Glypicans define unique roles for the Hedgehog co-receptors boi and ihog in cytoneme-mediated gradient formation
Figures
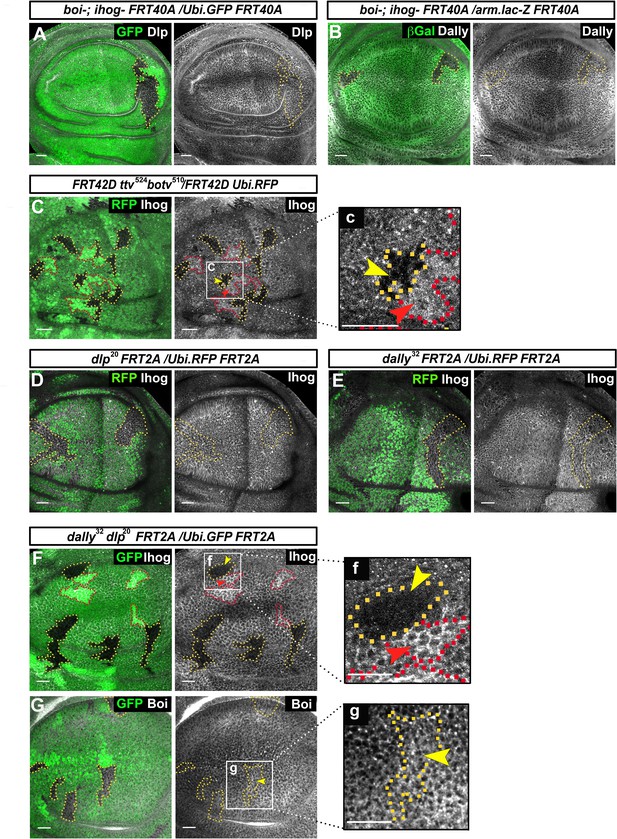
Glypicans regulate Ihog but not Boi presence at the plasma membrane.
(A, B) boi-; ihogZ23 double mutant clones (labeled by the lack of GFP) do not affect the expression of the glypicans Dlp (A) and Dally (B) (gray channels). (C) Ihog levels (Bac Ihog:GFP, gray channel) decrease in ttv524 botv510 double mutant clones (labeled by the lack of GFP, yellow arrowhead); (c) enlargement of one of the clones. (D, E) dlp20 (D) and dally32 (E) single mutant clones (labeled by the lack of RFP in green) do not affect Ihog levels (α-Ihog antibody, gray channel). (F) Ihog levels (labeled with α-Ihog antibody, gray channel) decrease in dally32 dlp20 double mutant clones (labeled by the lack of GFP, yellow arrowhead); (f) enlargement of one clone. (G) Boi levels (labeled with anti Boi antibody, gray channel) are not affected in dally32 dlp20 double mutant clones (labeled by the lack of GFP, yellow arrowhead); (g) enlargement of one clone. Note that the homozygous wild-type sister clones (more intense green) positively modulate Ihog levels (red arrowheads in C and F). The discs shown in panels are representative of four to eight discs containing clones in at least three experiments. Scale bar: 20 μm.

Different roles of Boi and Ihog in glypican recruitment.
(A, B) Apico-basal distribution of glypicans in wings discs after apGal4/+; UAS.boi/+ (A) and apGal4/+; UAS ihog/+ (B) expression. Note that while Boi ectopic expression preferentially accumulates Dally and Dlp in the apical side of the wing disc cells, Ihog accumulates Dally and Dlp at the basal side. These accumulations of Dally and Dlp allow the visualization of cytonemes in the basal confocal sections (blue arrowheads). The transversal sections shown in panels are representative of at least five discs in three independent experiments.
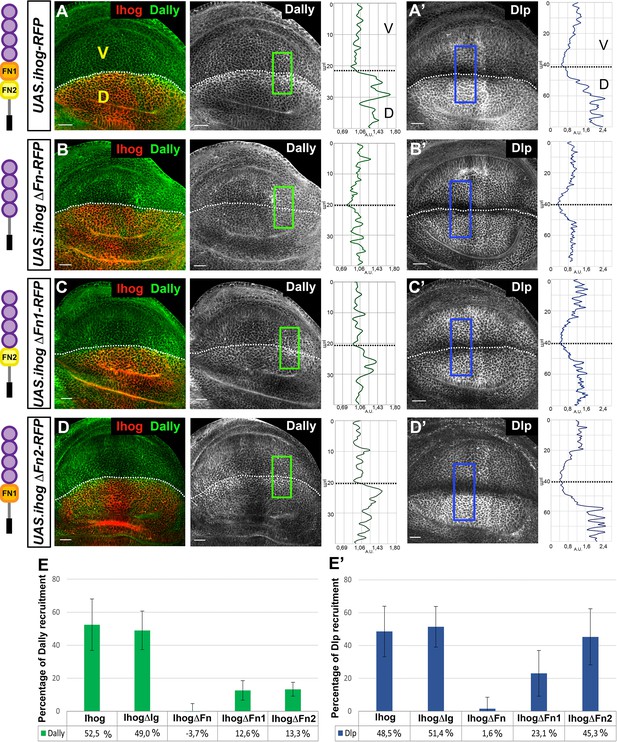
Interaction of Ihog mutant forms with glypicans.
(A–D) Glypicans accumulation in: apGal4 tubGal80ts/+; UAS.ihog-RFP/+ (A–A’), apGal4 tubGal80ts/+; UAS.ihogΔFn-RFP/ + (B–B’), apGal4 tubGal80ts/+; UAS.ihogΔFn1-RFP/+ (C–C’), apGal4tubGal80ts/+; UAS.ihogΔFn2-RFP/+ (D–D’) wing discs after 30 hr at the restrictive temperature. (E–E’) Percentage of Dally (E) and Dlp (E’) recruitment in the dorsal compartment relative to the endogenous control (ventral compartment). Each image incorporates at its side a plot profile (taken from the framed area in each image), indicating the relative intensity fluorescence for each glypican. Note that IhogΔFn-RFP does not increase either Dally (B, E) or Dlp (B’, E’). However, in ectopic expression of a partial deletions of the Ihog FN-type III domains (IhogΔFn1-RFP and IhogΔFn2-RFP) an accumulation of Dally (C–E) is observed although at lower levels than those of the ectopic Ihog-RFP (A, E). Curiously, IhogΔFn2-RFP recruits Dlp (D’, E’) at the same levels as Ihog-RFP (A’, E’) does. Discs are oriented with dorsal part down (D), ventral up (V), and posterior (P) right. Plot profiles were done over a ROI of size of 40 μm X 20 μm for Dally and 80 μm × 20 μm for Dlp. Data of glypicans recruitment is available at Figure 2—source data 1 for Dally and Figure 2—source data 2 for Dlp measure; p-values of the statistical analysis are shown in Tables 1 and 2 (Materials and methods). The discs shown in panels are representative of at least four discs in three independent experiments. Scale bar: 20 μm.
-
Figure 2—source data 1
Dally recruitment measures ussing different Ihog mutant constructs.
- https://cdn.elifesciences.org/articles/64581/elife-64581-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Dlp recruitment measures ussing different Ihog mutant constructs.
- https://cdn.elifesciences.org/articles/64581/elife-64581-fig2-data2-v2.xlsx
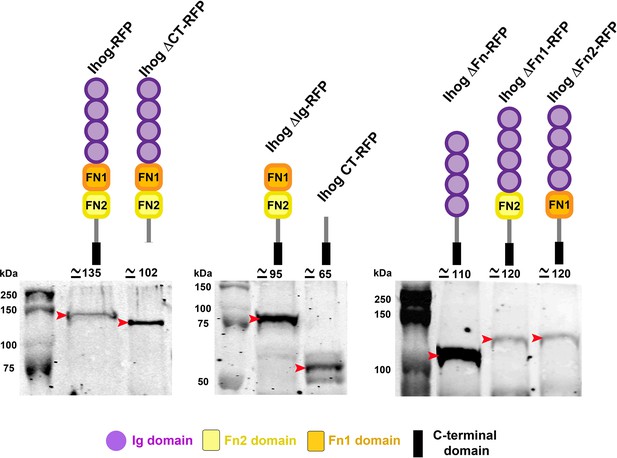
Scheme of the Ihog mutant constructs and the molecular weight of their proteins.
Scheme of Ihog-RFP, IhogΔCT-RFP, IhogΔIg-RFP, IhogCT-RFP IhogΔFn-RFP, IhogΔFn1-RFP, IhogΔFn2-RFP, and their respective molecular weight shown by western blot analysis.
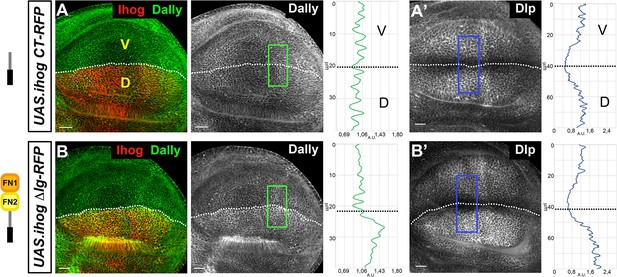
Role of Ihog ΔIg and Ihog CT domains in glypican retention.
(A, B) Wing discs after 30 hr induction at the restrictive temperature of apGal4TubGal80ts/+; UAS.ihogΔIg-RFP/+ (A) and after 22 hr induction at the restrictive temperature of apGal4TubGal80ts/+; UAS.ihogCT-RFP/+ (B). Plot profiles were done over ROI of size 80 μm × 30 μm for Hh, 40 μm × 20 μm for Dally and 80 μm × 20 μm for Dlp. Note the accumulation of both glypicans after the induction of IhogΔIg-RFP (A) and the lack of accumulation after the induction of IhogCT-RFP. Discs are oriented with dorsal (D) compartment down ventral (V) up and posterior (P) right. The discs shown in panels are representative of at least four discs in three independent experiments. Scale bar: 20 μm.
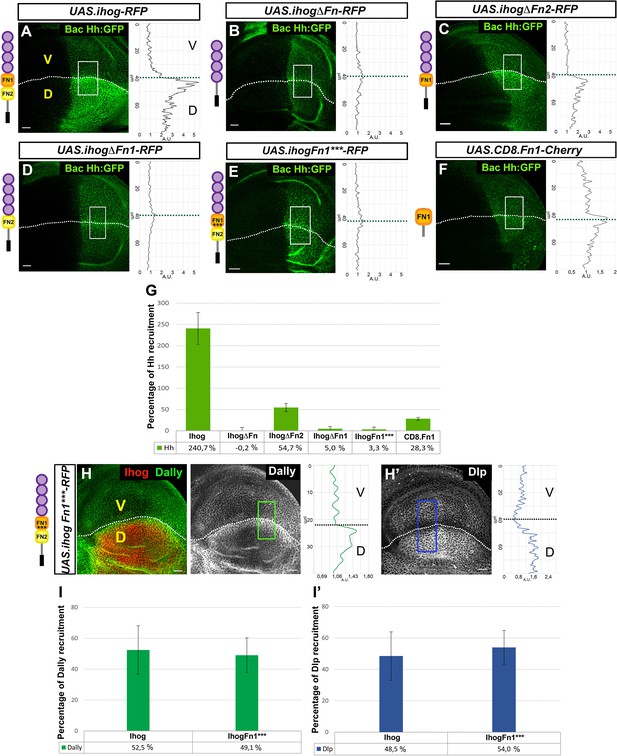
Effects of Ihog mutant forms and Ihog Fn point mutations on Hh and glypican interactions.
(A–G) Hh (BacHh:GFP) expression in wing discs after 30 hr at the restrictive temperature in apGal4 BacHh:GFP/+; UAS.ihog-RFP/tubGal80ts (A), apGal4 BacHh:GFP/+; UAS.ihogΔFn-RFP/tubGal80ts (B), apGal4 BacHh:GFP/+; UAS.ihogΔFn2-RFP/tubGal80ts (C), apGal4 BacHh:GFP/+; apGal4 BacHh:GFP/+; UAS.ihogΔFn1-RFP/tubGal80ts (D), UAS.ihogFn1***-RFP/tubGal80ts (E), apGal4 BacHh:GFP/+; UAS.cd8.Fn1-Cherry/tubGal80ts (F), percentage of Hh recruitment in the dorsal compartment relative to the endogenous control (ventral compartment) (G). (H, I) Increase of glypicans after 30 hr at the restrictive temperature in apGal4 tubGal80ts/+; UAS.ihogFn1***-RFP/+ (H, H’) and percentage of Dally (I) and Dlp (I’) recruitment in the dorsal compartment relative to the endogenous control (ventral compartment). Images incorporate at their side a plot profile (taken from the framed area in each image), indicating the modulation of glypican levels. Plot profiles were done over ROI of size 80 μm × 30 μm for Hh, 40 μm × 20 μm for Dally and 80 μm × 20 μm for Dlp. Note that Ihog Fn1*** does not interact with Hh (E, G) while increasing Dally (H, I) and Dlp (H’, I’) at same levels that Ihog-RFP does. Discs are oriented with dorsal part down (D), ventral up (V) and posterior (P) right. Data is available at Figure 3—source data 1 for Hh recruitment and p-values of the statistical analysis are shown in Table 3 (Materials and methods). Data is available at Figure 2—source data 1 for Dally recruitment and Figure 2—source data 2 for Dlp recruitment; p-values of the statistical analysis are shown in Tables 1 and 2 (Materials and methods). The discs shown in panels are representative of at least four discs from three independent experiments. Scale bar: 20 μm.
-
Figure 3—source data 1
Hh recruitment measures ussing different Ihog mutant constructs.
- https://cdn.elifesciences.org/articles/64581/elife-64581-fig3-data1-v2.xlsx
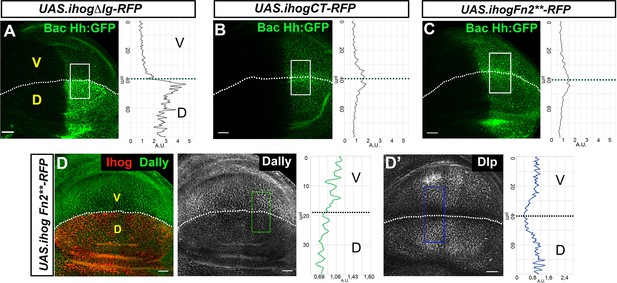
Effects of Ihog mutant forms on Hh and glypicans interactions.
(A–D) Hh (BacHh:GFP) expression in wing discs after 30 hr at the restrictive temperature in apGal4 BacHh:GFP/+; UAS.ihogΔIg-RFP/tubGal80ts (A), apGal4 BacHhGFP/+; UAS.ihogCT-RFP/tubGal80ts (B) and apGal4 BacHh:GFP/+; UAS.ihogFn2**-RFP/tubGal80ts (C). Increase of glypicans after 30 hr at the restrictive temperature in apGal4 tubGal80ts/+; UAS.ihogFn2**-RFP/+ (D, D’) recruitment in the dorsal compartment. Images incorporate at their side a plot profile (taken from the framed area in each image), indicating the modulation of glypican levels. Plot profiles were done over ROI of size 80 μm × 30 μm for Hh, 40 μm × 20 μm for Dally, and 80 μm × 20 μm for Dlp. Note that Ihog Fn2** does not interact with Dlp (D’) and remove Dally (D). Discs are oriented with dorsal part down (D), ventral up (V), and posterior (P) right. The discs shown in panels for Hh retention are representative of at least four discs from three independent experiments and panels for glypicans retention are representative of at least four discs in three independent experiments. Scale bar: 20 μm.
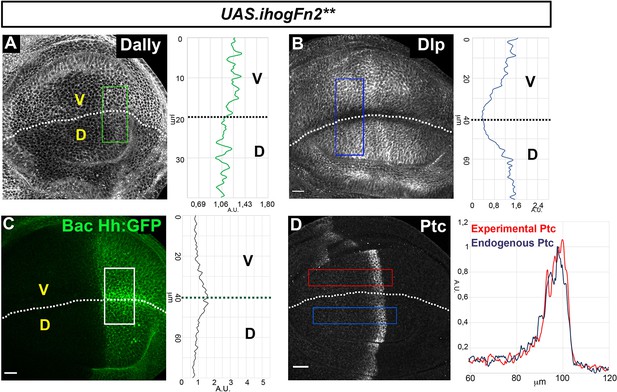
Effects of the ectopic expression of IhogFn2**.
(A–D) Glypicans accumulation after 30 hr at the restricted temperature in apGal4 tubGal80ts/+; UAS.ihogFn2**/+ wing discs (A, B), Hh expression (BacHh:GFP) in apGal4 BacHh:GFP/+; UAS.ihogFn2**-RFP/tubGal80ts wing discs (C); Ptc expression in LexAopGal80; apLexA ptcGal4/+; tubGal80ts/UAS.ihogFN2** wing discs (D). Plot profiles were done over ROI of size 80 μm × 30 μm for Hh, 40 μm × 20 μm for Dally, 80 μm × 20 μm for Dlp, and 120 μm × 20 μm for Ptc. Note that in all cases, its behavior is the same as that of IhogFn2**RFP. The discs shown in panels are representative of at least five discs in three independent experiments. Scale bar: 20 μm.
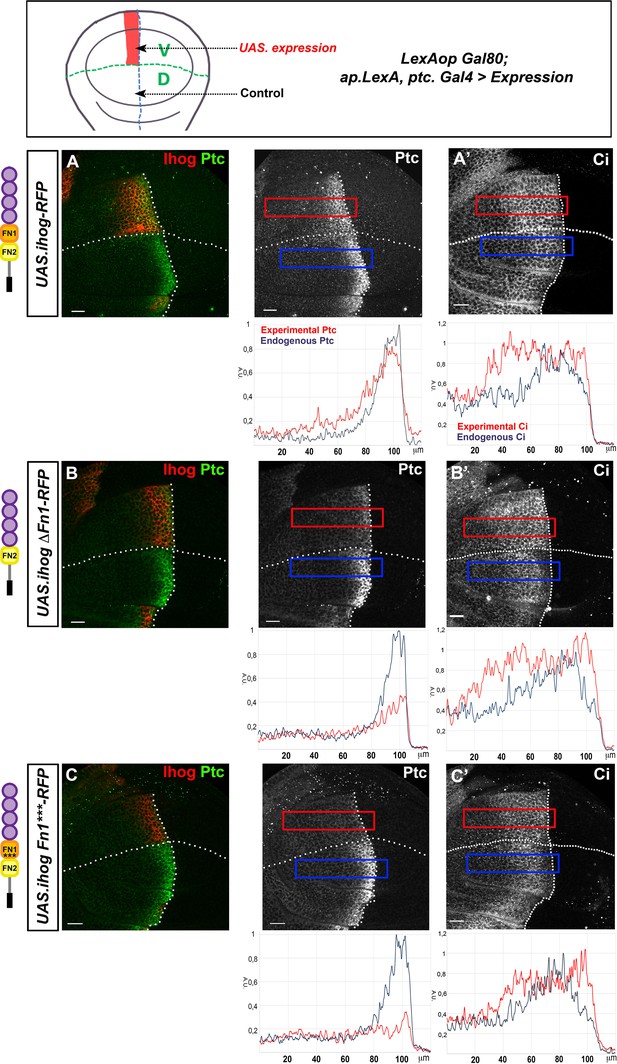
Effect of the ectopic expression of Ihog mutant forms on Hh gradient formation.
(A–C) Ptc (A–C) and Ci (A’–C’) expression in wing discs after 28 hr induction of: UAS.ihog-RFP (A), UAS.ihogΔFn1-RFP (B), and UAS.ihogFn1***RFP (C) using the multiple driver LexAopGal80; apLexA ptcGal4/+; tubGal80ts/+. Images incorporate underneath plots of the fluorescence intensity of Ptc and Ci expression in the ventral experimental side (red) compared with the dorsal control side (blue) of the wing disc. Note that both IhogΔFn1 and IhogFn1*** strongly reduce Hh reception. Discs are oriented with dorsal (D) part down and posterior (P) right. Plot profiles were performed over a ROI of size 120 μm × 20 μm. The discs shown in panels are representative of at least five discs in three independent experiments. Scale bar: 20 μm.

Effects of the ectopic expression of IhogΔFn2-RFP and IhogFn2**-RFP in Hh signaling.
(A, B) Ptc and Ci expression in wing discs after 28 hr induction at the restrictive temperature of the transgenes: UAS.ihogΔFn2-RFP (A) and UAS.ihogFn2**-RFP (B) using the multiple driver LexAopGal80; apLexA ptcGal4/+; tubGal80ts/+. Each image incorporates below a plot of the fluorescence intensity of either Ptc or Ci expression (red) in the ventral (V) compartment of the wing disc with their internal controls for wild-type expression in the dorsal (D) compartment (blue). Note that while IhogΔFn2 flattened the Hh signaling gradient, IhogFn2** do not have any effect. Discs are oriented with the D compartment down and the P compartment right. Plot profiles were performed over a ROI of size 120 μm × 20 μm. The discs shown in panels are representative of at least five discs in three independent experiments. Scale bar: 20 μm.
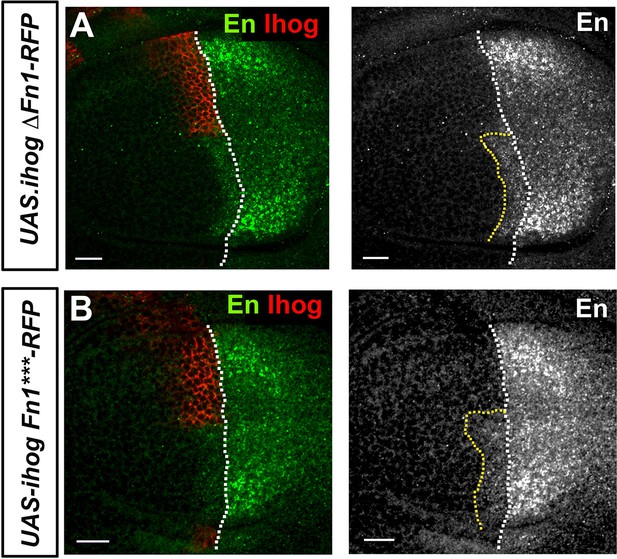
Effect of the Ihog Fn1 domain on high-threshold targets of Hh.
(A, B) En expression in wing discs after 25 hr induction of the transgenes: UAS.ihogΔFn1-RFP (A); and UAS.ihogFn1***RFP (B) using the multiple driver LexAopGal80; apLexA ptcGal4/+; tubGal80ts/+. Note that both Ihog constructs eliminate the ventral En expression in the A compartment cells. Discs are oriented with D compartment down and P compartment right. Endogenous control is framed with a yellow dotted line. Scale bar: 20 μm. The discs shown in panels are representative of at least five discs in three independent experiments.
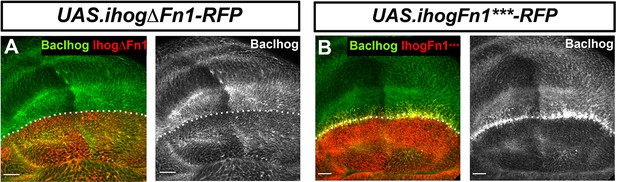
Endogenous Ihog protein decreases by the expression of IhogΔFn1 and IhogFn1***.
(A, B) Basal side of BacIhog:GFP discs after 30 hr induction at the restrictive temperature of apGal4 TubGal80ts/+; UAS.ihogΔFn1-RFP/+ (A) and of apGal4 TubGal80ts/+; UAS.ihogFn1***-RFP/+ (B). Note that both Ihog mutant constructs remove the endogenous Ihog (BacIhog:GFP) specifically in the basal part of the disc epithelium with clearer effect expressing the IhogFn1***-RFP. The discs shown in panels are representative of at least three discs in two independent experiments. Discs are oriented with D compartment down and P compartment right. Scale bar: 20 μm.
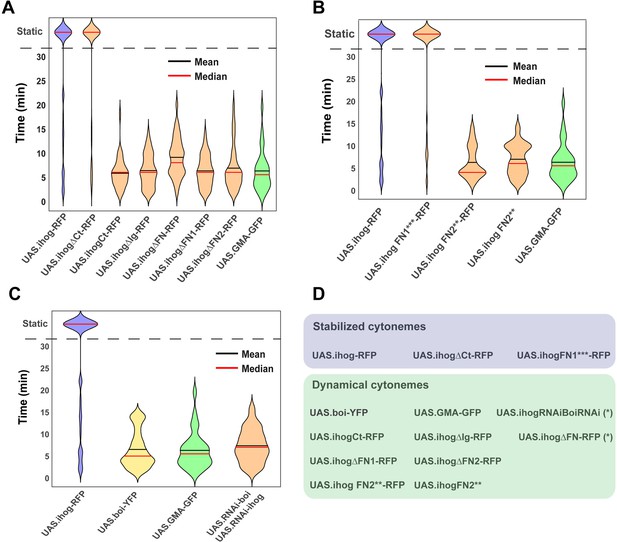
Effect of Ihog and its mutant forms in the regulation of cytoneme dynamics.
(A) Violin plots of lifetime distributions of dynamic cytonemes in different Ihog overexpression genotypes: UAS.ihog-RFP (blue), wild-type control UAS.GMA-GFP (green), and different Ihog deletion mutants (orange). (B) Comparison between the effect of the ectopic expression of different Ihog point mutations: Control UAS.ihog-RFP (blue), wild-type control UAS.GMA-GFP (green); mutants of Ihog (orange). (C) Comparison between different levels of Ihog and Boi: UAS.ihog-RFP (blue), UAS.boi-YFP (yellow), UAS.GMA-GFP (green), and UAS.RNAi-Boi/UAS.RNAi-ihog (orange). (D) Table summarizing the dynamic of cytoneme in different genotypes: Stabilized cytonemes in blue and dynamical cytonemes in green. (*) Statistically significant lifetime differences compared to wild-type GMA-GFP cytonemes. Data is available at Figure 5—source data 1 and p-values of the statistical analysis are shown in Tables 4 and 5 (Material and methods).
-
Figure 5—source data 1
Cytonemes lifetime measures under different experimental conditions.
- https://cdn.elifesciences.org/articles/64581/elife-64581-fig5-data1-v2.xlsx

Ihog and Boi RNAis effects.
Boi protein visualization using α-Boi antibody in wing disc (A); Boi protein visualization using α-Boi antibody in apGal4/+; UAS Boi-RNAi/+ wing disc (B); Ihog (BacIhog:GFP) expression wing disc (C) and Ihog (BacIhog:GFP) in apGal4/+; UAS ihog-RNAi/+ wing discs (D). Note the effect of both RNAi in diminish the endogenous protein levels. Discs are oriented with dorsal (D) part down and posterior (P) right. The discs shown in panels are representative of at least three discs in three independent experiments. Scale bar: 20 μm.
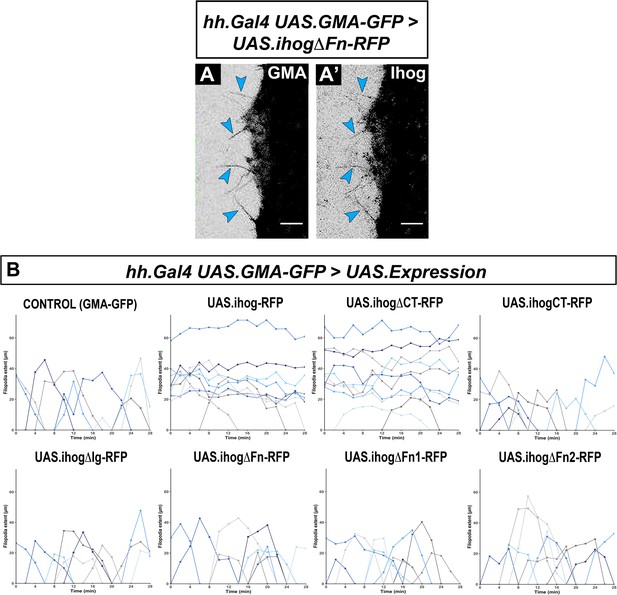
Cytoneme dynamics after expressing the Ihog mutant constructs.
Time-frames from Video 5 of wild-type histoblasts showing the GMA-GFP signal (A) and the ectopic IhogΔFn -RFP signal (A’). Representation of cytoneme extension values over time are the results of each Ihog construct dynamics from Video 5. (B) Variation of cytoneme extension over time are represented in different shades of blue. Note that cytonemes are labeled with both GFP (A) and IhogΔFn-RFP (A’) (arrowheads). Stabilized cytonemes do not show graphical alteration (Ihog-RFP and IhogΔCT-RFP constructs), which indicates no changes in cytoneme extension or retraction. Scale bar: 10 μm.
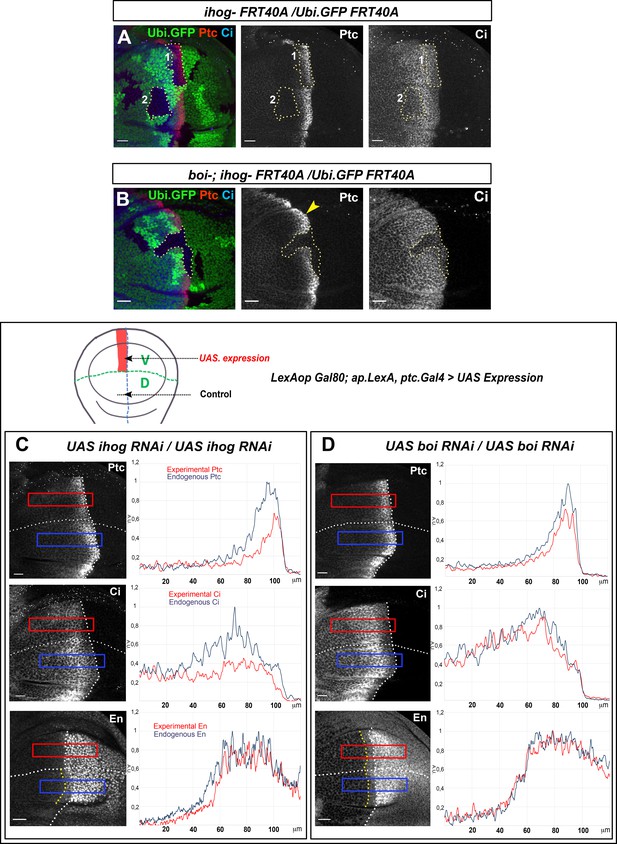
Ihog and Boi have differential roles on Hh gradient formation.
(A) Homozygous ihogZ23 mutant clones (labeled by the lack of GFP) in boi wild-type background (A) and in boi−/− mutant background (B). Both experiments are immunostained for Ptc and Ci. Ptc expression is significantly reduced in ihogZ23 mutant clones located in the A compartment abutting the A/P compartment border (clone one in A) and Ci is reduced in clones close to the A/P border in the Hh signaling zone (clone two in A). However, Ptc and Ci expressions are maintained in a boi- mutant background (B, yellow arrowhead). Note that there is not Hh signaling in the absence of both Ihog and Boi (B, dotted line area). (C, D) Ptc, Ci, and En expression in wing discs after 48 hr induction of UAS.ihog-RNAi (C) and UAS.boi-RNAi (D) using the multiple driver LexAopGal80; apLexA ptcGal4/+; tubGal80ts/+ (the scheme above shows the ventral/anterior domain of induction using this multiple driver). The UAS.ihog-RNAi expression reduces Ptc, Ci, and En levels (C), while extends Hh gradient slightly (Ptc and Ci show a more flattened pattern) after UAS.boi-RNAi expression (D). Discs are oriented with dorsal (D) part down and posterior (P) right. Plot profiles were performed over a ROI of size 120 μm × 20 μm for Ptc, Ci and En. The discs shown in panels are representative of at least five discs in three independent experiments. Scale bar: 20 μm.
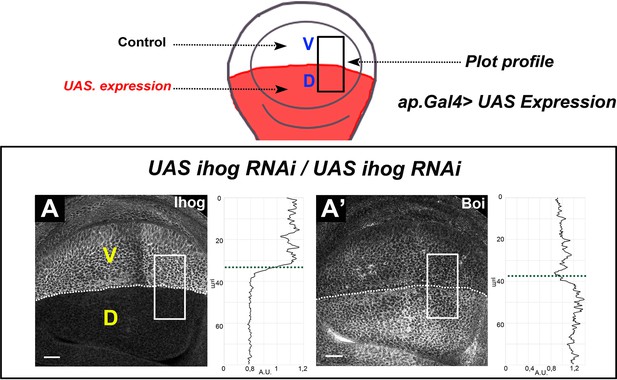
The lack of Ihog result in a Boi increase.
(A–A’) Ihog (BacIhog:GFP) (A) and Boi (using α-Boi antibody) (A’) expression in apGal4/+; UAS ihog-RNAi/+ wing discs. The scheme above represents the expression domain of Ihog RNAi in the dorsal compartment (apGal4) in red; the ventral side of the disc is used as control. Note that Ihog RNAi induces a slight increase in Boi levels. Discs are oriented with dorsal (D) part down and posterior (P) right. Plot profiles were performed over a ROI of size 80 μm × 30 μm. The discs shown in panels are representative of at least five discs in three independent experiments. Scale bar: 20 μm.
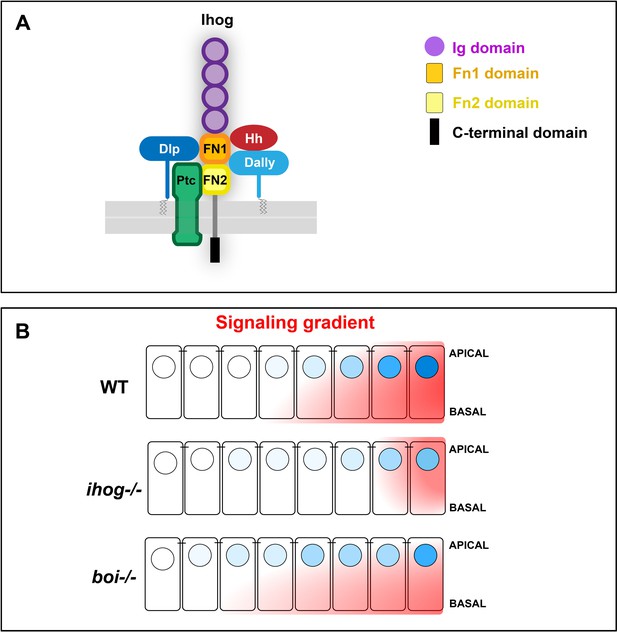
Scheme representatives of glypicans, Hh, and Ptc interaction with Ihog and its specific requirement for long-range Hh gradient.
(A) Scheme depicting the Ihog domains and their inetractions of the two FNIII domains with Hh (Yao et al., 2006b), Ptc (Zheng et al., 2010), and the glypicans Dally and Dlp. (B) Ihog and Boi loss of function differentially affect the formation of the Hh signaling gradient: while Ihog seems to be required for the short- and long range, boi is needed for the short-range Hh gradient.
Videos
Cytoneme dynamics under different levels of Ihog and Boi in histoblasts.
Abdominal histoblasts of pupae expressing UAS.GMA-GFP (black) under the control of Hh.Gal4; tubGal80ts and also expressing UAS.Ihog-RFP (after 24 hr of expression) (A). Note the stabilization of cytonemes compared with the very dynamic cytonemes of the control (expression of only GMA-GFP) (B). Abdominal histoblasts of a pupa continuously expressing UAS.RNAi-Ihog and UAS.RNAi-Boi in the P compartment during development (C) show cytonemes with similar dynamics to that of the control expressing only GMA-GFP (B). Movies are 30 min long. Frames correspond to the projection of a Z-stack at 2 min intervals. The A compartment is on the left. Pupae were around 30 hr after puparium formation. Scale bar: 10 μm.
Cytoneme dynamics of abdominal histoblasts expressing UAS.boi-YFP.
Abdominal histoblasts of a UAS.boi-YFP/+; hh.Gal4 tubGal80ts pupa expressing Boi-YFP (green) during 24 hr. (A) Z-stack from apical to basal, showing cytonemes in the most basal part. (B) 30 min movie of the same pupa showing dynamic cytonemes. Frames correspond to the projection of a Z-stack at 2 min intervals between frames. The A compartment is on the left. Pupa was around 30 hr after puparium formation. Scale bar: 10 μm.
Cytoneme dynamics of histoblasts expressing both UAS.boi-YFP and UAS.ihog-RFP.
Abdominal histoblasts of a UAS.boi-YFP/UAS.ihog-RFP; hh.Gal4 tubGal80ts pupa. Boi-YFP (green) and Ihog-RFP (gray) were expressed during 24 hr before recording. (A) Z-stack from apical to basal showing basal cytonemes. (B) 30 min movie of the same pupa showing stabilized cytonemes. Frames correspond to the projection of a Z-stack at 2 min intervals. The A compartment is on the left. Pupa was around 30 hr after puparium formation. Scale bar: 10 μm.
Cytoneme dynamics of histoblasts expressing UAS.ihog-RFP and UAS-RNAi-boi.
Abdominal histoblasts of a UAS.Ihog-RFP/UAS.RNAi-Boi; Hh.Gal4 tubGal80ts/+ pupa. Ihog-RFP (gray) and RNAi-Boi were expressed during 24 hr before recording. (A) Projection of a Z-stack from apical to basal show basal location of cytonemes. (B) 30 min movie of the same pupa showing stabilized cytonemes. Frames correspond to the projection of a Z-stack at 2 min intervals. The A compartment is on the left. Pupa was around 30 hr after puparium formation. Scale bar: 10 μm.
Roles of Ihog domains on cytoneme stability.
Abdominal histoblasts expressing UAS.GMA-GFP (black) under the control of Hh.Gal4 tubGal80ts pupae (24 hr of expression) (A) and also expressing UAS.Ihog-RFP (B); UAS.Ihog-ΔCT-RFP (C); UAS.IhogCT-RFP (D); UAS.Ihog-ΔIg-RFP (E); UAS.Ihog-ΔFn-RFP (F); UAS.Ihog-ΔFn1-RFP (G) and UAS.Ihog-ΔFn2-RFP (H). Note that cytonemes are stable in (B and C), while they are dynamic in (D–H). Movies are 30 min long. Frames correspond to the projection of a Z-stack at 2 min intervals. The A compartment is on the left. Pupae were around 30 hr after puparium formation. Scale bar: 10 μm.
Different effects of Ihog-FN1*** and IhogFN2** on cytoneme stability.
Abdominal histoblasts of pupae expressing UAS.GMA-GFP (in black) and UAS.ihog-RFP (A); UAS.ihog-Fn1***-RFP (B); UAS.ihogFn2**-RFP (C); UAS.ihogFn2** (D) under the Hh.Gal4 tubGal80ts control (24 hr of induction). Note that cytonemes from cells expressing Ihog-Fn1***-RFP, that alters its interaction with Hh but not with glypicans (B), are as stable as those from the control cells expressing Ihog-RFP (A), while IhogFn2** presents normal cytoneme dynamics (C, D). Movies are 30 min long. Frames correspond to the projection of a Z-stack at 2 min intervals. The A compartment is on the left. Pupae were around 30 hr after puparium formation. Scale bar: 10 μm.
Tables
Statistical analysis of the Dally recruitment for different Ihog mutants.
p-values obtained from pairwise T test to statistically compare the Dally recruitment for different Ihog mutants (gray: n.s = not significant; orange: significant, with the corresponding p-value in scientific notation).
| Pairwise.t.test | UAS.ihogDFN-RFP | UAS.ihogDFN1-RFP | UAS.ihogDFN2-RFP | UAS.ihogFN1***-RFP | UAS.ihogDIg-RFP |
|---|---|---|---|---|---|
| UAS.ihogDFN1-RFP | 0.027 | ||||
| UAS.ihogDFN2-RFP | 0.023 | n.s | |||
| UAS.ihogDIg-RFP | 4.7e-10 | 6.1e-07 | 8.1e-07 | ||
| UAS.ihogFN1***-RFP | 1.0e-09 | 1.0e-06 | 1.3e-06 | n.s | |
| UAS.ihog-RFP | 9.9e-11 | 1.1e-07 | 1.5e-07 | n.s | n.s |
Statistical analysis of the Dlp recruitment for different Ihog mutants.
p-values obtained from pairwise T test to statistically compare the Dlp recruitment for different Ihog mutants (gray: n.s = not significant; orange: significant, with the corresponding p-value in scientific notation).
| Pairwise.t.test | UAS.ihogDFN-RFP | UAS.ihogDFN1-RFP | UAS.ihogDFN2-RFP | UAS.ihogFN1***-RFP | UAS.ihogDIg-RFP |
|---|---|---|---|---|---|
| UAS.ihogDFN1-RFP | 0.0324 | ||||
| UAS.ihogDFN2-RFP | 6.4e-06 | 0.0288 | |||
| UAS.ihogDIg-RFP | 6.4e-07 | 0.0037 | n.s | ||
| UAS.ihogFN1***-RFP | 1.3e-06 | 0.0037 | n.s | n.s | |
| UAS.ihog-RFP | 1.8e-06 | 0.0094 | n.s | n.s | n.s |
Statistical analysis of the Hh recruitment for different Ihog mutants.
p-values obtained from pairwise T test to statistically compare the Hh recruitment for different Ihog mutants (gray: n.s = not significant; orange: significant, with the corresponding p-value in scientific notation).
| Pairwise.t.test | UAS.ihogDFN-RFP | UAS.ihogDFN1-RFP | UAS.ihogDFN2-RFP | UAS.ihogFN1***-RFP | UAS.ihogDIg-RFP |
|---|---|---|---|---|---|
| UAS.ihogDFN1-RFP | n.s | ||||
| UAS.ihogDFN2-RFP | 3.3e-06 | 9.6e-06 | |||
| UAS.ihogDIg-RFP | 0.012 | 0.027 | 0.027 | ||
| UAS.ihogFN1***-RFP | n.s | n.s | 9.6e-06 | 0.027 | |
| UAS.ihog-RFP | <2e-16 | <2e-16 | <2e-16 | <2e-16 | <2e-16 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Transgenic construct (Drosophila melanogaster) | UAS.GMA-GFP | Bloor and Kiehart, 2001 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihog-RFP | Callejo et al., 2011 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihog-CFP | Bilioni et al., 2013 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogΔCT-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogΔFN-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogΔIg-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transfgenic construct (Drosophila melanogaster) | UAS.ihogΔFn1-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogΔFn2-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogFn1***-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogFn2**-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogFn2** | Zheng et al., 2010 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihogCT-RFP | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.CD8.Fn1-mCherry | Available in Isabel Guerrero's laboratory, Severo Ochoa Molecular Biology Center, Madrid, Spain | ||
| Transgenic construct (Drosophila melanogaster) | UAS.boi-YFP | Bilioni et al., 2013 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.ihog-RNAi | Vienna Drosophila Resource Center (VDRC), ref: 102602 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.boi-RNAi | Vienna Drosophila Resource Center (VDRC), ref: 108265 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.dlp-RNAi | Vienna Drosophila Resource Center (VDRC), ref: 10299 | ||
| Transgenic construct (Drosophila melanogaster) | UAS.dally-RNAi | Vienna Drosophila Resource Center (VDRC), ref: 14136 | ||
| Genetic reagent (Drosophila melanogaster) | hs-FLP | Golic and Lindquist, 1989 | ||
| Genetic reagent (Drosophila melanogaster) | dally32 | Franch-Marro et al., 2005 | ||
| Genetic reagent (Drosophila melanogaster) | dlp20 | Franch-Marro et al., 2005 | ||
| Genetic reagent (Drosophila melanogaster) | ttv524 | Takei et al., 2004 | ||
| Genetic reagent (Drosophila melanogaster) | botv510 | Takei et al., 2004 | ||
| Genetic reagent (Drosophila melanogaster) | boi | Zheng et al., 2010 | ||
| Genetic reagent (Drosophila melanogaster) | ihogZ23 | Zheng et al., 2010 | ||
| Genetic reagent (Drosophila melanogaster) | Bac Hh:GFP | Chen et al., 2017 | ||
| Genetic reagent (Drosophila melanogaster) | Bac ihog:GFP | Hsia et al., 2017 | ||
| Genetic reagent (Drosophila melanogaster) | dally-trap-YFP | Lowe et al., 2014 | ||
| Genetic reagent (Drosophila melanogaster) | hh.Gal4 | Tanimoto et al., 2000 | ||
| Genetic reagent (Drosophila melanogaster) | ptc.Gal4 | Hinz et al., 1994 | ||
| Genetic reagent (Drosophila melanogaster) | ap.Gal4 | Calleja et al., 1996 | ||
| Genetic reagent (Drosophila melanogaster) | ap.LexA | Bloomington Stock Center ref:54268 | ||
| Genetic reagent (Drosophila melanogaster) | LexO.TubGal80 | Bloomington Stock Center ref:32217 | ||
| Genetic reagent (Drosophila melanogaster) | act>y+>Gal4 | Pignoni and Zipursky, 1997 | ||
| Antibody | Mouse monoclonal α-Ptc | Capdevila and Guerrero, 1994 | 1:30 | |
| Antibody | Mouse monoclonal α-Dlp | Lum et al., 2003b | 1:30 | |
| Antibody | Rabbit polyclonal α-GFP | Molecular Probes | 1:1000 | |
| Antibody | Rabbit polyclonal α-Ihog | Bilioni et al., 2013 | 1:100 | |
| Antibody | Mouse monoclonal α-En | Patel et al., 1989 | 1:100 | |
| Antibody | Rabbit polyclonal α-Boi | Bilioni et al., 2013 | 1:30 | |
| Antibody | Rat monoclonal α-Ci | Motzny and Holmgren, 1995 | 1:20 | |
| Antibody | Rabbit polyclonal α-RFP | Chromoteck | 1:5000 WB | |
| Antibody | 680RD fluorescent α-rabbit | Li-Cor | 1:10,000 WB |
Statistical analysis of the lifetime for different Ihog levels.
p-values obtained from Wilcoxon rank sum test to statistically compare cytoneme lifetimes for different levels of Ihog (n.s, not significant, in gray; significant with the corresponding p-value in scientific notation, in orange).
| Wilcoxon rank sum test | UAS.GMA-GFP | UAS.RNAi-boi UAS.RNAi-ihog |
|---|---|---|
| UAS.boi-YFP | n.s | n.s |
| UAS.GMA-GFP | 5.00E-02 | |
| UAS.RNAi-boi;UAS.RNAi-ihog |
Statistical analysis of the lifetime for different Ihog mutants.
p-values obtained from Wilcoxon rank sum test to statistically compare cytoneme lifetimes for different Ihog mutants with the control (UAS.GMA-GFP) (gray: n.s = not significant; orange: significant, with the corresponding p-value in scientific notation).
| Wilcoxon rank sum test | UAS.ihogCt-RFP | UAS.ihogDIg-RFP | UAS.ihogDFN-RFP | UAS.ihogDFN1-RFP | UAS.ihogDFN2-RFP | UAS.GMA-GFP |
|---|---|---|---|---|---|---|
| UAS.ihogCt-RFP | n.s | 7.23E-06 | n.s | n.s | n.s | |
| UAS.ihogDIg-RFP | 6.06E-04 | n.s | n.s | n.s | ||
| UAS.ihogDFN-RFP | 9.30E-04 | 2.05E-05 | ||||
| UAS.ihogDFN1-RFP | n.s | n.s | ||||
| UAS.ihogDFN2-RFP | n.s | |||||
| UAS.GMA-GFP |
Additional files
-
Source code 1
R-Studio code of the statistical analysis of the Dally, Dlp, and Hh recruitment under the expression of different Ihog mutant constructs.
The normality of the data was tested employing Shapiro test, and the differences between means was done using the T-test for normal data and the Wilcox test for non-normal data.
- https://cdn.elifesciences.org/articles/64581/elife-64581-code1-v2.pdf.zip
-
Source code 2
Matlab script was designed to organize the data, to compute the statistical analysis, and to represent the results in different violin plots.
For genotypes showing only dynamic cytonemes, the statistical analysis was done using their numerical lifetimes. To examine the normality of the data distribution, we performed a Shapiro–Wilk test. Since the results showed a non-parametric distribution of the experimental data, we selected a Wilcoxon rank sum test to compare the numerical lifetimes between two genotypes. For genotypes showing both static and non-static cytonemes we defined a no numerical case to quantify the frequency of static cytonemes. As a result, we obtained ‘mixed’ violin plots representing the distribution of the lifetime of the whole cytoneme population.
- https://cdn.elifesciences.org/articles/64581/elife-64581-code2-v2.pdf.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/64581/elife-64581-transrepform-v2.docx






