Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record updated
- Version of Record published
- Accepted Manuscript published
- Accepted
- Received
Decision letter
-
Hao YuReviewing Editor; National University of Singapore & Temasek Life Sciences Laboratory, Singapore
-
Detlef WeigelSenior Editor; Max Planck Institute for Developmental Biology, Germany
-
Chae EunyoungReviewer
-
Blake C MeyersReviewer; Donald Danforth Plant Science Center, United States
Our editorial process produces two outputs: i) public reviews designed to be posted alongside the preprint for the benefit of readers; ii) feedback on the manuscript for the authors, including requests for revisions, shown below. We also include an acceptance summary that explains what the editors found interesting or important about the work.
Acceptance summary:
In this study, the authors examined the function of the RNA-binding protein FPA through analyzing its protein interactome and its global impact on gene expression using a combined approaches of Nanopore DRS, Helicos DRS, and short-read Illumina RNA-Seq. The combined datasets and new computational approaches developed by the authors showed a predominant role of FPA in promoting poly(A) site choice. The authors further revealed that FPA mediates widespread premature cleavage and polyadenylation of the transcripts of NLR genes, which act as important plant immune regulators. Overall, this study suggests that control of transcription termination processes mediated by FPA provides an additional layer of the regulatory dynamics of NLRs in plant immune responses.
Decision letter after peer review:
Thank you for submitting your article "Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA" for consideration by eLife. Your article has been reviewed by 3 peer reviewers, and the evaluation has been overseen by Hao Yu as the Reviewing Editor and Detlef Weigel as the Senior Editor. The following individuals involved in review of your submission have agreed to reveal their identity: Chae Eunyoung (Reviewer #1); Blake C Meyers (Reviewer #3).
Essential Revisions:
While there was agreement that the topic is timely and the findings relevant, there were some concerns regarding manuscript structure, inconsistency among some results, and interpretation of biological relevance of the data, as listed below, that need to be addressed to support the conclusions.
1. The manuscript presents an extensive body of studies in analyzing FPA interacting proteins and its potential RNA targets including NLRs. Although the overall results cover a series of observations, many of them are descriptive and divert the audience's attention from understanding the novelty and significance of the findings. Thus, we suggest that the authors re-organize the manuscript into a more coherent story and focus on the most important data pertaining to NLR control as shown in the title and the abstract.
2. The authors should address or explain some inconsistencies in the results as mentioned by reviewers. For example, the authors found that at least for some pathogens, "loss of FPA function does not reduce plant resistance". This is not consistent with the hypothesis that FPA is important for regulating NLR immune response genes, and the observation that premature exonic termination of RPP7 caused by FPA has a functional consequence for Arabidopsis immunity against Hpa-Hiks1.
3. The significance of this study will be strengthened by analysis of the biological relevancy of the alternative polyadenylation events mediated by FPA pertaining to NLR functions. We suggest that the authors consider either providing new experimental data or clearly interpreting existing results, such as those relevant to regulation of RPP7, to provide better insights into biological significance of the data presented in this manuscript.
Please also take into consideration the other specific comments from the reviewers below to revise the manuscript.
Reviewer #1 (Recommendations for the authors (required)):
Comments on reference, figure/legend and typesets
1. The reference style shall be uniform. Currently, there seem to be two different formats with two author presentation in the parenthesis earlier in the document and with one author later in the document (e.g. Takagi, Iwamoto et al., 2020 vs Takagi et al., 2020). In addition, there are several articles missing full citation information (e.g. Parker MT et al., 2020a (not found by search, supposedly hen2-2 data according to main text), 2020b; Nat Communications citation format to be checked).
2. L844: Barragan et al., 2020 MBE is a good one to add as this work identifies a truncated NLR as the culprit of autoimmunity in an A. thaliana hybrid.
3. L846: One of highly appreciated works on TIR-only protein is by Nishimura et al., 2017 PNAS 114 (10): E2053-62 on RBA1, which should be cited.
4. L919: RPP7 locus spawning recurrent risk alleles was exemplified with later cases reported by Chae et al., 2014 (Cell) and Barragan et al., 2019 (PLoS Genetics). A proper term for RPP7 is "NLR genes at the RPP7 locus" as to consider allelic diversity. L924: hybrid "necrosis" should be used in this case instead of weakness.
5. It will be of interest how many of NLRs with alternative 3'-end processing detected in this study reside in a multi-gene cluster. As tandem repeats of highly similar genes tend to create problems in informatic analysis, this notion shall be carefully visited. Not only the technical side but also related to discussion on evolutionary dynamics, this notion can be related to the authors' proposition of cryptic transcript variation affecting evolutionary dynamics.
6. In all figures, the proportion of the panel pointer text (e.g. A, B) and actual text shall be modified. As compared to the panel pointer, actual texts seem disproportionately small and sometimes hard to read.
7. In the insets in Figures 6 and 8, there are transcripts arrow-pointed to indicate alternative 3'UTR or non-stop transcripts. I believe those are also present in other genotypes, most importantly in Col-0. Please be advised to point all the affected transcripts instead of pointing the ones in mutant genotypes. Using asterisk heads would be an option.
8. AGI locus identifier (At2gXXXXX) is conventionally not italicized. The authors may check with the journal for the final typesetting.
9. Data and code availability shall be updated on the XXX marked areas.
10. L73-74: italicize the gene name in full and abbreviation, as was done for RPP7 in L78-79.
11. L460: add hyphen (-) between nucleotide and binding.
12. L460: the wording should be genes encoding NB-ARC, Rx-like CC, OR LRR. Otherwise, it indicates genes encoding the three domains all together, which is not the data presented in Figure 5.
13. Figure 5 A, B: what is the difference between Rx-like CC and Rx N-terminal? If the HMM analysis picked up ZAR1 CC (latest structure of CNL), using ZAR1 will attract a broad audience in the NLR field.
14. L470: CCR shall be properly abbreviated or replaced with RPW8-like CC.
15. L474: indicate how many out of 206 NLRs were reannotated instead of using "some". Suggested table would help (see above comments).
16. Figure 5-Sup1: fix the typos in AGI identifiers in the bolded faced title (typos in both).
17. L883: meant for trials or trailing?
Reviewer #2 (Recommendations for the authors (required)):
This is an interesting piece of work. However, there are some essential data and analyses required to support the conclusions.
1. If the authors would like to tie the story with NLR regulation, the physiological functions of FPA in relation to plant defense response should be shown. Right now the disease resistance-related phenotypes of the loss of function mutant fpa-8 and the overexpressor 35S::FPA:YFP are quite weak.
2. The authors should clarify whether there could be additional mutations in fpa-8 as they have suggested.
3. For the MS experiments, a YFP-only control is essential to reduce the noise due to false positives. Validation of interactions to selected key target interacting partners are important to confirm the accuracy of the findings.
4. If the authors really want to discuss the functional relationship between FPA and IBM, IBM2 could be more relevant in this study.
5. The authors should provide a better explanation/model for the observation that a quantitative shift toward the selection of distal poly(A) sites in the loss-of-function fpa-8 mutant and a strong shift to proximal poly(A) site selection when FPA is overexpressed (35S::FPA:YFP) in some cases (Figure 3, Figure 5, Figure 8). But the situation could be kind of reversed in other cases (Figure 6).
[Editors' note: further revisions were suggested prior to acceptance, as described below.]
Thank you for resubmitting your work entitled "Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA" for further consideration by eLife. Your revised article has been evaluated by Detlef Weigel (Senior Editor), Hao Yu (Reviewing Editor) and three reviewers who reviewed the last version of the manuscript.
We feel that this revised manuscript has been significantly improved, but there are some remaining issues that need to be addressed, as indicated in the following comments given by Reviewers 1 and 2.
Reviewer #1 (Recommendations for the authors):
The authors made great efforts to reorganize the manuscript to address comments from all three reviewers. Current manuscript supports the main claim on FPA modulating the NLR regulation with a series of graphic illustration as main figures with supporting supplements. These encompass the breadth of regulatory roles of FPA on different NLR genes, in particular. Their quantitative assessment of the FPA effects on clustered or hypervariable NLR genes have been performed in a sound way, taking on the latest research outcomes (2020-2021 publications) on NLR diversity and evolution.
Reviewer #2 (Recommendations for the authors):
Overall, it is a piece of interesting research supported with rich data. The authors have addressed much of the concerns in the revised version and through further explanations. Some remaining questions could be addressed via clarification, strengthened comparison, and additional discussions.
1. In relation to my original Question 1. Since the title of this manuscript is "Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA" and some NLR gene expressions are responsive to pathogen attack, the readers may be interested to know the changes in NLR genes under pathogen attack conditions that are regulated by FPA. If the authors have these data, it will be great to share.
2. In relation to my original Question 2 and Question 5. Since overexpression of FPA only partially reduces the level of functional RPP7 transcripts, is it possible that FPA overexpression also acts on other NLR transcripts that leading to loss of resistance?
3. In relation to my original Question 4. Is it possible to make a comparison directly between the 35S::FPA:YFP line versus the fpa-8 mutant to investigate see whether all disappeared pre-mature transcriptional terminations have returned to the level of Col-0 or even more?
4. In relation to my original Question 6. The authors showed that overexpression FPA will decrease the overall FLC transcripts. Is the FPA acting on the pre-mature transcriptional termination of FLC too? Any data to support this?
5. In relation to my original Question 7. Does the anti-FPA chip data match well with the proximal APA in Col-0?
6. In relation to my original Question 9 and Question 10. IBM1 is a common target of FPA and EDM2, indicating the possible coordination of the FPA and EDM2 functions. There have been several studies on EDM2, could the authors compare the target of FPA and EDM2, and also address whether FPA also targets TEs in introns of function genes similar to that of EDM2?
Reviewer #3 (Recommendations for the authors):
I am satisfied with the authors' response to the reviewers, including the valuable points raised by the other reviewers. The extensive changes that the authors made to the manuscript have substantially improved the work.
https://doi.org/10.7554/eLife.65537.sa1Author response
Essential Revisions:
While there was agreement that the topic is timely and the findings relevant, there were some concerns regarding manuscript structure, inconsistency among some results, and interpretation of biological relevance of the data, as listed below, that need to be addressed to support the conclusions.
1. The manuscript presents an extensive body of studies in analyzing FPA interacting proteins and its potential RNA targets including NLRs. Although the overall results cover a series of observations, many of them are descriptive and divert the audience's attention from understanding the novelty and significance of the findings. Thus, we suggest that the authors re-organize the manuscript into a more coherent story and focus on the most important data pertaining to NLR control as shown in the title and the abstract.
We have restructured the manuscript to address this concern. Following the specific comments of the reviewers we have:
– Carried out a major-re-write of the Results section.
– Reduced the detailed descriptions in the proteomics and Illumina RNA-seq sections, so as to reach the NLR focus of the study more quickly.
– Created a new main text Figure explaining RNA processing around RPS6.
– Summarized the different FPA-dependent effects on NLRs into 3 new tables to provide a broader view that complements the examples detailed in the text and the detailed data provided as supplementary materials.
– We have removed non-NLR examples of FPA regulation from the manuscript, to focus on the NLR aspect.
– We have changed the text-based descriptions of FPA-NLR control, to rationalize the logic and focus of illustrated examples.
– We have moved the Figure on IBM1 from the main text to a Supplementary Figure.
– We have edited the section on RPP7-dependent pathogen testing, to clarify apparent misunderstanding.
2. The authors should address or explain some inconsistencies in the results as mentioned by reviewers. For example, the authors found that at least for some pathogens, "loss of FPA function does not reduce plant resistance". This is not consistent with the hypothesis that FPA is important for regulating NLR immune response genes, and the observation that premature exonic termination of RPP7 caused by FPA has a functional consequence for Arabidopsis immunity against Hpa-Hiks1.
There is a straightforward misunderstanding here, possibly because our text in the relevant section was not sufficiently clear.
We tested the impact of different activity levels of Arabidopsis FPA on NLR function by investigating the NLR, RPP7. We chose RPP7 because features of its function and regulation are relatively well characterised. RPP7 provides disease resistance to the oomycete pathogen Hyaloperonospora arbidopsidis (Hpa) strain Hiks1. The reference Arabidopsis accession, Col-0, encodes a functional RPP7 gene and hence is resistant to Hpa-Hiks1 infection. Not all Arabidopsis accessions are resistant to all Hpa strains. For example, the Duc-1 and Ksk-1 accessions have been reported as having susceptibility to Hpa-Hiks1 infection, likely due to the lack of a functional RPP7 gene (Lai et al., 2019). It was for this reason that we incorporated the Ksk accession as an infectionsensitive positive control accession in our pathogen tests.
The question we were addressing was: Does FPA-dependent premature cleavage and polyadenylation in RPP7 exon 6 compromise RPP7 function? To address this question, we therefore applied Hpa-Hiks to our different genetic lines. Neither Col-0 nor the fpa-8 mutant (which is in the Col-0 genetic background) were sensitive to infection. This is consistent with our hypothesis because the poly(A) site used in exon 6 in Col-0, is used significantly less in fpa-8. Hence, there is no compromise in the expression of full-length RPP7 in fpa-8 mutants. As Col-0 is already resistant to Hpa-Hiks1, we would therefore expect fpa-8 to also be resistant and indeed, this is what we found.
This was also true when we tested an independent allele, fpa-7, that is also in the Col-0 background. However, when we tested the line that was over-expressing FPA, which was introduced into an fpa-8 background (and hence, ultimately Col-0), we found that resistance was lost and Hpa-Hiks1 was able to infect these plants.
Therefore, the findings from this experiment are completely consistent “with the hypothesis that FPA is important for regulating NLR immune response genes, and the observation that premature exonic termination of RPP7 caused by FPA has a functional consequence for Arabidopsis immunity against Hpa-Hiks1.” We have clarified the text in this section to make our hypothesis and findings clearer.
3. The significance of this study will be strengthened by analysis of the biological relevancy of the alternative polyadenylation events mediated by FPA pertaining to NLR functions. We suggest that the authors consider either providing new experimental data or clearly interpreting existing results, such as those relevant to regulation of RPP7, to provide better insights into biological significance of the data presented in this manuscript.
To more clearly address the predicted consequences of FPA regulated alternative polyadenylation, we have added tables for the three classes of FPA-regulated alternative polyadenylation events to the main text and made predictions of the functional consequences for intronic and exonic polyadenylation events. Since there are several documented examples of TIR-only or LRR-truncated NLRs regulating resistance and cell death, we have also changed some of the example genes used to focus on those proximal polyadenylation events which can alter protein coding potential.
Reviewer #1 (Recommendations for the authors (required)):
Comments on reference, figure/legend and typesets
1. The reference style shall be uniform. Currently, there seem to be two different formats with two author presentation in the parenthesis earlier in the document and with one author later in the document (e.g. Takagi, Iwamoto et al., 2020 vs Takagi et al., 2020). In addition, there are several articles missing full citation information (e.g. Parker MT et al., 2020a (not found by search, supposedly hen2-2 data according to main text), 2020b; Nat Communications citation format to be checked).
We have addressed referencing issues throughout the document. Parker MT et al., 2020a referenced a preprint which is now published in Genome Biology, the citation has been updated accordingly.
2. L844: Barragan et al., 2020 MBE is a good one to add as this work identifies a truncated NLR as the culprit of autoimmunity in an A. thaliana hybrid.
Indeed, this is a very interesting article. We have found that FPA regulates DM10 (the truncated NLR causing hybrid necrosis in the paper), resulting in transcripts predicted to encode a protein with fewer LRR repeats. This truncation is intriguingly similar to the predicted product of the truncated DM10 allele and raises the question of whether over-expression of FPA or other RNA-binding proteins could cause hybrid necrosis in some Arabidopsis ecotypes. We have incorporated this idea, and the reference to the paper, into the Discussion.
3. L846: One of highly appreciated works on TIR-only protein is by Nishimura et al., 2017 PNAS 114 (10): E2053-62 on RBA1, which should be cited.
We thank the reviewer for this suggestion and after reading it we have incorporated reference to it in the Discussion.
4. L919: RPP7 locus spawning recurrent risk alleles was exemplified with later cases reported by Chae et al., 2014 (Cell) and Barragan et al., 2019 (PLoS Genetics). A proper term for RPP7 is "NLR genes at the RPP7 locus" as to consider allelic diversity. L924: hybrid "necrosis" should be used in this case instead of weakness.
We thank the reviewer for their insight. We have modified the text in this section to refer to RPP7 risk alleles as “Alleles of RPP7-like NLR genes”.
5. It will be of interest how many of NLRs with alternative 3'-end processing detected in this study reside in a multi-gene cluster. As tandem repeats of highly similar genes tend to create problems in informatic analysis, this notion shall be carefully visited. Not only the technical side but also related to discussion on evolutionary dynamics, this notion can be related to the authors' proposition of cryptic transcript variation affecting evolutionary dynamics.
We thank the reviewer for this suggestion. To test whether genes with FPA regulated 3’-processing are specifically located in multi-gene clusters, we used the major/minor cluster and singleton classifications of NLRs from the supplemental information of a recent study (Lee and Chae, 2020). We found 44 of the 74 expressed NLRs which are located in major clusters are regulated by FPA-dependent alternative polyadenylation, which is more than is expected by chance. More recently, protein-level sequence variation data has been used to identify rate of evolution of individual NLR genes (Prigozhin and Krasileva, 2021). The authors find that 30 of the NLRs present in Col-0 are under high levels of diversifying selection and indicate that this variability is due to the role of these NLRs in direct recognition of effectors. Hypervariable NLRs come with a risk of autoimmunity, however, and so might be expected to be controlled at the level of full-length RNA expression. We therefore looked to see whether hyper variable NLRs (hvNLRs) are enriched amongst those genes regulated by FPA. This analysis revealed that 20 of the 28 expressed Col-0 hvNLRs are regulated by FPA-dependent alternative polyadenylation, indicating that FPA regulation may be associated with rapidly evolving NLR genes. Finally, we used the synteny diversity scores generated from 7 chromosome-scale Arabidopsis assemblies to assess whether FPA-regulated NLRs are more likely to be found in regions which are hotspots for rearrangements. We show that FPA-regulated NLRs are more likely to be in regions with higher levels of synteny diversity (Figure 4—figure supplement 1), although this relationship was not significant. We have included these new results in the first paragraph of the section entitled “Widespread premature transcription termination of NLRs…”.
6. In all figures, the proportion of the panel pointer text (e.g. A, B) and actual text shall be modified. As compared to the panel pointer, actual texts seem disproportionately small and sometimes hard to read.
We have changed the size of the panel pointer text to 16pt (14pt for inset pointers). All other figure text is uniformly 8pt.
7. In the insets in Figures 6 and 8, there are transcripts arrow-pointed to indicate alternative 3'UTR or non-stop transcripts. I believe those are also present in other genotypes, most importantly in Col-0. Please be advised to point all the affected transcripts instead of pointing the ones in mutant genotypes. Using asterisk heads would be an option.
We thank the reviewer for this suggestion. We have revised the figure to use asterisks to denote non-stop/alternative 3’UTR transcripts on all affected genotypes in the relevant Insets.
8. AGI locus identifier (At2gXXXXX) is conventionally not italicized. The authors may check with the journal for the final typesetting.
For genes which have been named and AGI identifiers are provided additionally in brackets, we have removed italicisation. For genes which have not yet been named, we consider the AGI identifier to be the gene/protein name, and so we have italicised these to denote that they refer to the gene and not the protein.
9. Data and code availability shall be updated on the XXX marked areas.
All data and code is available from ProteomeXchange, ENA, Zenodo, or GitHub, and is declared correctly in the Code and Data Availability sections.
10. L73-74: italicize the gene name in full and abbreviation, as was done for RPP7 in L78-79.
We have rephrased this section to make it clear that we are referring to the RPS4 gene (not protein), and have added italics.
11. L460: add hyphen (-) between nucleotide and binding.
Hyphens have been added between nucleotide and binding
12. L460: the wording should be genes encoding NB-ARC, Rx-like CC, OR LRR. Otherwise, it indicates genes encoding the three domains all together, which is not the data presented in Figure 5.
We have changed the wording to “and/or” to address the reviewer’s suggestion.
13. Figure 5 A, B: what is the difference between Rx-like CC and Rx N-terminal? If the HMM analysis picked up ZAR1 CC (latest structure of CNL), using ZAR1 will attract a broad audience in the NLR field.
Rx-like CC (IPR038005) is an Interpro entry derived from the Conserved Domains database (CDD) entry cd14798, whilst Rx N-terminal (IPR041118) is an Interpro entry derived from the Pfam database entry PF18052. Both are meant to identify the same domain architectures but there may be differences in the specific sequences that each model detects.
14. L470: CCR shall be properly abbreviated or replaced with RPW8-like CC.
We now refer to CCR domains simply as RPW8 domains.
15. L474: indicate how many out of 206 NLRs were reannotated instead of using "some". Suggested table would help (see above comments).
We have removed this ambiguous statement, since we have not carried out a thorough reannotation of all NLR genes using our data.
16. Figure 5-Sup1: fix the typos in AGI identifiers in the bolded faced title (typos in both).
We have corrected the typo in the figure legend, which is now Figure 3—figure supplement 1.
17. L883: meant for trials or trailing?
This sentence has been removed during the rewrite of the Discussion section.
Reviewer #2 (Recommendations for the authors (required)):
This is an interesting piece of work. However, there are some essential data and analyses required to support the conclusions.
1. If the authors would like to tie the story with NLR regulation, the physiological functions of FPA in relation to plant defense response should be shown. Right now the disease resistance-related phenotypes of the loss of function mutant fpa-8 and the overexpressor 35S::FPA:YFP are quite weak.
We have shown that overexpression of FPA has a functional consequence for immunity using the case study of RPP7 expression and resistance to Hpa-hiks1. In future we would like to explore whether loss or overexpression of FPA might alter the resistance of Arabidopsis (or other species) to other pathogens, however these experiments are beyond the scope of the current work.
2. The authors should clarify whether there could be additional mutations in fpa-8 as they have suggested.
There is a possibility that this is the case and we have changed the language of the relevant section on pathogen susceptibility to state this as clearly as possible. It is important to emphasise that most, if not all mutants generated using EMS, T-DNA, or other mutagens, are likely to have additional mutations besides those that are causative for the observed phenotypes, and not all can be removed by back crossing (if the mutants are closely linked, for example). The controls we performed in the pathogen susceptibility experiment safeguard our conclusions against this issue: fpa-7 does not have the same variability in resistance as fpa-8, indicating that loss of FPA function is not the cause of variability; this is also demonstrated by the inability of pFPA::FPA to rescue variability. 35S::FPA:YFP has significantly higher susceptibility than Col-0, fpa-7, fpa-8, and pFPA::FPA, indicating that overexpression of FPA causes a reduction in resistance to Hpa-hiks1.
3. For the MS experiments, a YFP-only control is essential to reduce the noise due to false positives. Validation of interactions to selected key target interacting partners are important to confirm the accuracy of the findings.
This issue is addressed in our response to point 7 of reviewer 3’s public review.
4. If the authors really want to discuss the functional relationship between FPA and IBM, IBM2 could be more relevant in this study.
This issue is addressed in our response to point 10 of reviewer 3’s public review.
5. The authors should provide a better explanation/model for the observation that a quantitative shift toward the selection of distal poly(A) sites in the loss-of-function fpa-8 mutant and a strong shift to proximal poly(A) site selection when FPA is overexpressed (35S::FPA:YFP) in some cases (Figure 3, Figure 5, Figure 8). But the situation could be kind of reversed in other cases (Figure 6).
This issue is addressed in our response to point 4 of reviewer 3’s public review.
[Editors' note: further revisions were suggested prior to acceptance, as described below.]
Reviewer #2 (Recommendations for the authors):
Overall, it is a piece of interesting research supported with rich data. The authors have addressed much of the concerns in the revised version and through further explanations. Some remaining questions could be addressed via clarification, strengthened comparison, and additional discussions.
We thank the reviewer for these remarks. We have addressed their questions below.
1. In relation to my original Question 1. Since the title of this manuscript is "Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA" and some NLR gene expressions are responsive to pathogen attack, the readers may be interested to know the changes in NLR genes under pathogen attack conditions that are regulated by FPA. If the authors have these data, it will be great to share.
The question of whether FPA (or other RNA binding proteins) alter the 3’ processing of NLR transcripts during infection is something that we would like to explore. Whilst some microarray and RNA sequencing datasets collected during infection conditions are available, these are generally designed for analysing expression changes at the gene level. As a result, they are underpowered for the analysis of RNA processing, which generally requires higher sequencing depth and longer reads – for example 50-100 million 2´150bp reads per replicate, with 6 or more biological replicates, as was used in our Illumina experiment. As far as we are aware, no experiments using nanopore direct RNA sequencing to identify RNA processing changes during infection have yet been generated or published. It is clear that the absence of detailed analyses of NLR transcript processing and fate during pathogen infection represents a gap in understanding for the immunity field as a whole, and so generating these data is a goal for our future enquiries.
2. In relation to my original Question 2 and Question 5. Since overexpression of FPA only partially reduces the level of functional RPP7 transcripts, is it possible that FPA overexpression also acts on other NLR transcripts that leading to loss of resistance?
We cannot rule out the possibility that FPA-dependent proximal polyadenylation at other NLR loci besides RPP7 may contribute quantitatively to the loss of immunity to Hpa-hiks1 seen in the 35S::FPA:YFP line. We have therefore reworded our conclusions in the relevant Results section. We now state: “We conclude that FPA control of poly(A) site selection can modulate NLR function, with a functional consequence for immunity.”
3. In relation to my original Question 4. Is it possible to make a comparison directly between the 35S::FPA:YFP line versus the fpa-8 mutant to investigate see whether all disappeared pre-mature transcriptional terminations have returned to the level of Col-0 or even more?
We compared fpa-8 and 35S::FPA:YFP nanopore DRS data directly using the Earth mover distance (EMD) method, as suggested by the reviewer. We found that 80.0% of the loci with significantly increased distal poly(A) site choice in fpa-8 when compared with Col-0, were also significant when compared to 35S::FPA:YFP (hypergeometric p = 2.2´10-172). This indicates that the 35S::FPA:YFP transgene is able to reverse the readthrough at these loci displayed in fpa-8. Furthermore, 77.2% of loci with significantly increased proximal poly(A) site choice in 35S::FPA:YFP when compared to Col0, were also significant when compared to fpa-8 (hypergeometric p = 1.9´10-119). Of the loci with altered poly(A) site choice when comparing 35S::FPA:YFP to either Col-0 or fpa-8, 85.9% had a larger EMD when compared to fpa-8, suggesting that there are reciprocal changes beyond Col-0 levels in 35S::FPA:YFP and fpa-8 at these loci.
4. In relation to my original Question 6. The authors showed that overexpression FPA will decrease the overall FLC transcripts. Is the FPA acting on the pre-mature transcriptional termination of FLC too? Any data to support this?
There is no evidence that sense FLC transcripts are targeted by FPA-dependent proximal polyadenylation (Duc et al., 2013; Hornyik et al., 2010). Instead, there is a significant body of literature on the role of FPA and other 3’ processing factors influencing long non-coding antisense RNAs at the FLC locus (Hornyik et al., 2010). The ratio of proximal to distal antisense RNAs correlates negatively with sense FLC expression (Duc et al., 2013; Hornyik et al., 2010).
5. In relation to my original Question 7. Does the anti-FPA chip data match well with the proximal APA in Col-0?
To test whether there was an overlap between the sites of FPA-associated chromatin and loci with FPA-sensitive poly(A) site choice, we called peaks from the FPA ChIP-seq data using MACS2 (Zhang et al., 2008). This resulted in the identification of 1120 unstranded peaks. We then assigned peaks to loci by identifying the closest (or overlapping) transcribed locus in an upstream orientation to each peak, using bedtools (Quinlan and Hall, 2010). Where there were multiple tied loci that could be assigned to a single peak (e.g. at convergent terminators or overlapping loci), peaks were assigned to all tied loci. We then compared the loci with identified FPA peaks to those loci with FPA-dependent alternative polyadenylation, identified using Nanopore DRS sequencing. We found that of the 222 loci with increased distal polyadenylation in fpa-8, 38 were also associated with an FPA ChIP-seq peak (hypergeometric p = 3.5´10-4). Of the 166 loci with increased proximal polyadenylation in 35S::FPA:YFP, 27 were also associated with an FPA ChIP-seq peak (hypergeometric p = 3.3´10-3). The lack of FPA ChIP-seq peaks on many genes with FPA-dependent RNA processing may be explained by low Pol II occupancy. In agreement with this, loci with FPA-dependent alternative polyadenylation, which did not have an associated FPA peak, were more weakly expressed than those that did have an FPA peak (Mann-Whitney U p=7.2´10-4). Notably, 95.6% of genes which were associated with FPA ChIP-seq peaks did not have FPA-sensitive alternative polyadenylation under our experimental conditions, and global FPA ChIP-seq signal at 3’ ends was well correlated with Pol II Ser2 signal (Spearman’s ρ = 0.67, p < 2´10-308, 95% CIs [0.66, 0.68]). This suggests that FPA is able to associate with terminating Pol II at most loci but is necessary for poly(A) site choice at a relatively smaller number of loci in our experimental conditions. We have added our findings on the correlation of FPA and Pol II ChIP-seq signal to the relevant Results section and updated the Discussion section “Uncovering protein assemblies that mediate 3ʹ end processing in living plant cells”. We now state:
“Such interactions [between FPA and Pol II Ser2] could account for the global correlation between FPA and Pol II Ser2 occupancy and explain how FPA is able to associate with terminating Pol II at the 3’ ends of most expressed genes.”
6. In relation to my original Question 9 and Question 10. IBM1 is a common target of FPA and EDM2, indicating the possible coordination of the FPA and EDM2 functions. There have been several studies on EDM2, could the authors compare the target of FPA and EDM2, and also address whether FPA also targets TEs in introns of function genes similar to that of EDM2?
Previous studies have indicated that FPA and the EDM2/IBM2/AIPP1 complex act antagonistically to regulate the expression of IBM1 (Deremetz et al., 2019). As a result, mutations disrupting FPA were identified in a genetic screen to isolate suppressors of the ibm2 mutation, the phenotype of which is caused by dysregulation of IBM1. This finding is consistent with our previous discovery that FPA controls the proximal polyadenylation of IBM1 (Duc et al., 2013). Other genes regulated by EDM2/IBM2/AIPP1 include RPP7, RPP4, AT3G05410, and AT1G11270 (Lai et al., 2020; Wang et al., 2013). We have shown in this manuscript that FPA-dependent alternative polyadenylation of RPP7 occurs via an independent exon 6 poly(A) site to the one regulated by EDM2 (which is in intron 1). At AT3G05410, loss of EDM2 function causes proximal polyadenylation near the 5’ splice site of intron 3, resulting in complete loss of expression of the distal exon. We find that loss of FPA causes a slight increase in distal polyadenylation of AT3G05410, including at a poly(A) site in a cryptic final exon in intron 3 (Author response image 1A). This may explain the findings of Deremetz et al., who showed that loss of FPA function partially rescued the proximal polyadenylation of AT3G05410 in ibm2 mutants (Deremetz et al., 2019). At AT1G11270, loss of EDM2 function causes proximal polyadenylation near the 5’ splice site of intron 3, also resulting in complete loss of expression of the distal exon (Duan et al., 2017). In comparison, loss of FPA function does not have a strong effect on AT1G11270, though it may cause a slight increase in intronic proximal polyadenylation at a cryptic final exon in intron 3 (Author response image 1B). This suggests that any control of AT1G11270 by FPA occurs via an independent mechanism to the regulation by EDM2/IBM2/AIPP1. Finally, at RPP4, loss of EDM2 function suppresses readthrough into the downstream COPIA retrotransposon (Lai et al., 2020). Similar suppression of readthrough is seen in the fpa-8 mutant (Figure 5) indicating that at RPP4, EDM2 and FPA do not act antagonistically. FPA also controls poly(A) site choice a large number of genes which have no evidence of intronic transposons or intragenic heterochromatin. In addition, FPA predominantly co-purifies with RNA 3’ processing factors rather than histone readers in our proteomics dataset. None of the known interactors of EDM2 and IBM2, which include AIPP1, AIPP2, AIPP3, and CPL2 (Duan et al., 2017), were found associated with FPA. We conclude that regulation of 3’ processing by FPA occurs independently to EDM2 regulated 3’ processing, at a different set of poly(A) sites, and using different mechanisms.
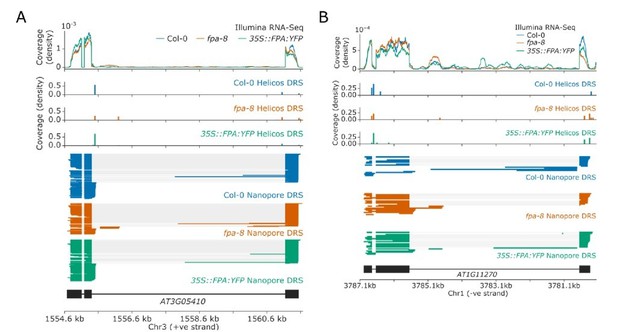
Alternative polyadenylation of genes with intronic heterochromatin in fpa-8 and 35S::FPA:YFP lines.
(A-B) Gene track showing poly(A) site choice of (A) AT3G05410 and (B) AT1G11270 in fpa-8 and 35S::FPA:YFP lines..





