R7 photoreceptor axon targeting depends on the relative levels of lost and found expression in R7 and its synaptic partners
Figures
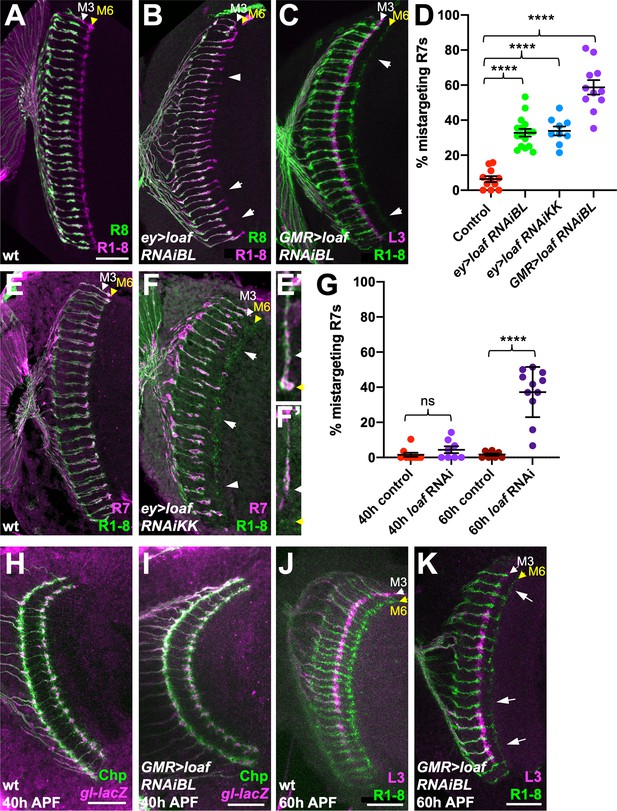
loaf RNAi in photoreceptors causes R7 mistargeting.
(A–C, E, F) cryostat sections of adult heads stained for Chaoptin (Chp) to label all photoreceptor axons (magenta in A, B, green in C, E, F), Rh5-GFP and Rh6-GFP to label R8 (green in A, B), 22E09-LexA driving LexAop-myr-tdTomato to label lamina neuron L3, which projects to the M3 layer (magenta in C) or panR7-lacZ to label R7 (magenta in E, F). (A, E) wild type; (B) ey3.5-FLP, Act>CD2>GAL4; UAS-dcr2; UAS-loaf RNAiBL (P{TRiP.JF03040}attP2); (C) GMR-GAL4, UAS-dcr2; UAS-loaf RNAiBL; (F) ey3.5-FLP, Act>CD2>GAL4; UAS-dcr2; UAS-loaf RNAiKK (P{KK112220}VIE-260B). Arrows show examples of R7 mistargeting. White arrowheads indicate the M3 layer and yellow arrowheads the M6 layer. (E’, F’) show enlargements of single R7 axons from (E, F). (D) Quantification of the percentage of R7 axons that failed to reach the M6 layer in the same genotypes. n = 11 (control, GMR > RNAiBL), 16 (ey>RNAiBL), or 9 (ey>RNAiKK). ****p<0.0001 by unpaired t-test. Error bars show mean ± standard error of the mean (SEM) in this and all other graphs. (H–K) Pupal brains stained for Chp (green) and glass (gl)-lacZ, which labels all photoreceptor axons (magenta in H, I) or 22E09-LexA driving LexAop-myr-tdTomato to label L3 neuronal processes in the M3 layer (magenta in J, K). (H, I) Forty hr after puparium formation (APF); (J, K) 60 hr APF. (H, J) wild type (GMR-GAL4, UAS-dcr2/+); (I, K) GMR-GAL4, UAS-dcr2; UAS-loaf RNAiBL. Loss of loaf does not prevent the initial targeting of R7 axons to their temporary layer at 40 hr APF, but many axons fail to project beyond that layer at 60 hr APF. (G) Quantification of the percentage of R7 axons that did not reach the appropriate layer for these genotypes and stages. n = 9 (40 h control), 8 (40 h RNAi, 60 hr control), or 11 (60 h RNAi). ****, p<0.0001 by unpaired t-test with Welch’s correction; ns, not significant. Scale bars, 20 μm.
-
Figure 1—source data 1
Data shown in Figure 1D and G.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig1-data1-v2.xlsx
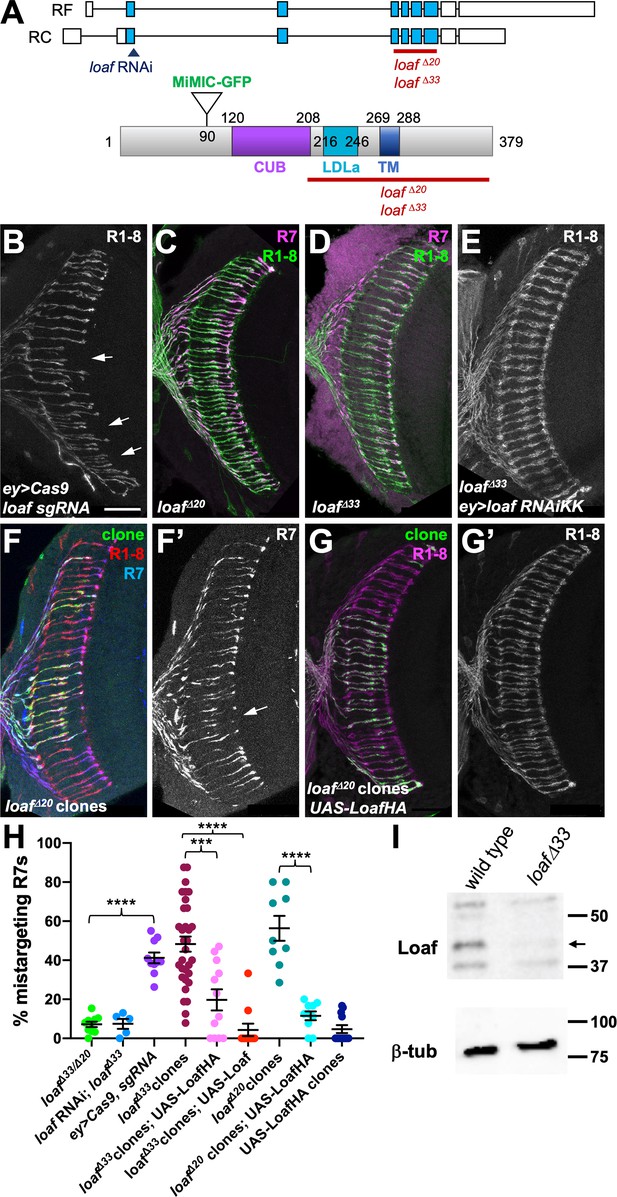
R7 is only affected by eye-specific loss of loaf.
(A) Diagrams of the loaf gene and protein. Coding exons, which are identical for the two isoforms, are shown as blue boxes and non-coding exons as white boxes. The region targeted by both RNAi lines, the MiMIC GFP insertion and the extent of the loafΔ20 and loafΔ33 deletions are indicated. These two deletions were independently generated and have minor sequence differences around the cut site. TM, transmembrane domain. (B–G) cryostat sections of adult heads stained for Chp (B, E, G’, green in C, D, red in F, magenta in G), panR7-lacZ (F’, magenta in C, D, blue in F), and GFP (green in F, G). (B) ey3.5-FLP, Act>CD2>GAL4; loaf sgRNAs; UAS-Cas9P2; (C) loafΔ20 homozygote; (D) loafΔ33 homozygote; (E) ey3.5-FLP, Act>CD2>GAL4; UAS-dcr2/UAS-loaf RNAiKK; loafΔ33; (F) loafΔ20 clones positively labeled with lGMR-GAL4, UAS-GFP; (G) loafΔ20 clones expressing UAS-LoafHA with lGMR-GAL4, positively labeled with GFP. Scale bar, 20 μm. (H) quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 10 (loafΔ33/loafΔ20; ey>Cas9, sgRNA; loafΔ20 clones, UAS-LoafHA), 5 (loafRNAi; loafΔ33), 32 (loafΔ33 clones), 12 (loafΔ33 clones, UAS-LoafHA; wild type clones, UAS-LoafHA), 11 (loafΔ33 clones, UAS-Loaf), or 9 (loafΔ20 clones). Error bars show mean ± SEM. ***, p<0.0005; ****, p<0.0001 by unpaired t-test, with Welch’s correction when variances are significantly different. loaf homozygotes show little R7 mistargeting, but are resistant to the effect of loaf RNAi. R7 mistargeting is observed when loaf sgRNAs and Cas9 are expressed in the eye, and in clones homozygous for loaf alleles. This clonal phenotype is rescued by expressing UAS-LoafHA or UAS-Loaf in the mutant cells. (I) Western blot of extracts from wild type and loafΔ33 larval brains using an antibody to the cytoplasmic domain of Loaf and β-tubulin antibody as a loading control. Loaf protein (arrow) is absent in loafΔ33 mutants.
-
Figure 2—source data 1
Data shown in Figure 2H.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig2-data1-v2.xlsx
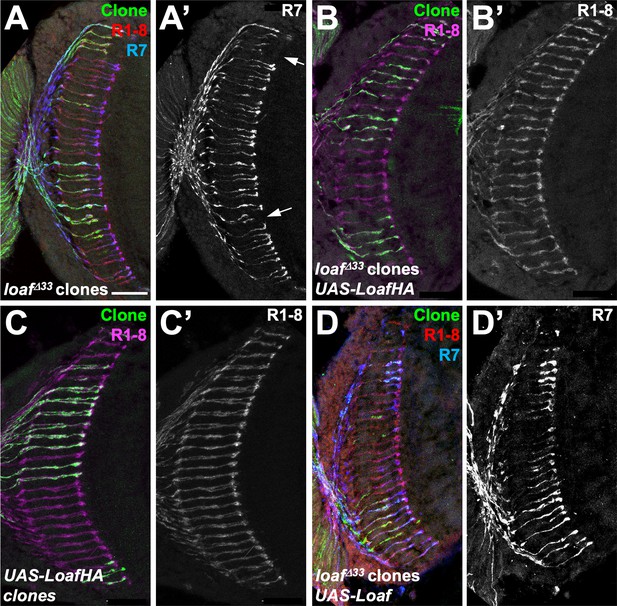
R7 mistargeting in loaf mutant clones is rescued by tagged or untagged Loaf.
Cryostat sections of adult heads with clones positively labeled with GFP, stained for Chp (B’, C’, red in A, D, magenta in B, C), GFP (green), and panR7-lacZ (A’, D’, blue in A, D). (A) loafΔ33 clones; (B) loafΔ33 clones expressing UAS-LoafHA with lGMR-GAL4; (C) clones expressing UAS-LoafHA with lGMR-GAL4; (D) loafΔ33 clones expressing UAS-Loaf with lGMR-GAL4. Scale bar, 20 μm.
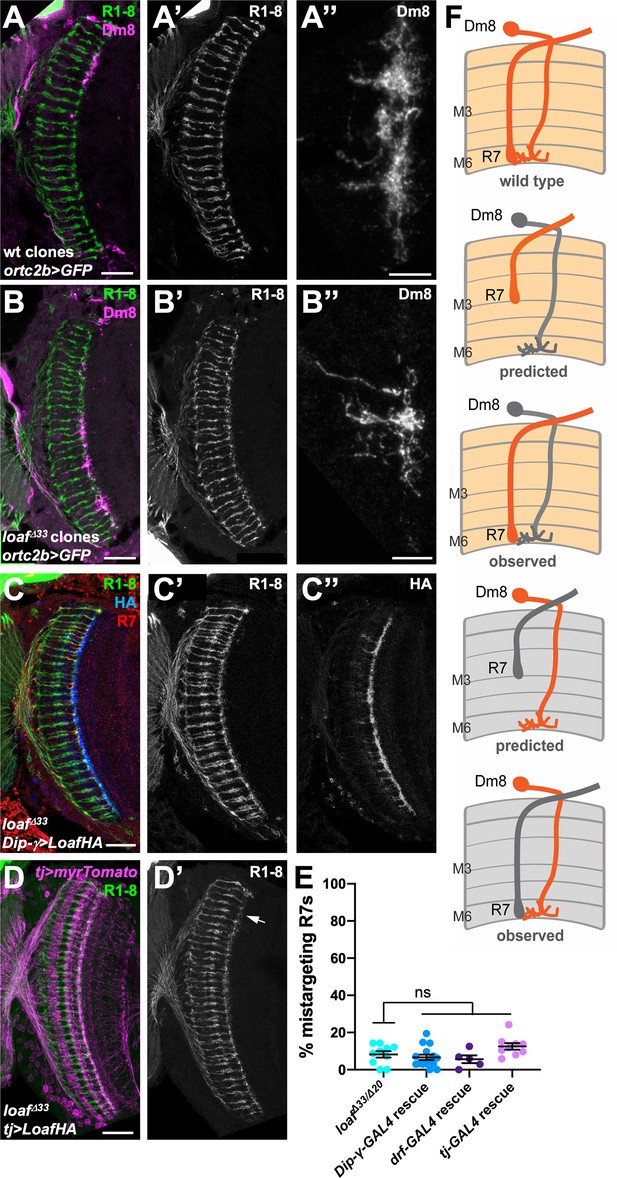
Changing the level of Loaf in Dm8 does not affect R7 targeting.
(A–D) cryostat sections of adult heads stained for Chp (A’-D’, green in A-D), GFP (A’’, B’’, magenta in A, B), panR7-lacZ (red in C), HA (C’’, blue in C), or myrTomato (magenta in D). (A) wild type clones in which Dm8 is labeled with ortc2b-GAL4, UAS-CD8GFP; (B) loafΔ33 clones in which Dm8 is labeled with ortc2b-GAL4, UAS-CD8GFP. A’’ and B’’ show enlargements of labeled Dm8 dendrites. loaf mutant Dm8 dendrites and the R7 axons that target them have the normal position and morphology. (C) UAS-LoafHA; DIP-γ-GAL4, loafΔ33/loafΔ33; (D) tj-GAL4/UAS-LoafHA; loafΔ33/loafΔ33. The arrow in (D’) indicates minor R7 mistargeting that was not statistically significant. Scale bars, 20 μm (A–D), 5 μm (A’’, B’’). (E) quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 10 (loafΔ33/loafΔ20; DIP-γ -GAL4 rescue; tj-GAL4 rescue), or 5 (drf-GAL4 rescue). Error bars show mean ± SEM. ns, not significant by unpaired t-test. Expressing Loaf in Dm8 neurons in a loaf mutant does not cause R7 mistargeting. (F) diagrams explaining the predicted results if Loaf expression in R7 has to match its expression in Dm8. R7 and Dm8 both express Loaf (orange), which is also present in other cells in the brain. Removing loaf from Dm8 (gray) or expressing Loaf in Dm8 in a loaf mutant (gray in R7 and brain) would cause a mismatch and is predicted to result in R7 mistargeting. However, (A–E) show that there is no mistargeting in these situations (observed), indicating that Loaf does not act in Dm8 to regulate R7 targeting.
-
Figure 3—source data 1
Data shown in Figure 3E.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig3-data1-v2.xlsx
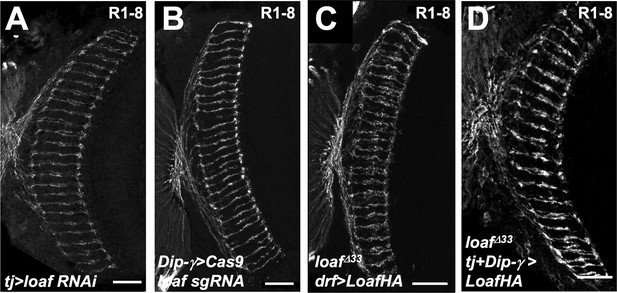
Changing Loaf levels in Dm8 has no effect.
Cryostat sections of adult heads stained for Chp. (A) tj-GAL4; loaf RNAiBL; (B) loaf sgRNAs; DIP-γ-GAL4/UAS-Cas9P2; (C) UAS-LoafHA; drf-GAL4, loafΔ33/loafΔ33; (D) tj-GAL4/UAS-LoafHA; DIP-γ-GAL4, loafΔ33/loafΔ33. Scale bar, 20 μm.
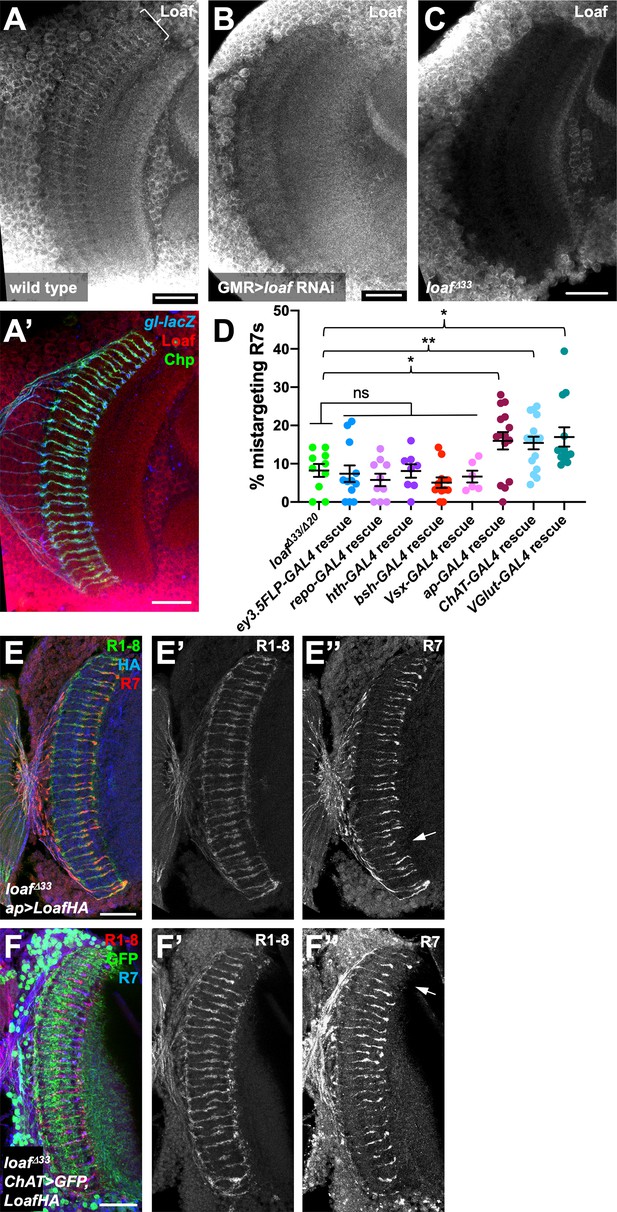
Loaf levels in cholinergic and glutamatergic neurons influence R7 targeting.
(A–C) Pupal brains at 60 hr APF stained for Loaf (A-C, red in A’), Chp (green in A’) and gl-lacZ (blue in A’). (A) wild type; (B) GMR-GAL4, UAS-dcr2; UAS-loaf RNAiBL; (C) loafΔ33. Loaf antibody staining in the medulla neuropil is absent in the loaf mutant (C). Enriched staining in R7 axons (bracket in A) is lost when loaf is knocked down in photoreceptors (B). The antibody appears to cross-react with a protein present in medulla cell bodies, as this staining is still present in loaf mutant brains (C). (D) quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 10 (loafΔ33/loafΔ20; repo-GAL4 rescue), 12 (ey3.5-FLP, Act>CD2>GAL4 rescue), 8 (hth-GAL4 rescue), 11 (bsh-GAL4 rescue), 6 (Vsx-GAL4 rescue), 15 (ap-GAL4 rescue), 16 (ChAT-GAL4 rescue), or 13 (vGlut-GAL4 rescue). Error bars show mean ± SEM. *, p<0.05; **, p<0.01; ns, not significant by unpaired t-test. Expressing Loaf in cholinergic or glutamatergic neurons or in the precursors of cholinergic neurons with ap-GAL4 in a loaf mutant causes R7 mistargeting. (E, F) cryostat sections of adult heads stained for Chp (E’, F’, green in E, red in F), panR7-lacZ (E’’, F’’, red in E, blue in F), HA (blue in E), or GFP (green in F). (E) ap-GAL4/UAS-LoafHA; loafΔ33; (F) ChAT-GAL4, UAS-CD8GFP/UAS-LoafHA; loafΔ33. Scale bars, 20 μm.
-
Figure 4—source data 1
Data shown in Figure 4D.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig4-data1-v2.xlsx
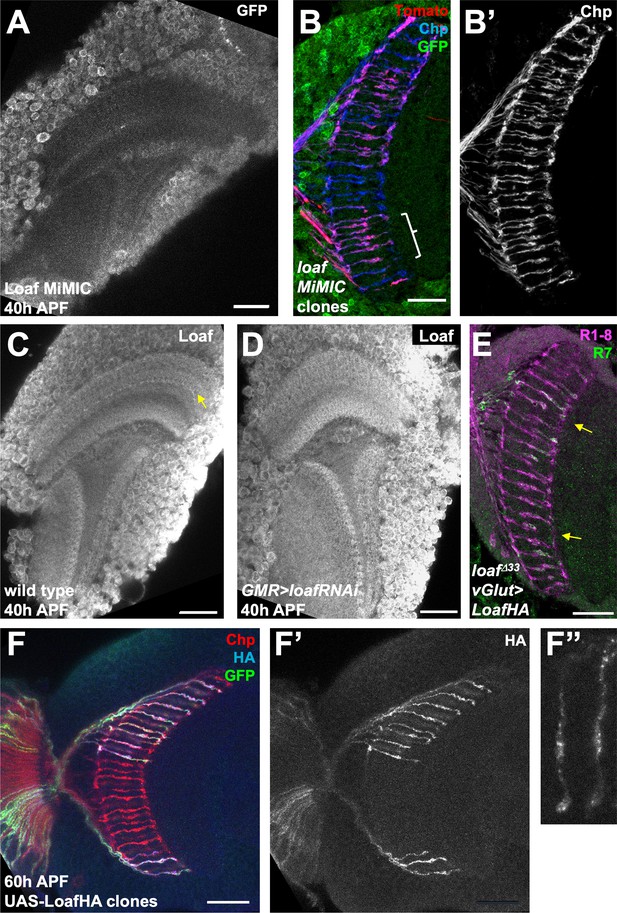
Loaf is expressed in many cells and enriched in R7 terminals.
(A) Mi{PT-GFSTF.1}CG6024MI00316-GFSTF.1 brain at 40 hr APF stained for GFP. GFP-tagged Loaf is mostly confined to cell bodies and only weakly present in the neuropil. (B) Cryostat section of an adult head in which Mi{PT-GFSTF.1}CG6024MI00316-GFSTF.1 homozygous clones are marked with UAS-myr-tdTomato (red) and their axons are stained for Chp (B’, blue in B). GFP-Loaf is stained in green. R7 mistargeting is observed in some clones homozygous for the insertion (bracket), indicating that it disrupts the function of the protein. (C, D) pupal brains at 40 hr APF stained for Loaf. (C) wild type; (D) GMR-GAL4, UAS-dcr2; UAS-loaf RNAiBL. Loaf is enriched in R7 terminals at this stage (yellow arrow in C) and this staining is absent when loaf is knocked down in photoreceptors. (E) Cryostat section of a vGlut-GAL4/UAS-LoafHA; loafΔ33 adult head stained for Chp (magenta) and panR7-lacZ (green). Arrows indicate mistargeted R7 axons. (F) 60 hr APF brain in which LoafHA is misexpressed in clones of photoreceptors with lGMR-GAL4, marked with GFP (green) and stained for Chp (red) and HA (F’, F’’, blue in F). (F’’) shows an enlargement of two R7 axons. LoafHA is transported to R7 axons and terminals. Scale bars, 20 μm.
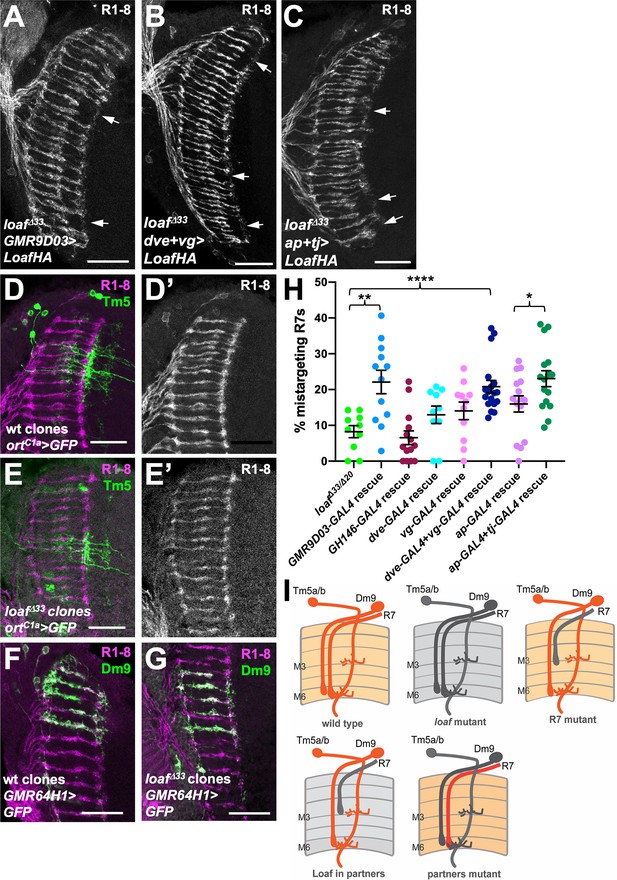
Expressing Loaf in synaptic partners of R7 in a loaf mutant causes mistargeting.
(A–C) cryostat sections of adult heads stained for Chp. (A) UAS-LoafHA; GMR9D03-GAL4, loafΔ33/loafΔ33; (B) dve-GAL4, vg-GAL4/UAS-LoafHA; loafΔ33; (C) ap-GAL4, tj-GAL4/UAS-LoafHA; loafΔ33. Expressing Loaf in populations of neurons that form synapses with R7 in a loaf mutant background causes R7 mistargeting. (D–G) cryostat sections of adult heads in which clones generated with hs-FLP are labeled in green with UAS-CD8-GFP and R1-8 are stained with anti-Chp (D’, E’, magenta in D-G). (D) wild type and (E) loafΔ33 mutant clones in which Tm5a/b/c and Tm20 are labeled with ortC1a-GAL4. The genotypes are (D) hsFLP, UAS-GFP; ortc1a-GAL4/CyO; FRT80/tub-GAL80, FRT80; (E) hsFLP, UAS-GFP; ortc1a-GAL4/CyO; loafΔ33, FRT80/tub-GAL80, FRT80. (F) wild type and (G) loafΔ33 mutant clones in which Dm9 cells are labeled with GMR64H1-GAL4. The genotypes are (F) hsFLP, UAS-GFP; GMR64H1-GAL4, FRT80/tub-GAL80, FRT 80; (G) hsFLP, UAS-GFP; GMR64H1-GAL4, loafΔ33, FRT80/tub-GAL80, FRT 80. The morphologies of wild type and loaf mutant Tm5 and Dm9 cells appear similar. (H) Quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 10 (loafΔ33/loafΔ20; dve-GAL4 rescue), 12 (GMR9D03-GAL4 rescue), 14 (GH146-GAL4 rescue), 11 (vg-GAL4 rescue), 18 (dve-GAL4 + vg GAL4 rescue), 15 (ap-GAL4 rescue), or 16 (ap-GAL4 +tj GAL4 rescue). Error bars show mean ± SEM. *p<0.05; **p<0.01; ****p<0.0001 by unpaired t-test. (I) model showing that the presence of Loaf in Dm9 or Tm5a/b when loaf is absent in R7 causes R7 mistargeting. Scale bars, 20 μm.
-
Figure 5—source data 1
Data shown in Figure 5H.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig5-data1-v2.xlsx
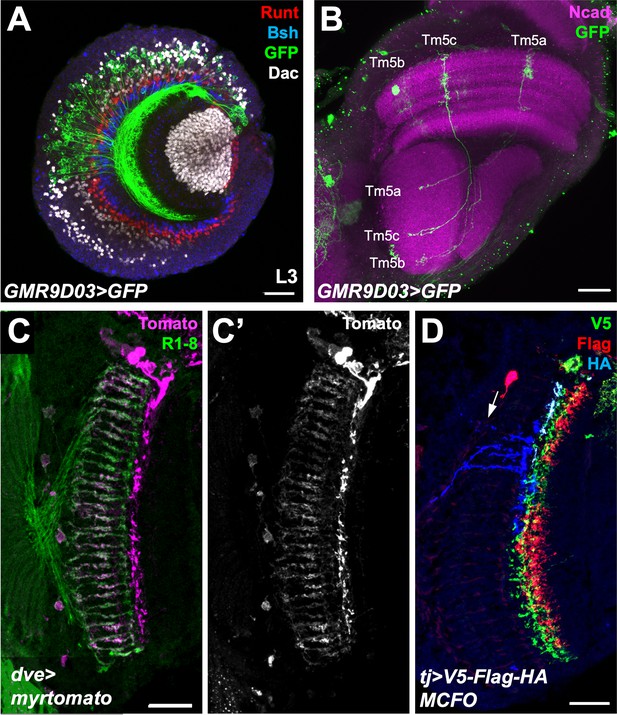
GAL4 drivers for Tm5a/b and Dm9.
(A) A third instar larval brain expressing UAS-CD8-GFP with GMR9D03-GAL4, stained for GFP (green), Dac (white), Runt (red) and Bsh (blue). GMR9D03-GAL4-expressing cell bodies do not express Bsh, Runt or Dac, consistent with Tm5a/b identity, and are restricted to the dorsal half of the medulla cortex. (B) Adult brain expressing UAS>stop> CD4-tdGFP with GMR9D03-GAL4 after a 7 min heat shock at late L3 to induce the expression of hs-FLP2PEST, stained for GFP (green) and Ncad (magenta). Cells with the morphology and projection pattern of Tm5a/b/c are labeled with GFP, showing that the GMR9D03 enhancer is expressed in these cell types. (C) Cryostat section of adult head expressing UAS-myr-tdTomato with dveNP3428-GAL4, stained for Tomato (C’, magenta in C) and Chp (green). dveNP3428-GAL4 labels cells with the morphology and projection pattern of Dm9. (D) tj-GAL4 expressing cells labeled with V5 (green), FLAG (red), and HA (blue) using the MultiColor FlpOut (MCFO) system. The cell labeled in blue (arrow) resembles a Dm11 cell. Scale bars, 20 μm.
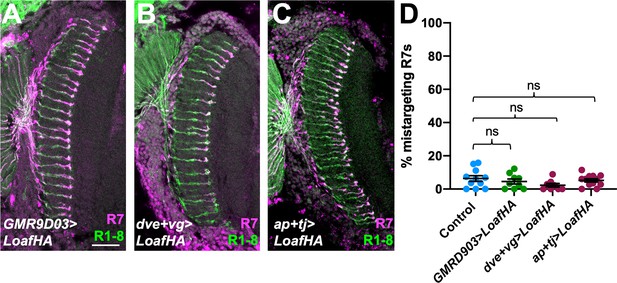
Overexpression of Loaf in the synaptic partners of R7 does not cause mistargeting.
(A–C) Cryostat sections of adult heads stained for Chp (green) and panR7-lacZ (magenta). (A) GMR9D03-GAL4; UAS-LoafHA; (B) dve-GAL4, vg-GAL4; UAS-LoafHA; (C) ap-GAL4, tj-GAL4; UAS-LoafHA. Overexpression of Loaf in these cell types does not cause significant mistargeting. Scale bar, 20 μm. (D) Quantification of the percentage of R7 axons that failed to reach the M6 layer in these genotypes. n = 11 (control), 9 (GMR9D03 > LoafHA), 3 (dve +vg > LoafHA), or 12 (ap +tj > LoafHA). ns, not significant by unpaired t-test.
-
Figure 5—figure supplement 2—source data 1
Data shown in Figure 5—figure supplement 2D.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig5-figsupp2-data1-v2.xlsx
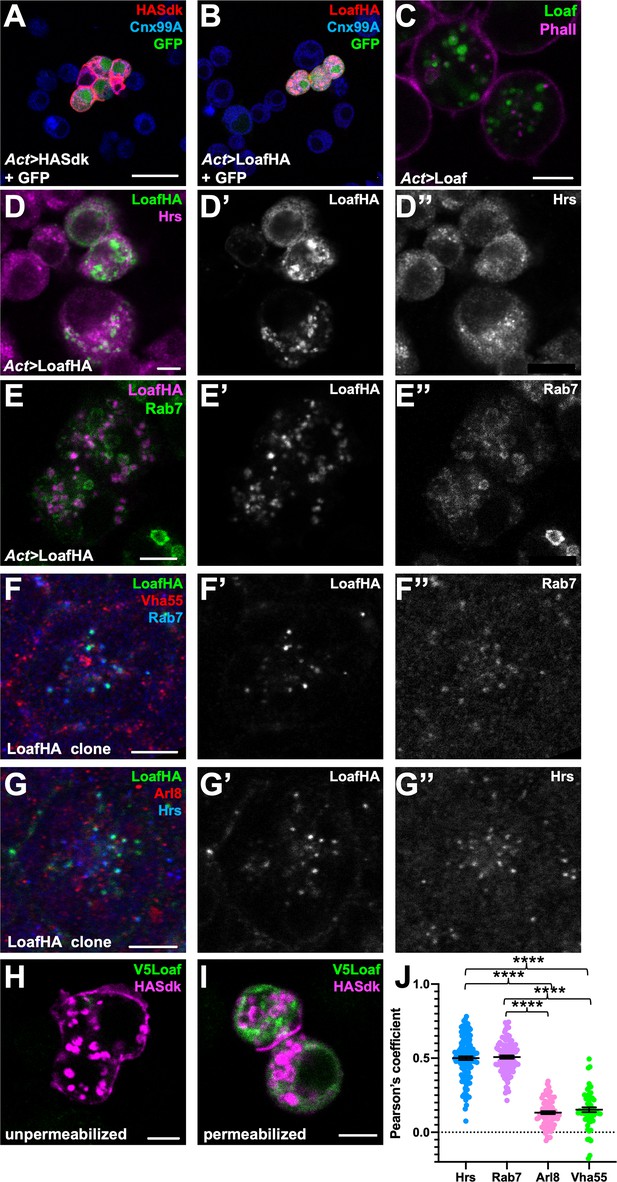
Loaf localizes to endosomes.
(A, B) S2 cells transfected with Act-GAL4, UAS-GFP, and UAS-HASdk (A) or UAS-LoafHA (B) and allowed to aggregate, stained for GFP (green), HA (red) and the ER marker Calnexin 99A (Cnx99A, blue). Sdk localizes to cell contacts and induces aggregation, but Loaf does not. (C) S2 cells transfected with Act-GAL4 and UAS-Loaf, stained with anti-Loaf (green) and Phalloidin (magenta). (D, E) S2 cells transfected with Act-GAL4 and UAS-LoafHA, stained for HA (D’, E’, green in D, magenta in E), Hrs (D’’, magenta in D), or Rab7 (E’’, green in E). Loaf localizes to intracellular vesicles that show some colocalization with Hrs and Rab7. (F, G) Ommatidia from 42 hr APF pupal retinas in clones expressing UAS-LoafHA, stained for HA (F’, G’, green in F, G) Rab7 (F’’, blue in F), Vha55 (red in F), Hrs (G’’, blue in G), and Arl8 (red in G). Loaf colocalizes with the endosomal markers Rab7 and Hrs, but not the lysosomal markers Vha55 and Arl8, in photoreceptors in vivo. (H, I) S2 cells transfected with Act-GAL4, UAS-HASdk, and UAS-V5Loaf and incubated with antibodies to HA (magenta) and V5 (green) at room temperature prior to fixation (H) or after fixation and permeabilization (I). Sdk is detected on the cell surface and in internalized vesicles without fixation, but Loaf is not. (J) Quantification of the colocalization of LoafHA with Hrs, Rab7, Arl8, and Vha55 in 42 hr APF retinas by Pearson’s correlation. n = 131 ommatidia from 19 retinas (Hrs), 100 ommatidia from 19 retinas (Rab7), 85 ommatidia from 16 retinas (Arl8) or 59 ommatidia from 11 retinas (Vha55). Error bars show mean ± SEM. ****p<0.0001. Scale bars, 20 μm (A) or 5 μm (C–I).
-
Figure 6—source data 1
Data shown in Figure 6J.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig6-data1-v2.xlsx
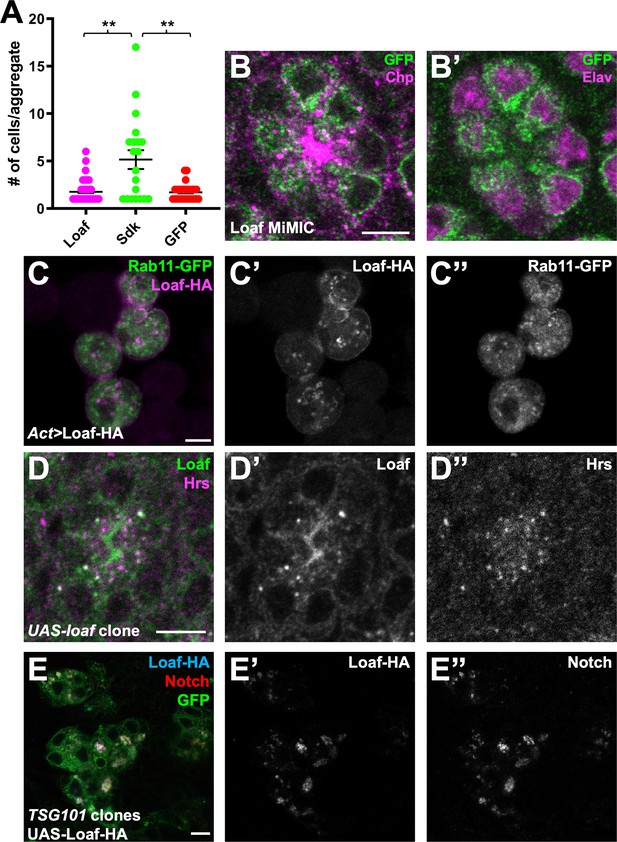
Loaf localizes to endosomes and does not mediate cell aggregation.
(A) Quantification of the number of cells per aggregate for S2 cells transfected with Act-GAL4 and UAS-LoafHA, UAS-HASdk, or UAS-GFP. n = 63 (Loaf), 20 (Sdk), or 27 (GFP). **, p<0.01 by unpaired t-test with Welch’s correction. (B) Ommatidium from a Loaf protein trap Mi{PT-GFSTF.1}CG6024MI00316-GFSTF.1 retina, stained for GFP (green), Chp (magenta in B), and Elav (magenta in B’). Endogenously tagged LoafGFP is present just inside the plasma membrane marked by Chp and in the cytoplasm close to the rhabdomere. (C) S2 cells transfected with Act-GAL4, UAS-LoafHA, and UAS-Rab11-GFP, stained for HA (C’, magenta in C) and GFP (C’’, green in C). Loaf does not colocalize with Rab11. (D) Ommatidium from a clone expressing UAS-Loaf in a 42 hr APF retina, stained for Loaf (D’, green in D) and Hrs (D’’, magenta in D). Untagged Loaf colocalizes with Hrs in vivo. (E) 42 hr APF retina with a TSG101 clone in which UAS-LoafHA was expressed with lGMR-GAl4, positively labeled with GFP and stained for GFP (green), HA (E’, blue in E), and Notch (E’’, red in E). Loss of TSG101 causes accumulation of enlarged endosomes in which Loaf colocalizes with internalized Notch. Scale bars, 5 μm.
-
Figure 6—figure supplement 1—source data 1
Data shown in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig6-figsupp1-data1-v2.xlsx
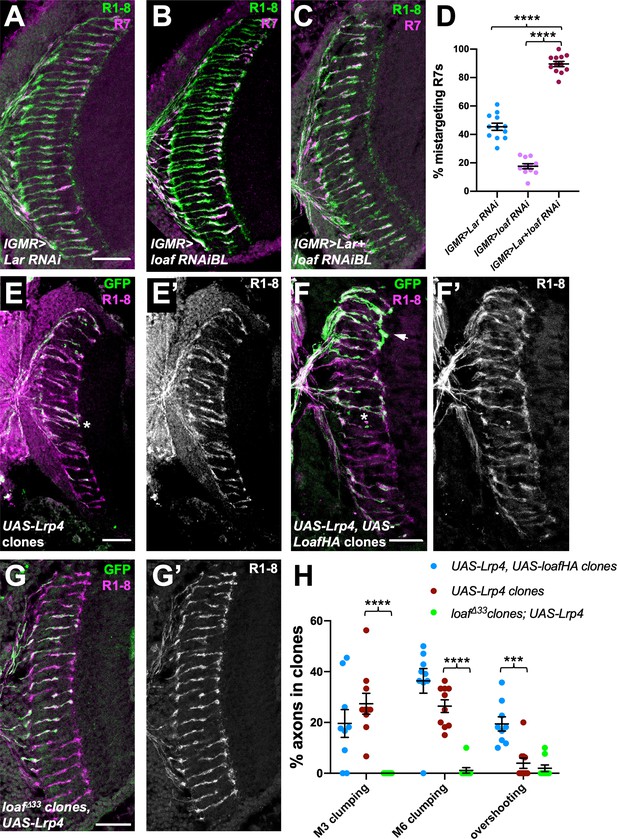
loaf genetically interacts with Lar and Lrp4.
(A–C) Cryostat sections of adult heads stained for Chp (green) and panR7-lacZ (magenta). (A) lGMR-GAL4, UAS-dcr2/UAS-Lar RNAi; (B) lGMR-GAL4, UAS-dcr2; UAS-loaf RNAiBL; (C) lGMR-GAL4, UAS-dcr2/UAS-Lar RNAi; UAS-loaf RNAiBL. With this driver, Lar RNAi induces moderate and loaf RNAi mild R7 mistargeting, but the combination has a severe phenotype. (D) Quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 12 (Lar RNAi, Lar RNAi +loaf RNAi) or 11 (loaf RNAi). ****, p<0.0001 by unpaired t-test. (E–G) Cryostat sections of adult heads with clones positively labeled with GFP, stained for Chp (E’, F’, G’, magenta in E-G) and GFP (green). (E) Clones expressing UAS-Lrp4 with lGMR-GAL4. (F) Clones expressing UAS-Lrp4 and UAS-LoafHA with lGMR-GAL4. (C) loafΔ33 clones in which UAS-Lrp4 is expressed with lGMR-GAL4. (H) Quantification of the percentage of labeled R7s of each genotype that show R7 axons clumping together in the M3 (asterisk in F) or M6 (asterisk in E) layers or overshooting the M6 layer (arrow in F). n = 9 heads (UAS-Lrp4, UASLoafHA; loafΔ33 clones, UAS-Lrp4) or 10 (UAS-Lrp4). Error bars show mean ± SEM. ***p<0.001; ****p<0.0001 by multiple t-tests with two-stage linear step-up procedure. Scale bars, 20 μm.
-
Figure 7—source data 1
Data shown in Figure 7D and H.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig7-data1-v2.xlsx
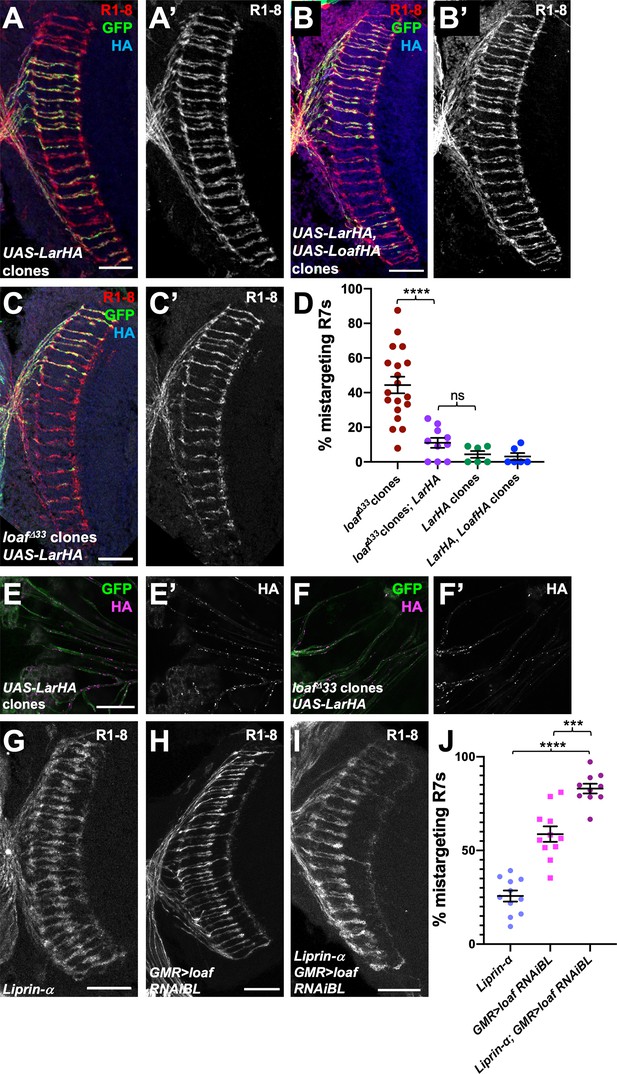
Lar overexpression can compensate for loss of Loaf.
(A–C) Cryostat sections of adult heads with clones positively labeled with GFP, stained for Chp (A’, B’, C’, red in A-C), GFP (green) and HA (blue). (A) Clones expressing UAS-LarHA with lGMR-GAL4. (B) Clones expressing UAS-LarHA and UAS-LoafHA with lGMR-GAL4. (C) loafΔ33 clones in which UAS-LarHA is expressed with lGMR-GAL4. (D) Quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. Expressing Lar rescues mistargeting in loaf mutant clones. n = 19 (loafΔ33 clones), 10 (loafΔ33 clones; LarHA), or 6 (LarHA clones; LarHA, LoafHA clones). Error bars show mean ± SEM. ****p<0.0001; ns, not significant by unpaired t-test with Welch’s correction. (E, F) photoreceptor axons from clones in 42 hr APF retinas positively labeled with GFP, stained for GFP (green) and HA (E’, F’, magenta in E, F). (E) Wild-type clones expressing UAS-LarHA with lGMR-GAL4; (F) loafΔ33 clones in which UAS-LarHA is expressed with lGMR-GAL4. Lar is transported into axons in wild type or loaf mutant photoreceptors. (G–I) Cryostat sections of adult heads stained for Chp. (G) Liprin-αoos; (H) GMR-GAL4/UAS-loafRNAiBL; (I) Liprin-αoos; GMR-GAL4/UAS-loafRNAiBL. (J) Quantification of the percentage of R7 axons that failed to reach the M6 layer in the indicated genotypes. n = 11 (Liprin-α, GMR > loafRNAiBL) or 10 (Liprin-α; GMR > loafRNAiBL). ****p<0.0001; ***p=0.0001. Scale bars, 20 μm.
-
Figure 7—figure supplement 1—source data 1
Data shown in Figure 7—figure supplement 1D and J.
- https://cdn.elifesciences.org/articles/65895/elife-65895-fig7-figsupp1-data1-v2.xlsx
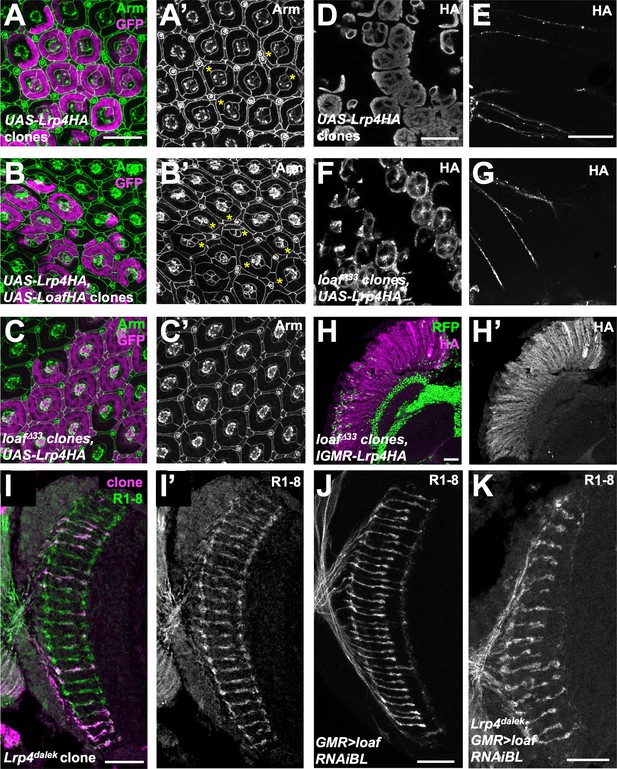
Lrp4 is not the only effector of Loaf.
(A–C) 42 hr APF retinas in which clones are positively labeled with GFP, stained for GFP (magenta) and Arm (A’-C’, green in A-C). (A) Clones expressing UAS-Lrp4 with lGMR-GAL4; (B) clones expressing UAS-Lrp4 and UAS-LoafHA with lGMR-GAL4; (C) loafΔ33 clones in which UAS-Lrp4HA is expressed with lGMR-GAL4. The ordered lattice structure of cone and pigment cells at the apical surface of the retina shows abnormalities (asterisks) when Lrp4 is overexpressed. These are more severe with Loaf coexpression and rescued in loaf mutant clones. (D–G) 42 hr APF pupal retinas in which UAS-Lrp4HA is expressed with lGMR-GAL4 in wild-type clones (D, E) or loafΔ33 mutant clones (F, G), stained for HA. (D, F) show photoreceptor cell bodies and (E, G) show their axons. Lrp4 is still transported into axons in loaf mutant clones, but has a different appearance in cell bodies. (H) Cryostat section of an adult head in which UAS-Lrp4HA is expressed in all cells with lGMR-GAL4, stained for HA (H’, magenta in H). loafΔ33 clones are marked by the absence of nuclear RFP (green). The level of Lrp4 protein is unaffected by loss of loaf. (I–K) Cryostat sections of adult heads stained for Chp (I’, J, K, green in I) and GFP to positively label clones (magenta in I). (I) Lrp4dalek clones do not show R7 mistargeting. (J, K) GMR-GAL4 driving UAS-loaf RNAiBL causes R7 mistargeting in an otherwise wild-type background (J) and in an Lrp4dalek background (K). Scale bars, 20 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | loaf | Flybase | FLYB: FBgn0036202 | CG6024 |
| Genetic reagent (D. melanogaster) | Rh5-GFP | Bloomington Drosophila Stock Center | BDSC:8600 FLYB: FBti0038634 | FlyBase Symbol: P{Rh5-EGFP.P} |
| Genetic reagent (D. melanogaster) | Rh6-GFP | Bloomington Drosophila Stock Center | BDSC:7461 FLYB: FBti0038637 | FlyBase Symbol: P{Rh6-EGFP.P} |
| Genetic reagent (D. melanogaster) | gl-lacZ | Moses and Rubin, 1991 | FLYB: FBtp0001226 | FlyBase Symbol: Ecol\lacZ5xglBS.38-1 |
| Genetic reagent (D. melanogaster) | R22E09-LexA | Pecot et al., 2013 | FLYB: FBtp0108724 | FlyBase Symbol: P{R22E09-nlsLexA::GADfl} |
| Genetic reagent (D. melanogaster) | LexAop:myrTomato | Pecot et al., 2013 | FLYB: FBtp0141256 | FlyBase Symbol: M{13xlexAop-tdTomato.IVS.Myr} |
| Genetic reagent (D. melanogaster) | GMR-GAL4 | Pecot et al., 2013 | FLYB: FBtp0001315 | FlyBase Symbol: P{GAL4-ninaE.GMR} |
| Genetic reagent (D. melanogaster) | ey3.5-FLP | Bloomington Drosophila Stock Center | BDSC:35542 FLYB: FBti0141243 | FlyBase Symbol: P{ey3.5-FLP.B} |
| Genetic reagent (D. melanogaster) | Act>CD2>GAL4 | Bloomington Drosophila Stock Center | BDSC:4780 FLYB: FBti0012408 | FlyBase Symbol: P{GAL4-Act5C(FRT.CD2).P}S |
| Genetic reagent (D. melanogaster) | lGMR-GAL4 | Bloomington Drosophila Stock Center | BDSC: 8605 FLYB: FBti0058798 | FlyBase Symbol: P{longGMR-GAL4} |
| Genetic reagent (D. melanogaster) | UAS-loaf RNAiBL | Bloomington Drosophila Stock Center | BDSC: 28625 FLYB: FBti0127178 | FlyBase Symbol: RNAiBL P{TRiP.JF03040}attP2 |
| Genetic reagent (D. melanogaster) | UAS-loaf RNAiKK | Viennna Drosophila Resource Center | VDRC: 102704 FLYB: FBst0474570 | FlyBase Symbol: P{KK112220}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-LarRNAi | Viennna Drosophila Resource Center | VDRC: 107996 FLYB: FBst0479809 | FlyBase Symbol: P{KK100581}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-dcr2 | Bloomington Drosophila Stock Center | BDSC: 24650 FLYB: FBti0100275 | FlyBase Symbol: P{UAS-Dcr-2.D} |
| Genetic reagent (D. melanogaster) | panR7-lacZ | Hofmeyer et al., 2006 | FLYB: FBtp0022109 | FlyBase Symbol: P{PanR7-lacZ} |
| Genetic reagent (D. melanogaster) | nos-Cas9 | Bloomington Drosophila Stock Center | BDSC: 54591 FLYB: FBti0159183 | FlyBase Symbol: M{nos-Cas9.P}ZH-2A |
| Genetic reagent (D. melanogaster) | UAS-Cas9-P2 | Bloomington Drosophila Stock Center | BDSC: 58986 FLYB: FBti0166500 | FlyBase Symbol: P{UAS-Cas9.P2}attP2 |
| Genetic reagent (D. melanogaster) | DIP-γ-GAL4 | Carrillo et al., 2015 | FLYB: FBal0319064 | FlyBase Symbol: DIP-γMI03222-GAL4 |
| Genetic reagent (D. melanogaster) | tj-GAL4NP1624 | Kyoto Drosophila Stock Center | Kyoto: 104055 FLYB: FBst0302922 | FlyBase Symbol: P{GawB}NP1624/CyO |
| Genetic reagent (D. melanogaster) | drf-GAL4 | Brody et al., 2012 | FLYB: FBal0270054 | FlyBase Symbol: GAL4vvl.43 |
| Genetic reagent (D. melanogaster) | loafMiMIC-GFSTF | Bloomington Drosophila Stock Center | BDSC: 64464 FLYB: FBti0181845 | FlyBase Symbol: Mi{PT-GFSTF.1}CG6024MI00316-GFSTF.1 |
| Genetic reagent (D. melanogaster) | ap-GAL4 | Bloomington Drosophila Stock Center | BDSC: 3041 FLYB: FBti0002785 | FlyBase Symbol: P{GawB}apmd544 |
| Genetic reagent (D. melanogaster) | ChAT-GAL4 | Bloomington Drosophila Stock Center | BDSC: 6798 FLYB: FBti0024050 | FlyBase Symbol: P{ChAT-GAL4.7.4} |
| Genetic reagent (D. melanogaster) | repo-GAL4 | Bloomington Drosophila Stock Center | BDSC: 7415 FLYB: FBti0018692 | FlyBase Symbol: P{GAL4}repo |
| Genetic reagent (D. melanogaster) | hth-GAL4 | Wernet et al., 2003 | FLYB: FBti0058519 | FlyBase Symbol: P{GawB}hthGAL4 |
| Genetic reagent (D. melanogaster) | bsh-GAL4 | Hasegawa et al., 2011 | FLYB: FBtp0069756 | FlyBase Symbol: P{bsh-GAL4.H} |
| Genetic reagent (D. melanogaster) | Vsx-GAL4 | Bloomington Drosophila Stock Center | BDSC: 29031 FLYB: FBti0037957 | FlyBase Symbol: P{GawB}MzVum |
| Genetic reagent (D. melanogaster) | VGlut-GAL4 | Bloomington Drosophila Stock Center | BDSC: 26160 FLYB: FBti0076967 | FlyBase Symbol: P{GawB}VGlutOK371 |
| Genetic reagent (D. melanogaster) | GMR9D03-GAL4 | Bloomington Drosophila Stock Center | BDSC: 40726 FLYB: FBti0152068 | FlyBase Symbol: P{GMR9D03-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | GH146-GAL4 | Bloomington Drosophila Stock Center | BDSC: 30026 FLYB: FBti0016783 | FlyBase Symbol: P{GawB}GH146 |
| Genetic reagent (D. melanogaster) | dveNP3428-GAL4 | Kyoto Drosophila Stock Center | Kyoto: 113273 FLYB: FBti0035416 | FlyBase Symbol: P{GawB}dveNP3428 |
| Genetic reagent (D. melanogaster) | vg-GAL4 | Bloomington Drosophila Stock Center | BDSC: 6819 FLYB: FBal0047077 | FlyBase Symbol: GAL4vg.PM |
| Genetic reagent (D. melanogaster) | ortC1a-GAL4 | Bloomington Drosophila Stock Center | BDSC: 56519 FLYB: FBti0161257 | FlyBase Symbol: P{ort-GAL4.C1a} |
| Genetic reagent (D. melanogaster) | ortC2b-GAL4 | Ting et al., 2014 | FLYB: FBtp0093983 | FlyBase Symbol: P{ort-GAL4.C2b} |
| Genetic reagent (D. melanogaster) | GMR64H01-GAL4 | Bloomington Drosophila Stock Center | BDSC: 39322 FLYB: FBti0137495 | FlyBase Symbol: P{GMR64H01-GAL4}attP2 |
| Genetic reagent (D. melanogaster) | UAS-LarHA | Hofmeyer and Treisman, 2009 | FLYB: FBal0193546 | FlyBase Symbol: LarUAS.Tag:HA |
| Genetic reagent (D. melanogaster) | UAS-Lrp4HA | Mosca et al., 2017 | FLYB: FBal0326704 | FlyBase Symbol: Lrp4UAS.Tag:HA,Tag:FLAG |
| Genetic reagent (D. melanogaster) | Lrp4dalek | Mosca et al., 2017 | FLYB: FBal0326703 | FlyBase Symbol: Lrp4dalek |
| Genetic reagent (D. melanogaster) | Liprin-αoos | Hofmeyer et al., 2006 | FLYB: FBal0193553 | FlyBase Symbol: Liprin-αoos |
| Genetic reagent (D. melanogaster) | GMR9D03-DBD | Bloomington Drosophila Stock Center | BDSC: 68766 FLYB: FBti0192157 | FlyBase Symbol: P{R9D03-GAL4.DBD}attP2 |
| Genetic reagent (D. melanogaster) | GMR38H04-AD | Bloomington Drosophila Stock Center | BDSC: 75758 FLYB: FBti0188278 | FlyBase Symbol: P{R38H04-p65.AD}attP40 |
| Genetic reagent (D. melanogaster) | MCFO-1 | Nern et al., 2015 | FLYB: FBti0169283 | FlyBase Symbol: PBac{10XUAS(FRT.stop)myr::smGdP-HA}VK00005-P{10xUAS(FRT.stop)myr::smGdP-V5-THS-10xUAS(FRT.stop)myr::smGdP-FLAG}su(Hw) |
| Genetic reagent (D. melanogaster) | TSG1012 | Moberg et al., 2005 | FLYB: FBal0212938 | FlyBase Symbol: TSG1012 |
| Genetic reagent (D. melanogaster) | loafΔ33 | This paper | CRISPR deletion allele; see Materials and methods | |
| Genetic reagent (D. melanogaster) | loafΔ20 | This paper | CRISPR deletion allele; see Materials and methods | |
| Genetic reagent (D. melanogaster) | UAS-LoafHA | This paper | Inserted at VK1 attP site | |
| Genetic reagent (D. melanogaster) | UAS-Loaf | This paper | Inserted at VK1 attP site | |
| Genetic reagent (D. melanogaster) | pCFD4-loaf sgRNAs | This paper | Inserted at attP40 site | |
| Cell line (D. melanogaster) | S2 | Laboratory of Ruth Lehmann | FLYB:FBtc0000181; RRID:CVCL_Z992 | FlyBase symbol: S2-DRSC. |
| Antibody | Anti-Chp (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 24B10, RRID:AB_528161 | IF(1:50) |
| Antibody | Anti-GFP (Chicken polyclonal) | Life Technologies | Cat# A10262, RRID:AB_2534023 | IF(1:400) |
| Antibody | Anti-HA (Rat monoclonal) | Sigma (Roche 3F10) | Cat# 11 867 423 001, RRID:AB_390918 | IF(1:400) |
| Antibody | Anti-β-galactosidase (Rabbit polyclonal) | Fisher | Cat# A11132, RRID:AB_221539 | IF(1:100) |
| Antibody | Anti-Ncad (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# DN-Ex #8, RRID:AB_528121 | IF(1:50) |
| Antibody | Anti-DsRed (Rabbit polyclonal) | Takara Bio | Cat# 632496, RRID:AB_10013483 | IF(1:50) |
| Antibody | Anti-Loaf (Guinea pig polyclonal) | Proteintech (this paper) | IF(1:400) WB(1:1000) See Materials and methods | |
| Antibody | Anti-Cnx99A (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# Cnx99A 6-2-1, RRID:AB_2722011 | IF(1:10) |
| Antibody | Anti-Hrs (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# Hrs 27–4, RRID:AB_2618261 | IF(1:10) |
| Antibody | Anti-Rab7 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# Rab7, RRID:AB_2722471 | IF(1:10) |
| Antibody | Anti-ATP6V1B1 (Rabbit polyclonal) | Abgent | Cat# AP11538C, RRID:AB_10816749 | IF(1:200) |
| Antibody | Anti-Arl8 (Rabbit polyclonal) | Developmental Studies Hybridoma Bank | Cat# Arl8, RRID:AB_2618258 | IF(1:200) |
| Antibody | Anti-Elav (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# Rat-Elav-7E8A10 anti-elav, RRID:AB_528218 | IF(1:100) |
| Antibody | Anti-Notch (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# C17.9C6, RRID:AB_528410 | IF(1:10) |
| Antibody | Anti-Arm (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# N2 7A1 Armadillo, RRID:AB_528089 | IF(1:10) |
| Antibody | Anti-GFP (Sheep polyclonal) | BioRad | Cat# 4745–1051, RRID:AB_619712 | IF(1:200) |
| Antibody | Anti-RFP (Rabbit polyclonal) | MBL International | Cat# PM005, RRID:AB_591279 | IF(1:500) |
| Antibody | Anti-V5 (Rabbit polyclonal) | Abcam | Cat# ab9116, RRID:AB_307024 | IF(1:1000) |
| Antibody | Anti-FLAG (Mouse monoclonal) | Sigma | Cat# F3165, RRID:AB_259529 | IF(1:500) |
| Antibody | Anti-Dac (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# mAbdac2-3, RRID:AB_528190 | IF(1:40) |
| Antibody | Anti-Bsh (Rabbit polyclonal) | Özel et al., 2021 | IF(1:1800) | |
| Antibody | Anti-Runt (Guinea pig polyclonal) | Genscript (this paper) | IF(1:600) See Materials and methods | |
| Antibody | Anti-β-tubulin (Mouse monoclonal) | Sigma | Cat# T4026, RRID:AB_477577 | WB (1:10,000) |
| Recombinant DNA reagent | UAS-HASdk | Astigarraga et al., 2018 | In UASt-attB | |
| Recombinant DNA reagent | UAS-LoafHA | Drosophila Genomics Resource Center | Clone UFO07678 | In UASt-attB |
| Recombinant DNA reagent | UAS-Loaf | This paper | See Materials and methods | |
| Recombinant DNA reagent | UAS-V5Loaf | This paper | See Materials and methods | |
| Sequence-based reagent | Loaf_F | This paper | PCR primer | CGCACGAACTTTGTGACACT |
| Sequence-based reagent | Loaf_R | This paper | PCR primer | CTCAAGTCAATCGGTCCTTCC |
| Commercial assay or kit | SuperSignal WestPico | ThermoFisher | Cat # 34579 | |
| Software, algorithm | Fiji-ImageJ | NIH | https://fiji.sc/ |





