicken anti-Rabbit Alexa Fluor 488 | Thermo Fisher Scientific | A-21441 RRID:AB_2535859 | (1:100) |
| Sequence-based reagent | Sost | NCBI BLAST | NM_024449.6 | GGA ATG ATG CCA CAG AGG TCA T and CCC GGT TCA TGG TCT GGT T |
| Sequence-based reagent | Gapdh | NCBI BLAST | NG_007785.2 | CGT GTT CCT ACC CCC AAT GT and TGT CAT CAT ACT TGG CAG GTT TCT |
| Sequence-based reagent | Hprt | NCBI BLAST | NM_013556.2 | AGC AGT ACA GCC CCA AAA TGG and AAC AAA GTC TGG CCT GTA TCC AA |
| Sequence-based reagent | Rpl13 | NCBI BLAST | NM_016738.5 | CGA AAC AAG TCC ACG GAG TCA and GAG CTT GGA GCG GTA CTC CTT |
| Commercial assay or kit | Magic Red | Sigma | CS0370 | |
| Commercial assay or kit | siR-Lysosome | Spirochrome | SC016 | |
| Commercial assay or kit | Lysotracker | Thermo Fisher Scientific | L7528 | |
| Commercial assay or kit | High-Capacity RNA-to-cDNA Kit | Thermo Fisher Scientific | 4388950 | |
| Commercial assay or kit | Maxima SYBR Green/ROX qPCR Master Mix | Thermo Fisher Scientific | FERK0221 | |
| Commercial assay or kit | JetPrime Transfection kit | PolyPlus Transfection | 114–75 | |
| Chemical compound, drug | Bafilomycin A1 (in vitro studies) | Cell Signalling Technology | 54645 | |
| Chemical compound, drug | Bafilomycin A1 (in vivo studies) | Research Products International | 88899-55-2 | |
| Chemical compound, drug | Brefeldin A | Cell Signalling Technology | 9972 | |
| Chemical compound, drug | MG-132 | Cell Signalling Technology | 2194 | |
| Chemical compound, drug | Cycloheximide | Cell Signalling Technology | 2112 | |
| Chemical compound, drug | PTH (1–34) | US Biological Life Sciences | #P3109-24D | |
| Chemical compound, drug | Leupeptin | Millipore | EI8 | |
| Chemical compound, drug | Apocynin | Sigma | 178385 | |
| Chemical compound, drug | L-NAME | Millipore | N5751 | |
| Chemical compound, drug | SNAP | Sigma | N3398 | |
| Chemical compound, drug | Alizarin Red | Sigma | A3882 | |
| Chemical compound, drug | KN-93 | Sigma | K1358 | |
| Chemical compound, drug | Calcein | Sigma | C0875 | |
| Chemical compound, drug | Halt Protease and Phosphatase Inhibitor Cocktail (EDTA-free) | Thermo Fisher Scientific | 78440 | |
| Chemical compound, drug | SuperBlockPBS | Thermo Fisher Scientific | 37515 | |
| Software, algorithm | FIJI | ImageJ | RRID:SCR_002285 | |
| Software, algorithm | Nikon NIS Elements 5.2 | Nikon | RRID:SCR_014329 | |
| Other | REVERT Total Protein Stain | Licor | 827–15733 | |
| Other | Prolong Gold Antifade Reagent with DAPI | Cell Signalling Technology | 8961S | |
| Other | TRIzol | Sigma | T9424 | |
| Other | DCF | Invitrogen | D399 |
Additional files
-
Source data 1
This is the raw data for the entire manuscript.
Each tab of the excel references a different figure.
- https://cdn.elifesciences.org/articles/64393/elife-64393-data1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64393/elife-64393-transrepform-v3.docx





