Morphological and genomic shifts in mole-rat ‘queens’ increase fecundity but reduce skeletal integrity
Figures
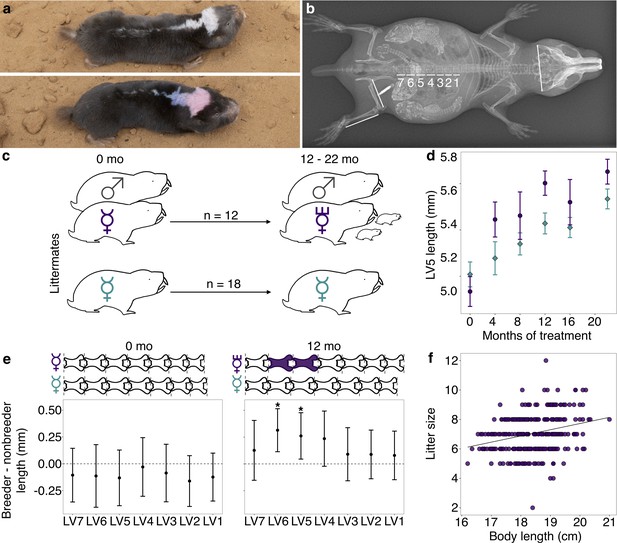
Transition to queen status leads to lumbar vertebral (LV) lengthening.
(a) A breeding ‘queen’ (top) and nonbreeding female (bottom) Damaraland mole-rat, shown at the same scale. Animals are dye marked to allow for individual identification. Photo credit James Bird. (b) An x-ray of a female breeder, with lines depicting the x-ray measurements taken (LV1–LV7, right femur, right tibia, and zygomatic arch). Three developing offspring are visible within the abdomen (highlighted by increased brightness and contrast), and span the length of the LV. (c) Experimental design: nonbreeding adult female ( ) littermates were randomly assigned to transition to queen status (purple
) littermates were randomly assigned to transition to queen status (purple  ) by being paired with an unrelated male (
) by being paired with an unrelated male ( ), or to remain in a nonbreeding treatment (cyan). Duration of treatment ranged from 12 to 22 months. (d) Queens (purple dots) show more rapid growth in LV5 in the first four months of the experiment, relative to nonbreeders (cyan diamonds; treatment by time point interaction: β = 0.078, n = 49, p=3.47×10−3). Dots show means ± standard errors (bars). (e) At the start of the experiment (0 months, left panel), the LV of breeders do not differ from those of nonbreeders (unpaired t-tests, all p>0.05). However, at 12 months (right panel), queens have longer LV relative to nonbreeders (unpaired t-tests, * indicates p<0.05). Dots show means ± standard errors (bars). Lengths of LV above the plots are scaled to indicate the mean lengths of queens (top) and nonbreeders (bottom) at each time point; vertebrae highlighted in purple are significantly longer in queens relative to nonbreeders. (f) Litter size is positively correlated with maternal body length in the Damaraland mole-rat colony (β = 0.353, n = 328 litters, p=1.35×10−3).
), or to remain in a nonbreeding treatment (cyan). Duration of treatment ranged from 12 to 22 months. (d) Queens (purple dots) show more rapid growth in LV5 in the first four months of the experiment, relative to nonbreeders (cyan diamonds; treatment by time point interaction: β = 0.078, n = 49, p=3.47×10−3). Dots show means ± standard errors (bars). (e) At the start of the experiment (0 months, left panel), the LV of breeders do not differ from those of nonbreeders (unpaired t-tests, all p>0.05). However, at 12 months (right panel), queens have longer LV relative to nonbreeders (unpaired t-tests, * indicates p<0.05). Dots show means ± standard errors (bars). Lengths of LV above the plots are scaled to indicate the mean lengths of queens (top) and nonbreeders (bottom) at each time point; vertebrae highlighted in purple are significantly longer in queens relative to nonbreeders. (f) Litter size is positively correlated with maternal body length in the Damaraland mole-rat colony (β = 0.353, n = 328 litters, p=1.35×10−3).
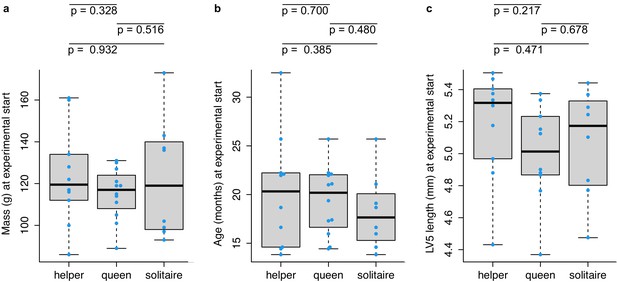
Mass, age, and lumbar vertebra (LV) 5 length of queen, helper, and solitaire female mole-rats at the start of the experiment.
Queen, helper, and solitaire female mole-rats do not differ in (a) mass, (b) age, or (c) lumbar vertebra (LV) 5 length at the start of the experiment. Each box represents the interquartile range, with the median value depicted as a horizontal bar. Whiskers extend to the most extreme values within 1.5× of the interquartile range. Dots represent individual animals. P-values are from unpaired t-tests between each pairwise comparison between helpers (n = 10), queens (n = 12), and solitaires (n = 8). For LV5 length, data were only available for 10 queens. Raw data values are provided in Supplementary file 1.
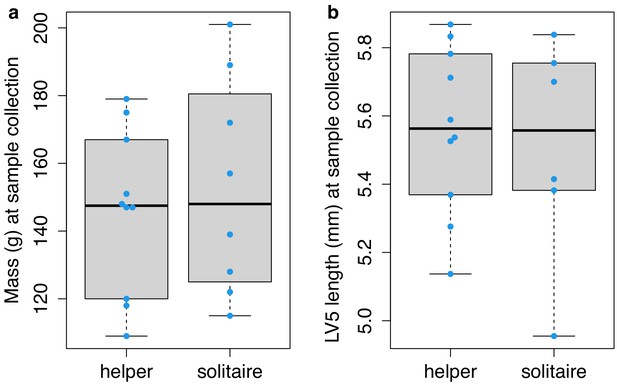
Mass and lumbar vertebra (LV) 5 length of helper and solitaire female mole-rats after 12–22 months of experimental treatment of social status.
Helper and solitaire female mole-rats do not differ in (a) mass (unpaired t-test; t = 0.496, df = 12.733, p=0.629) or (b) LV5 length (unpaired t-test; t = −0.358, df = 8.391, p=0.729) after 12–22 months of experimental treatment of social status. Each box represents the interquartile range, with the median value depicted as a horizontal bar. Whiskers extend to the most extreme values within 1.5× of the interquartile range. Dots represent individual animals. Raw data values are provided in Supplementary file 1.
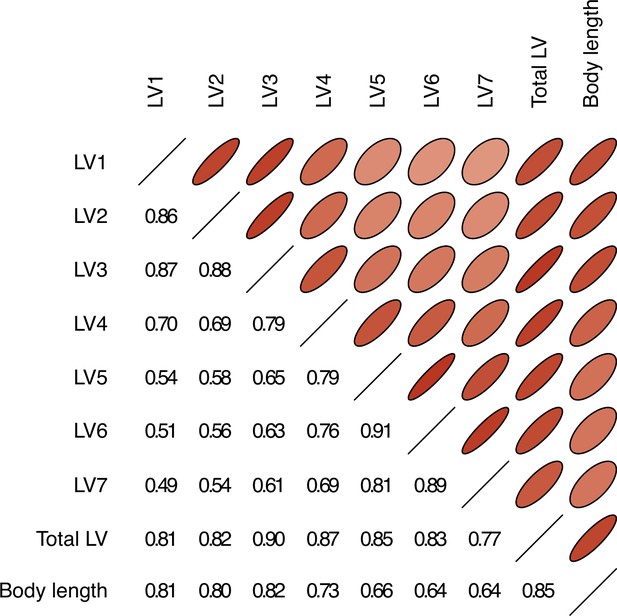
Body length is positively correlated with the lengths of lumbar vertebrae (LV) 1–7.
Pearson correlations between the length of each LV and body length from mole-rat x-ray data. Narrower ovals with darker shades of red indicate larger Pearson correlations; correlation values are also given in the lower left triangle.
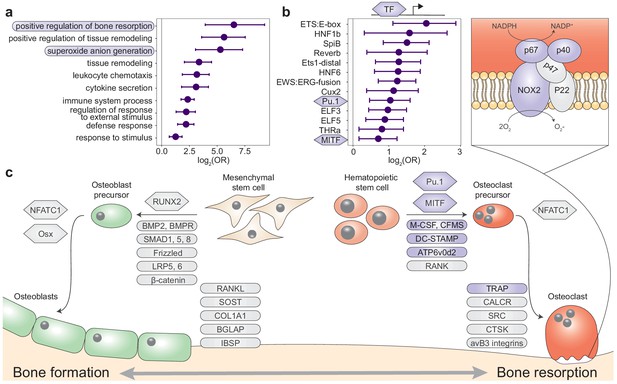
Queen status drives increased regulatory activity of bone resorption pathways.
(a) Gene Ontology (GO) terms enriched in queen upregulated genes, relative to the background set of all genes tested. Bars represent 95% confidence intervals. Processes highlighted in purple are also depicted in (c). Highest-level (most general) terms are shown; for full GO enrichment results, see Supplementary file 7. (b) Accessible transcription factor binding site motifs enriched near queen upregulated genes, relative to all genes tested. Bars represent 95% confidence intervals. Transcription factors highlighted in purple are also depicted in (c). (c) Schematic of the balance between bone formation and bone resorption, showing key regulators and markers for mesenchymal stem cell differentiation into osteoblasts and hematopoietic stem cell differentiation into osteoclasts (Redlich and Smolen, 2012; Segeletz and Hoflack, 2016). Note that not all genes or proteins in gray were tested for differential expression (e.g., because they were not annotated in the Damaraland mole-rat genome or were too lowly expressed in our sample); see Supplementary file 4 for full set of tested genes. Queen upregulated genes or corresponding proteins (false discovery rate [FDR] < 10%) are highlighted as purple ovals, and transcription factors with binding motifs enriched near queen upregulated genes are highlighted as purple hexagons. Inset for osteoclasts shows the NADPH oxidase system, which generates superoxide radicals (O2-) necessary for bone resorption and is highly enriched for queen upregulated genes (purple ovals).
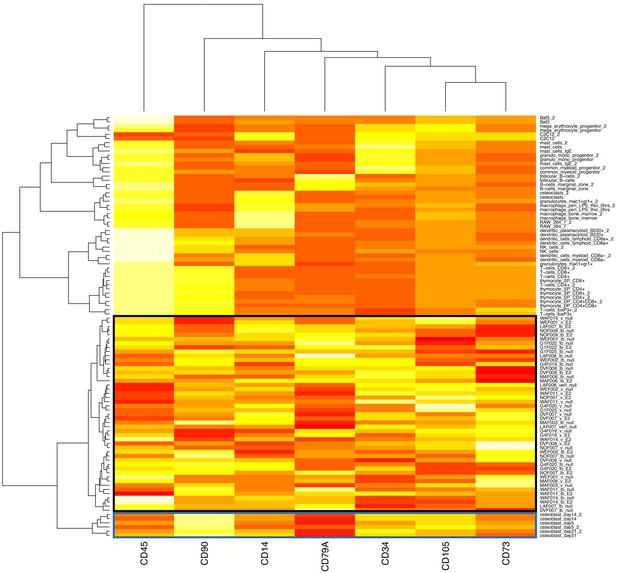
Mole-rat RNA-Seq samples cluster closest with purified mouse osteoblasts, based on canonical bone marrow-derived mesenchymal stromal cell (bMSC) markers.
Clustering was performed using Ward’s hierarchical clustering method on Euclidean distances of the quantile normalized expression of the seven bMSC markers (out of 11 described; Dominici et al., 2006) that were quantified in both the mole-rat and reference mouse (Hume et al., 2010) data sets. The black box indicates mole-rat samples, and the blue box indicates mouse osteoblasts, a bMSC lineage cell type.

Estimated cell proportions for the 47 mole-rat RNA-Seq samples.
Each box represents the interquartile range, with the median value depicted as a horizontal bar. Whiskers extend to the most extreme estimates within 1.5× the interquartile range. Cell proportions were estimated with CIBERSORT (Newman et al., 2015), based on reference gene expression levels for 412 marker genes in 27 purified mouse cell types (Hume et al., 2010). The predicted predominant cell type in the mole-rat samples is most similar to early stage osteoblasts, which are cells from the bone marrow-derived mesenchymal stromal cell lineage.
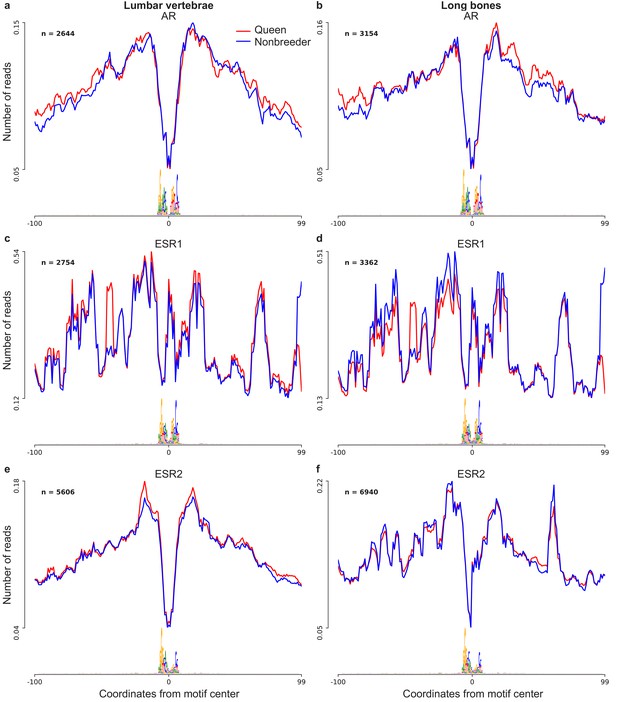
Footprint profiles of transcription factors androgen receptor (AR), estrogen receptor 1 (ESR1), and estrogen receptor 2 (ESR2).
Transcription factor footprints were profiled separately for (a, c, e) lumbar vertebrae (n = 4; two queens and two nonbreeders) and for (b, d, f) long bones (n = 4; two queens and two nonbreeders). Transcription factor activity was not significantly different between queens and nonbreeders in any of the three transcription factors, in either bone type (paired t-tests; all p>0.05).
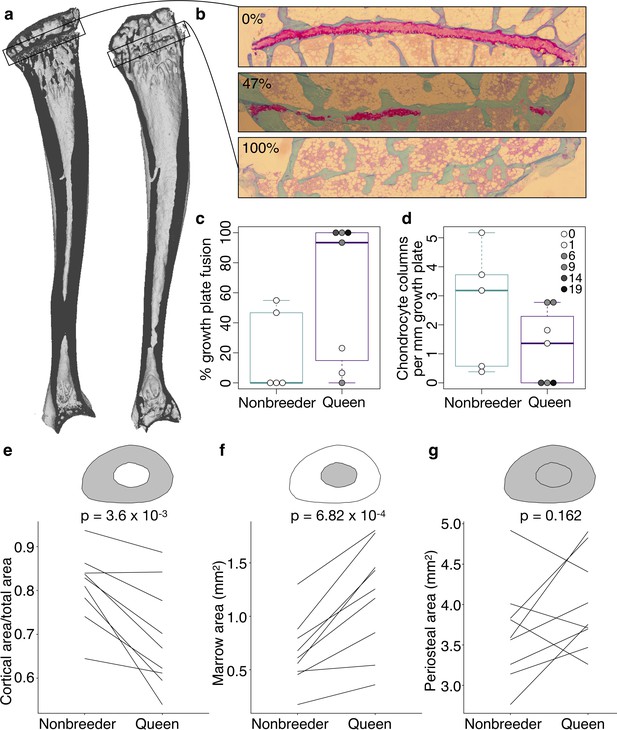
Queen status leads to reduced growth potential in the tibia and reduced cortical area at the femoral midshaft.
(a) Micro-computed tomography (μCT) scans of Damaraland mole-rat tibias. Boxes indicate the location of the proximal growth plate, which varies between unfused (left) to fully fused (right). (b) Example Safranin O-stained histological sections of the proximal tibia, in which the growth plate is unfused (top), partially fused (middle), or fully fused (bottom). Values indicate percent growth plate fusion across the width of the bone. The cartilaginous growth plate is stained deep pink, and calcified bone is stained green. (c) Queens, and specifically queens that gave birth to more offspring, show increased growth plate fusion (β = 0.050, p=4.51×10−3, n = 12, controlling for age) and (d) decreased number of chondrocyte columns within the remaining growth plate (β = −0.132, p=0.020, n = 12, controlling for age). Each box represents the interquartile range, with the median value depicted as a horizontal bar. Whiskers extend to the most extreme values within 1.5× of the interquartile range. In (c) and (d), dots represent individual animals, and shading indicates each animal’s total offspring number. Ages of queens and nonbreeders do not significantly differ (unpaired t-test, t = 0.489, n = 12, p=0.644). (e–g) Femoral cross-sections with area highlighted in gray show the measures represented in the corresponding plots below. Each line represents an age-matched, nonbreeder and queen littermate pair. (e) Queens have less cortical bone (relative to the total area of the femoral midshaft cross-section) compared to their paired nonbreeding littermates (paired t-test, t = −4.07, df = 8, p=3.60×10−3). (f) Queens also have enlarged marrow cavities (paired t-test, t = 5.36, df = 8, p=6.82×10−4) but (g) show no difference in overall periosteal area (paired t-test, t = 1.54, df = 8, p=0.162).

Growth potential in lumbar vertebra (LV) 7.
(a) Micro-computed tomography (μCT) scan of LV7 of a female Damaraland mole-rat. The boxes indicate the locations of the caudal and cranial growth plates. (b-c) The number of offspring produced by queens does not significantly predict (b) growth plate fusion (quantified as the average of the caudal and cranial growth plates; β = 2.745×10−4, p=0.970, n = 12, controlling for age) or (c) chondrocyte proliferation within the remaining growth plate (β = −0.033, p=0.293, n = 12, controlling for age). Each box represents the interquartile range, with the median value depicted as a horizontal bar. Whiskers extend to the most extreme values within 1.5× of the interquartile range. Dots represent individual animals, and shading indicates each animal’s total offspring number.
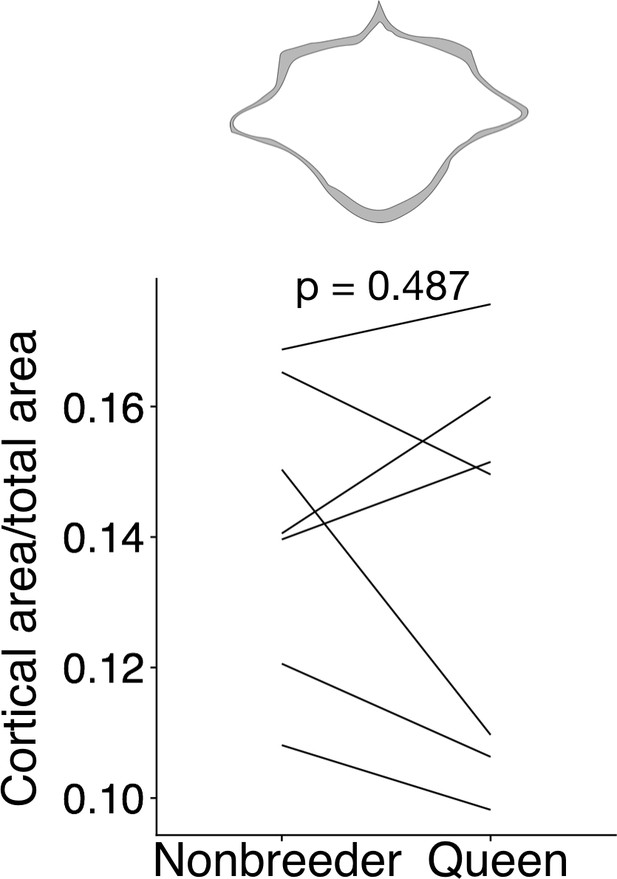
Queens do not exhibit reduced cortical area at the midsection of lumbar vertebra (LV) 6.
At top, cross-section with area highlighted in gray shows the measure represented in the plot. Each line represents an age-matched nonbreeder and queen littermate pair. Queens and nonbreeders show no difference in cortical area/total area at the LV6 midsection (paired t-test of cortical area/total area, t = −0.741, df = 6, p=0.487).
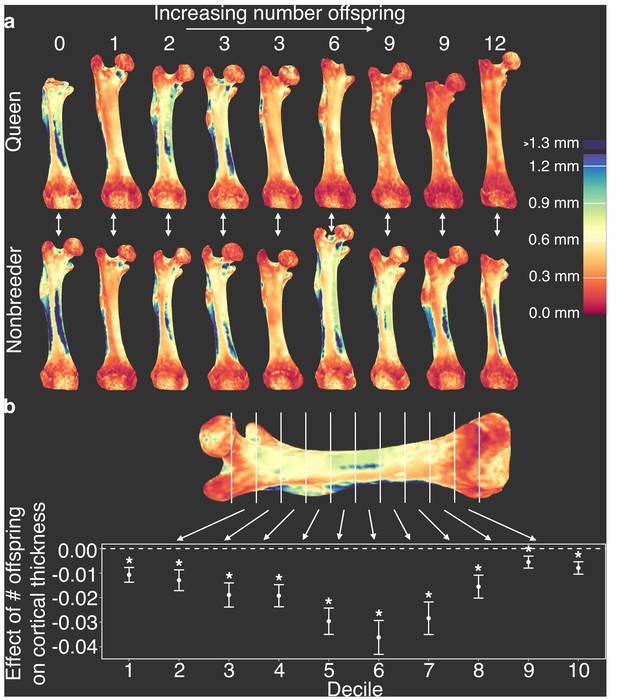
Offspring production in queens leads to cortical thinning across the femoral shaft.
(a) Queens (top row) relative to their same-aged female nonbreeding littermates (bottom row) present thinner cortical bone across the femur, particularly in females that have many offspring (top right). Number of offspring is indicated above each femur, and vertical arrows indicate littermate pairs. (b) Within each decile section across the femoral shaft, number of offspring is negatively correlated with average cortical thickness (linear mixed model with littermate pair as random effect). Full results are presented in Supplementary file 10. Asterisk indicates p<0.05.
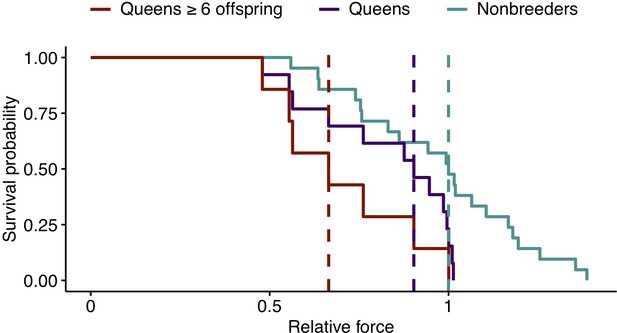
Effect of reproductive status on the probability of bone failure.
Survival curves for femurs from nonbreeders versus queens (Wald test, p=0.02, n = 34) and versus queens with ≥6 offspring (Wald test, p=0.006, n = 28), based on predictions from the midshaft cortical area and data from Jepsen et al., 2003. Vertical dashed lines indicate group medians, with the median failure time for nonbreeders fixed at a value of 1.
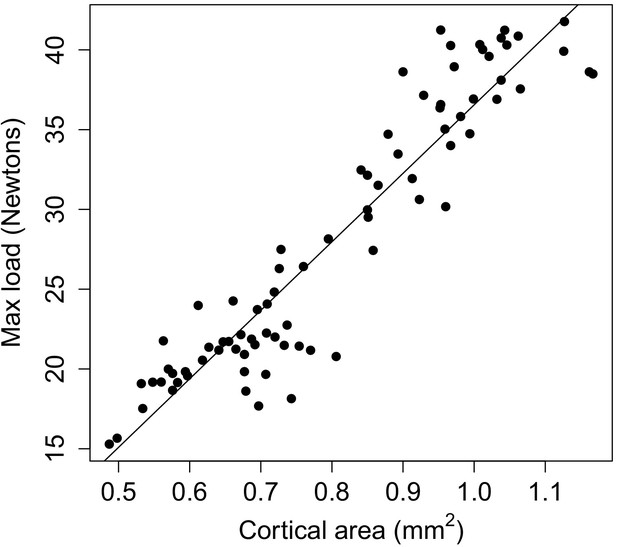
Relationship between max load and cortical area in mouse femurs.
Max load shows a highly linear relationship with cortical area across mouse femurs (R2 = 0.88, p=6.64×10−38). Each dot represents a single mouse femur. Solid line indicates the best fit line. Data are from Jepsen et al., 2003.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Peptide, recombinant protein | Recombinant human fibroblast growth factor-basic | Biocom Africa Biotech (http://www.biocombiotech.com) | Cat # 571504 | |
| Chemical compound, drug | Y-27632 Rock Inhibitor | Cayman Chemical | Cat # 10005583 | |
| Software, algorithm | R Project for Statistical Computing | R Project for Statistical Computing | RRID:SCR_001905 | |
| Software, algorithm | cutadapt | cutadapt | RRID:SCR_011841 | |
| Software, algorithm | HTSeq | HTSeq | RRID:SCR_005514 | |
| Software, algorithm | STAR | STAR | RRID:SCR_004463 | |
| Software, algorithm | LIMMA | LIMMA | RRID:SCR_010943 | |
| Software, algorithm | DESeq | DESeq | RRID:SCR_000154 | |
| Software, algorithm | G:Profiler | G:Profiler | RRID:SCR_006809 | |
| Software, algorithm | GATK | GATK | RRID:SCR_001876 | |
| Software, algorithm | VCFtools | VCFtools | RRID:SCR_001235 | |
| Software, algorithm | BEAGLE | BEAGLE | RRID:SCR_001789 | |
| Software, algorithm | CIBERSORT | CIBERSORT | RRID:SCR_016955 | |
| Software, algorithm | Trim Galore | Trim Galore | RRID:SCR_011847 | |
| Software, algorithm | BWA | BWA | RRID:SCR_010910 | |
| Software, algorithm | MACS | MACS | RRID:SCR_013291 | |
| Software, algorithm | HOMER | HOMER | RRID:SCR_010881 | |
| Software, algorithm | MATLAB | MATLAB | RRID:SCR_001622 | |
| Software, algorithm | ImageJ | ImageJ | RRID:SCR_003070 | |
| Software, algorithm | Avizo 3D Software | Avizo 3D Software | RRID:SCR_014431 | |
| Other | MEM-alpha | Sigma-Aldrich | Cat # M4526 | |
| Other | Hyclone Research Grade Fetal Bovine Serum (FBS), South American (Colombia) origin, IRRADIATED | Separations (South Africa; http://separations.co.za) | SV30160.03IR |
Additional files
-
Supplementary file 1
Table summarizing study animals.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp1-v2.xlsx
-
Supplementary file 2
Table of results of mixed effects model of mole-rat gene expression data testing for effect of solitaire versus helper social status.
bone_0 refers to long bones; bone_1 refers to lumbar vertebrae.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp2-v2.xlsx
-
Supplementary file 3
Table of results of multivariate model explaining litter size (first model) or pup mass (second model).
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp3-v2.xlsx
-
Supplementary file 4
Table of results of mixed effects model of mole-rat gene expression data.
bone0 refers to long bones; bone1 refers to lumbar vertebrae.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp4-v2.xlsx
-
Supplementary file 5
Table of proportions of cell types estimated with CIBERSORT, based on reference gene expression levels for 412 marker genes in 27 purified mouse cell types (Hume et al., 2010).
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp5-v2.xlsx
-
Supplementary file 6
Table of 95% confidence intervals of mediation analysis testing for cell-type proportions as mediating the effect of queen status on gene expression (in long bones or in lumbar vertebrae).
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp6-v2.xlsx
-
Supplementary file 7
Table of Gene Ontology (GO) enrichment results of genes upregulated with queen status.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp7-v2.xlsx
-
Supplementary file 8
Table of transcription factor binding motifs enriched in open chromatin regions near genes upregulated with queen status.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp8-v2.xlsx
-
Supplementary file 9
Table of sample info for bone sections stained with Safranin O.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp9-v2.xlsx
-
Supplementary file 10
Table of effects of number of total offspring on mean cortical thickness per femur shaft decile.
- https://cdn.elifesciences.org/articles/65760/elife-65760-supp10-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65760/elife-65760-transrepform-v2.docx





