The recycling endosome protein Rab25 coordinates collective cell movements in the zebrafish surface epithelium
Figures
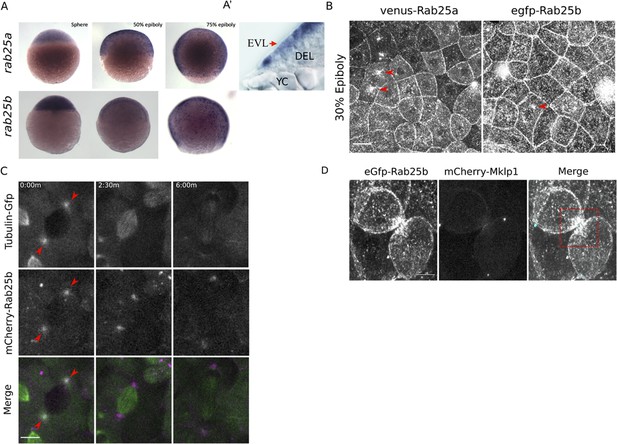
rab25a and rab25b expression pattern and subcellular localization.
(A) Bright field images of whole-mount in situ hybridizations for rab25a (top row) and rab25b (bottom row), in WT embryos; lateral views with animal pole positioned to the top. (A’) Section of a WT embryo showing rab25a expression restricted to the EVL (arrow, top row far right panel); transcripts absent from the deep cells (DEL) and yolk cell (YC). (B) Confocal z-projections of Venus-Rab25a and eGfp-Rab25b subcellular localization in WT embryos; red arrowheads denote perinuclear aggregates. Scale bar 20 μm. (C) Confocal z-projections of stills from time-lapse movies of transgenic Tubulin-GFP (green) embryos expressing mCherry-Rab25b (magenta). Arrowheads denote co-localization of mCherry-Rab25b at centrosomes. Scale bar 20 μm. (D) Confocal z-projections of eGfp-Rab25b (white) and mCherry-Mklp1 (teal) localization in WT embryos; box highlights enrichment of eGfp-Rab25b adjacent to the midbody. Scale bar 20 μm.

Sense probe controls for rab25a and rab25b in situ hybridizations.
(A) Lateral views of WT embryos at 6hpf hybridized with sense probes for rab25a (left panel) and rab25b (center panel), no staining was observed. Positive control using antisense ntla probe shown on the right. Scale bar 100 μm.
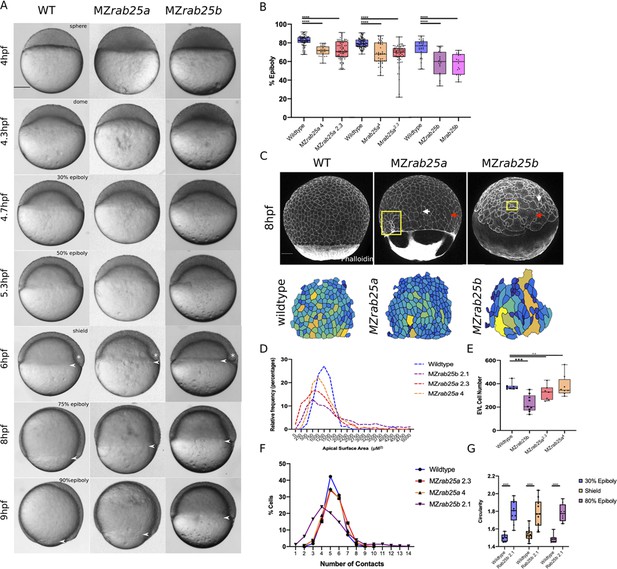
Epithelial spreading delays and heterogenous cell morphology, size and spatial arrangements in MZrab25a and MZrab25b embryos.
(A) Time-matched bright field images of lateral views of WT, MZrab25a and MZrab25b embryos during epiboly. Arrowheads indicate blastoderm margin, asterisks denote embryonic organizer (shield). (B) Quantification of epiboly progression after 8hpf in: WT (n = 80), MZrab25a (2.3, n = 87), MZrab25a (4,n = 28); WT (n = 97), Mrab25a (2.3,n = 73), MZrab25a (4,n = 49); WT (n = 29), MZrab25b (n = 21), Mrab25b (n = 18). Means: SEM; Two-Way ANOVA; ***p<0.001, ****p<0.0001.(N = 3). (C) Confocal z-projections of time-matched lateral views of WT, MZrab25a and MZrab25b embryos at 8hpf stained with phalloidin and corresponding apical surface area heat maps. Cooler colors represent smaller areas, warmer colors represent larger areas. Yellow boxes indicate cells with reduced apices. Red arrows denote cells with increased apical surface areas. White arrows indicate curved cell junctions. Scale bar 100 μm. (D) Frequency distribution of apical surface areas of WT (n = 817, N = 8), MZrab25a 2.3 (n = 651, N = 14), MZrab25a 4 (n = 654, N = 15) and MZrab25b (n = 503, N = 15) embryos at 6hpf. (E) EVL Cell number in WT (n = 8), MZrab25b (n = 8), MZrab25a 2.3 (n = 9) and MZrab25a 4 (n = 7) embryos at 6hpf. Means: SEM; Two-Way ANOVA; ***p<0.001. (F) Frequency distributions of EVL cellular contacts number at 6hpf in WT (n = 817, N = 8), MZrab25a 2.3 (n = 651, N = 14), MZrab25a 4 (n = 654, N = 15) and MZrab25b (n = 503, N = 15). (G) Circularity quantifications for WT and MZrab25b embryos during epiboly. 30% epiboly: WT (n = 7), Mrab25b (n=7). Shield: WT (n=14), Mrab25b (n=10). 80% epiboly: WT (n=8), Mrab25b (n=9). Means: SEM: One-way ANOVA, ****p<0.0001.
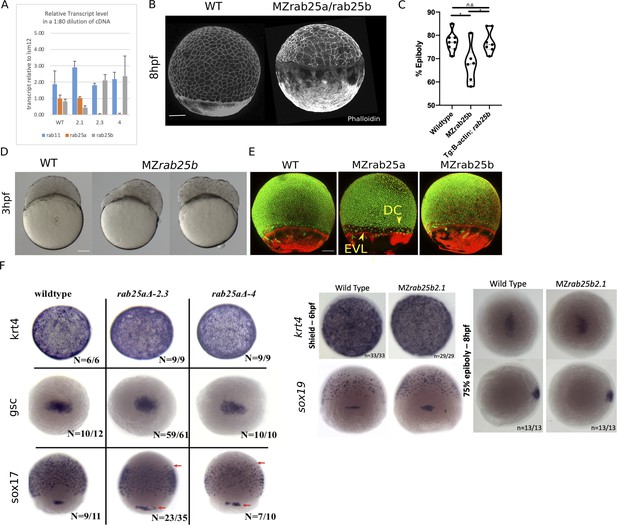
MZrab25a and MZrab25b phenotypic characterization.
(A) qPCR analysis of rab25a, rab25b, and rab11a transcripts levels in WT, MZrab25a, and MZrab25b embryos at shield stage. (B) Time-matched phalloidin stained WT and MZrab25b double mutants at 8hpf. Scale bar 100 μm. (C) Tg: MZrab25b (ß-actin;rab25b) epiboly movements rescued compared to MZrab25b mutants. WT (n = 8), MZrab25b (n = 6), and MZrab25b Tg (ß-actin; rab25b) (n = 7) time-matched at 8hpf. Means:SEM; One-Way ANOVA, *p<0.05. (D) Bright-field images of WT and MZrab25b embryos positioned laterally at 3hpf. (E) Stage-matched WT, MZrab25a and MZrab25b embryos at 75% epiboly showing phalloidin staining of F-actin (red) and sytox green labeling of nuclei (green). Embryos positioned laterally with animal pole to the top; Scale bar 100 μm. (F) Whole-mount in situ hybridizations using indicated probes at indicated stages. Scale bar 100 μm. Arrows indicate sox17 staining.
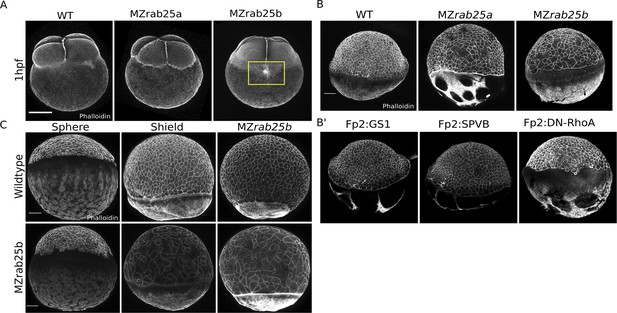
MZrab25a and MZrab25b phenotypic characterization.
(A) Time-matched phalloidin stained WT, MZrab25a, and MZrab25b embryos at 1hpf; Yellow box shows disorganized cortical actin network. Scale bar 100 μm. (B,B’) EVL morphology does not resemble mutant embryos when WT yolk cell F-actin networks are perturbed. Stage-matched embryos positioned laterally at 6hpf stained with phalloidin. Scale bar 100 μm. (C) EVL defects emerge over the duration of epiboly in MZrab25b mutant embryos. Stage-matched WT and MZrab25b embryos positioned laterally stained with phalloidin. Scale bar 100 μm.
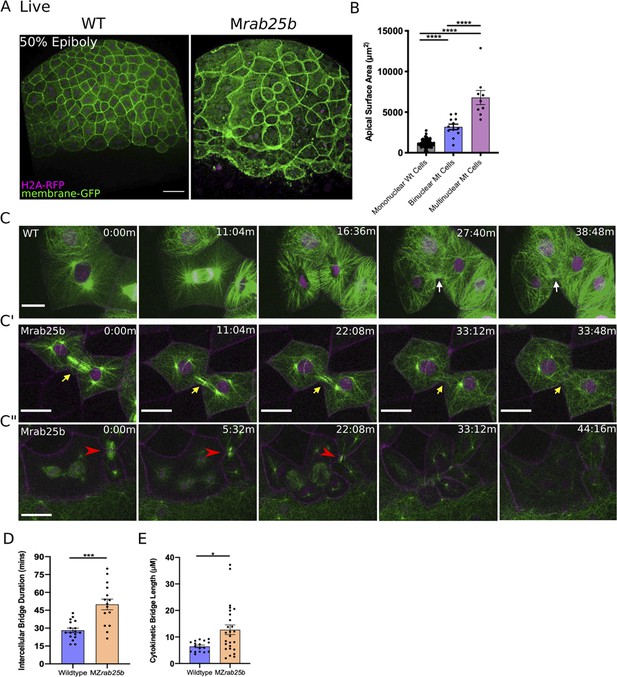
Unstable cytokinetic bridges and delayed abscission in MZrab25b embryos.
(A) Confocal z-projections of stage-matched WT and MZrab25b embryos at 50% epiboly expressing mGfp (green) and H2A-Rfp (magenta). Embryos positioned laterally. Scale bar 40 μm. (B) Apical surface area quantifications of WT mononucleate (n = 151, N = 6), MZrab25a binucleate (n = 10, N=6) and MZrab25b bi-(n = 3, N=4) and multinucleate (n = 9, N=4) at 80% epiboly. Mean: SEM; One-Way ANOVA; ****, p<0.0001. (C–C”) z-projection of stills from confocal time-lapses of WT and MZrab25b embryos labeled for microtubules (green), nuclei (magenta), and plasma membrane (magenta) starting at dome stage; white arrow marks WT bridge; yellow arrows marks bridge regression in MZrab25b cell. (C”) Red arrowheads indicate cytokinetic bridge torn open by neighboring morphogenic stress. Scale bar 20 μm. (D) Intercellular bridge duration in WT (n = 17, N = 3) and MZrab25b (n = 15, N = 4) embryos during epiboly initiation in EVL marginal regions. Mean: SEM; Mann- Whitney Test; ***, p<0.001. (E) Intercellular bridge length following formation in WT (n = 17,N = 3) and MZrab25b (n = 26,N = 4) embryos during epiboly initiation in EVL marginal regions. Mean: SEM; Mann-Whitney Test; *p<0.05.
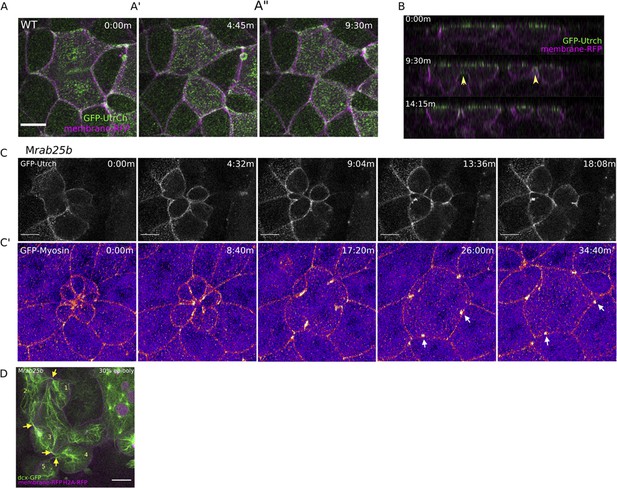
Multipolar cytokinesis failure in MZrab25b embryos.
(A–A”) Confocal z-projections of stills from time-lapse of a WT EVL cell during mitosis expressing Gfp-Utrophin (green) and mRfp (magenta). Scale bar 20 μm. (B) Lateral views with apical to the top of stills from single-plane confocal time-lapses of WT EVL cells during mitosis expressing Gfp-Utrophin (green) and mRfp (magenta). Arrowheads denote cleavage furrow ingression from basal to apical. (C) Confocal z-projections of stills from time-lapse of MZrab25b multipolar cleavage failure. F-actin labeled with Gfp-Utrophin. Scale bar 20 μm. (C’) Confocal z-projections of time-lapse of MZrab25b Tg (Myl1.1-Gfp) (Fire-LUT) during multipolar cytokinesis failure. White arrows indicate Myosin-Gfp foci. (D) Confocal z-projection of MZrab25b embryo showing an array of EVL cells interconnected via cytokinetic bridges at 30% epiboly. Microtubules (green), nuclei (magenta), and plasma membrane (magenta), arrows and numbers denote connected cells and cytokinetic bridges. Scale bar 20 μm.
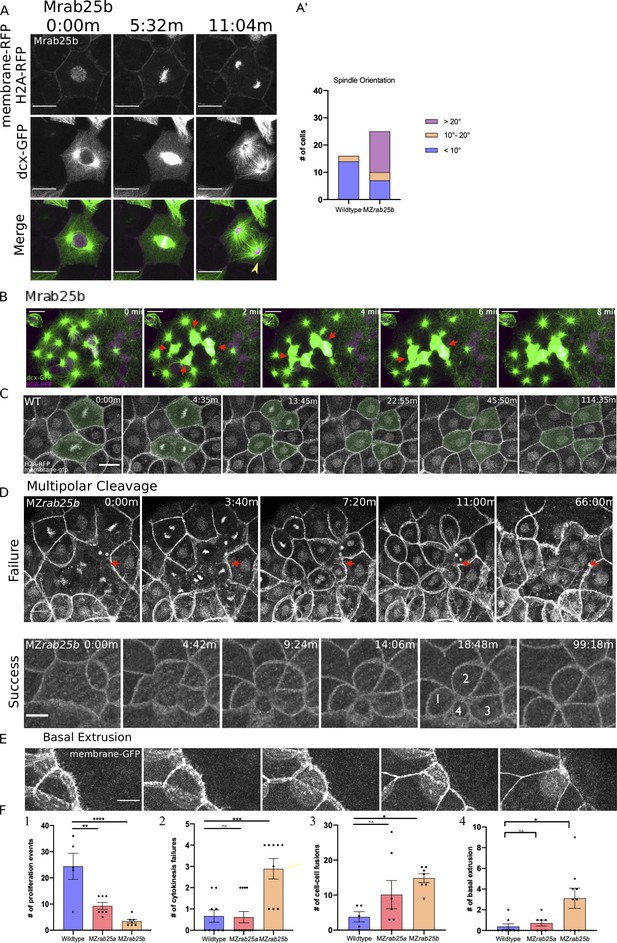
Cytokinesis-related defects in MZrab25b embryos.
(A) Confocal z-projections of stills from confocal time lapse of Mrab25b mononucleate EVL cell during mitosis at 30% epiboly. Microtubules (green), membrane (magenta), and nuclei (magenta). Arrowhead indicates reduced density of astral microtubules. Relative proportions of spindle orientations along the long axis of cells during mitosis. Scale bar 20 μm. (B) Confocal z-projections of stills from confocal time lapse of Mrab25b mitotic spindles fusing during multipolar cytokinesis. Time-lapses began at 30% epiboly in marginal regions; arrows denote fusing spindles: Microtubules (green), membrane (magenta), and nuclei (magenta). Scale bar 10 μm. (C) Confocal z-projections of stills from confocal time lapse of a WT embryo imaged beginning at dome stage. Shaded cells are dividing; Nuclei and membranes labeled. Scale bar 20 μm. (D) Confocal z-projections of stills from confocal time lapse of multipolar mitosis in MZrab25b embryos beginning at 30% epiboly. Failed cleavage indicated by arrows; Successful cleavage generates cells with reduced apices indicated by numerals; Nuclei and membranes labeled. Scale bar 10 μm. (E) Confocal z-projection of a MZrab25b EVL cell expressing mGfp undergoing basal cell extrusion. Scale bar 10 μm. (F) Quantifications of means of (1) proliferation events in WT (n = 5), MZrab25a (n = 7) and MZrab25b (n = 7);(2) Cytokinesis events in WT (n = 9), MZrab25a (n = 13) and MZrab25b (n = 9); (3) non-sister cell fusions in WT (n = 5), MZrab25ba (n = 7) and MZrab25b (n = 7); (4) Basal Cell Extrusion events in WT (n = 8), MZrab25a (n = 7) and MZrab25b (n = 9). Means: SEM; One-Way ANOVA; *, **, ***, ****, p<0.05, 0.01, 0.001, 0.0001.
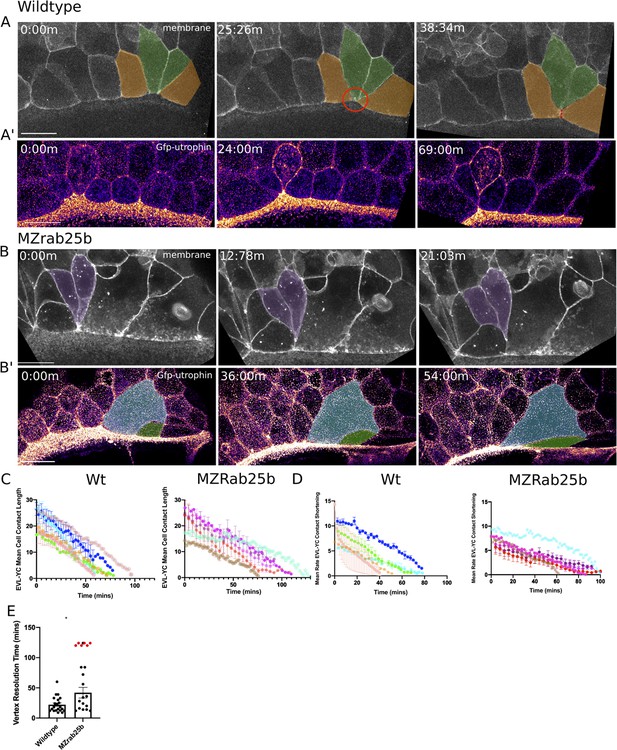
Aberrant EVL cell rearrangements in MZrab25b embryos.
(A) Confocal z-projections of stills from a time-lapse of a WT embryo expressing mGfp starting at 7hpf. Lateral view focused on the margin. Green shaded cells shrink the EVL-YC junction and intercalate into submarginal zones. Orange shaded cells establish new cell-cell contacts following intercalation events (denoted by red dotted line). Circle denotes shared vertex with underlying yolk cell. Scale bar 20 μm (A’) Confocal z-projection time-lapse of WT embryo labeled for F-actin; (Fire-LUT) (Gfp-Utrophin) starting at 7hpf; lateral view; scale bar 20 μm. (B) Confocal z-projection of stills from a confocal time-lapse starting at 7hpf of an MZrab25b embryo expressing membrane-Gfp. Purple shaded cells exit EVL marginal region. Scale bar 20 μm (B’) Confocal z-projection of stills from time-lapse starting at 7hpf of an MZrab25b embryo labeled for F-actin (Fire-LUT) (Gfp-Utrophin); scale bar 20 μm. Shaded cells denote an EVL circumferential stretching event. Scale bar 20 μm. (C–D) EVL-YC mean contact length or shortening rate over time in rearranging EVL marginal cells in WT (N = 5) and MZrab25b embryos (N = 5). Mean:SEM. Each color indicates a separate trial of a single embryo. Each line represents the average of the contact length or junction shrink rate in each trial (n = 2–5). (E) Resolution times following formation of EVL-YC multicellular vertices. Mean: SEM. WT (n = 20,N = 4) and MZrab25b (n = 12,N = 5), unresolved MZrab25b vertices (red) (n = 6,N = 5). Mann-Whitney, *p<0.05.

Marginal EVL cell behaviors during epiboly progression.
(A) Individual marginal cell EVL-YC contact length over time in WT (n = 8, N = 3) and MZrab25b (n = 13, N = 3) beginning at 7hpf. (B) Confocal z-projections of stills from confocal time lapse of stage-matched WT and MZrab25b embryos during late epiboly stained for F-actin. Arrowhead denotes uneven EVL margin in MZrab25b embryos; Scale bar 20 μm.
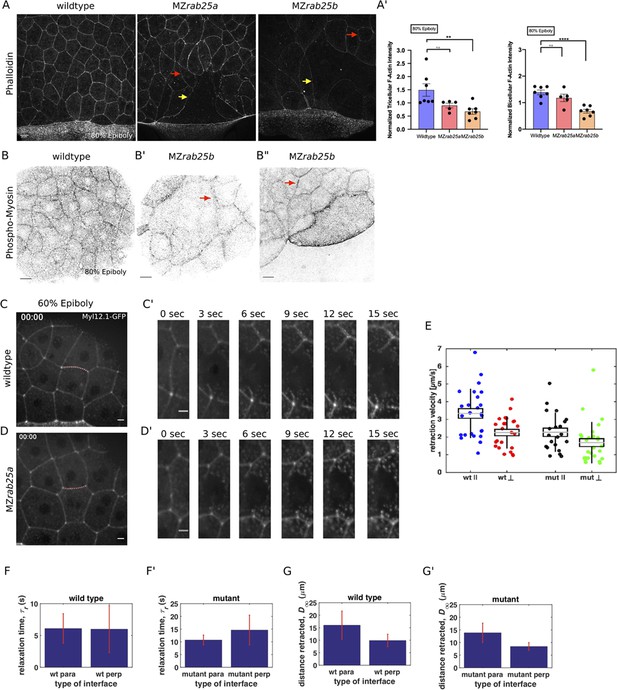
Reduced cortical actin and pMyosin associated with less tension and more viscous tissue responses in mutant embryos.
(A) Lateral views with animal pole to the top of z-confocal projections of phalloidin stained WT, MZrab25a and MZrab25b embryos stage-matched at 80% epiboly; red arrows show reduced cortical actin in normal sized cells; yellow arrows show reduced actin in large cells; Scale bar 20 μm. (A’) Quantification of normalized tricellular and bicellular F-actin intensity at 80% epiboly. WT (n = 90,N = 9), MZrab25a (n = 90,N = 9) and MZrab25b (n = 90,N = 9). Means: SEM; Mann-Whitney, **,p<0.001. (B) Confocal z-confocal projections of WT and MZrab25b embryos at 80% epiboly antibody stained for pMyosin. Red arrows denote uneven distribution of pMyosin along individual MZrab25b cellular junctions. Scale bar 20 μm. (C–D’) Confocal z-confocal projections of WT or MZrab25a Tg(Myl1.1-Gfp) at 60% epiboly; lateral positioned embryo focused on EVL margin; red line marks the ablated junction. Scale bar 5 μm. (E–G’) Initial recoil velocity, relaxation time and distance retracted in WT and MZrab25a embryos following junction laser cutting (see Materials and methods). WT and MZrab25a perpendicular and parallel cuts (n = 26,23);(n = 21,25).
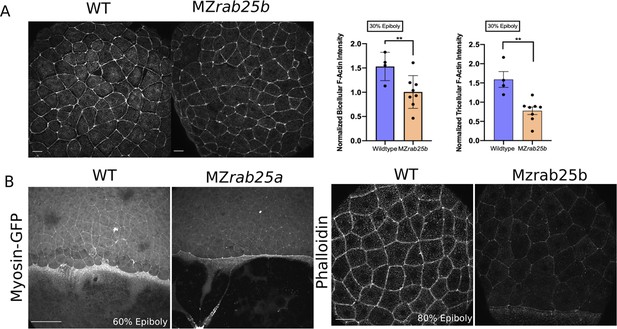
Actomyosin intensity in MZrab25a and MZrab25b epiboly.
(A) Confocal z-projections WT and MZrab25b embryos stage-matched at 30% epiboly and stained for f-actin (phalloidin). Quantifications of EVL tri-or bicellular f-actin intensity in WT (n = 40, N=4) and MZrab25b embryos (n = 70, N = 7). Means:SEM; t-test **, p<0.01. Scale bar 20 μm. (B) Confocal z-projections of stage-matched WT Tg(Myl1.1-gfp) and MZrab25a Tg(Myl1.1-Gfp) at 60% epiboly positioned laterally. Confocal z-projections of phalloidin stained stage-matched WT and MZrab25b embryos at 80% epiboly. Scale bars 20 μm.
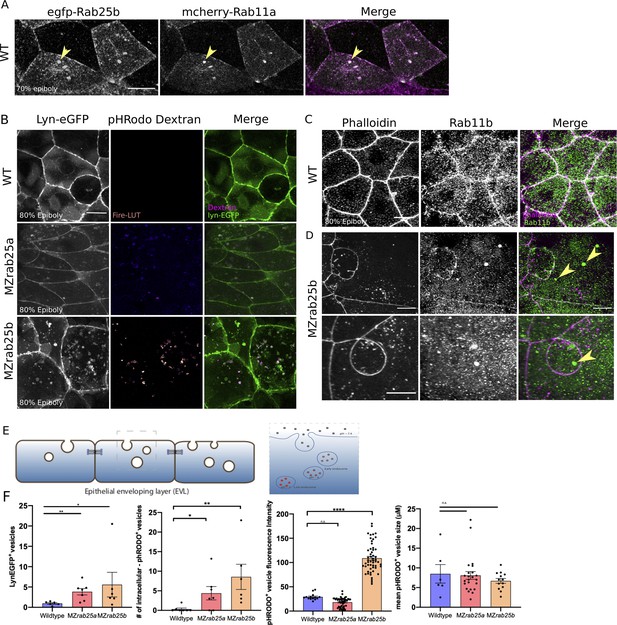
Co-localization of tagged-Rab25 with mCherry-Rab11a and MZrab25a and MZrab25b embryos exhibit trafficking defects.
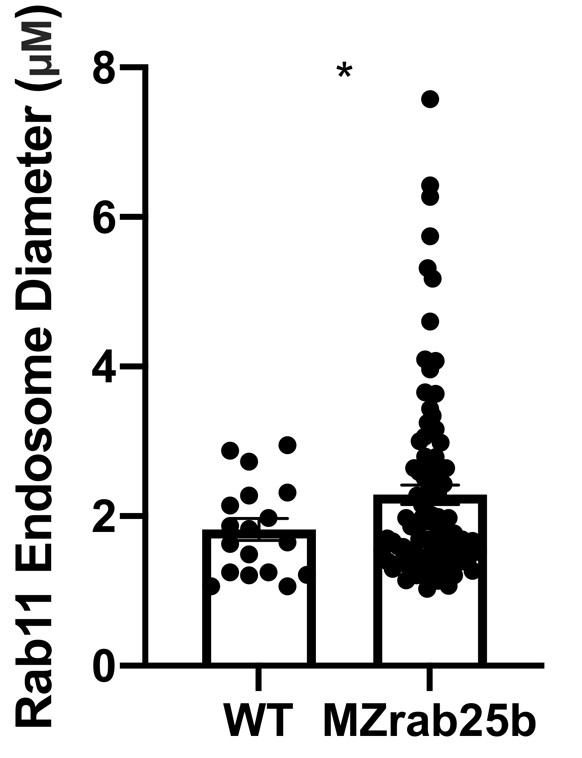
(A) z-projection stills of a wild-type embryo at 80% epiboly co-expressing mCherry-Rab11a (green) and eGfp-Rab25b (magenta); arrowhead denotes overlap. (B) Live WT, MZrab25a and MZrab25b embryos expressing Lyn-eGfp (green) and containing cytoplasmic pHRodo dextran puncta (magenta) following incubation. Scale bar 20 μm. (C,D) Rhodamine phalloidin stained (magenta) and Rab11b antibody (green) stained WT and MZrab25b embryos at 80% epiboly; arrowheads denote large Rab11b endosomes. Scale bar 20 μm (E) Schematic of pHRodo dextran apical endocytosis (F) Mean number of Lyn-eGfp- or pHRodo-positive vesicles/cell. WT, MZrab25a and MZrab25b embryos (N = 7,7,6). Fluorescence intensity measured over a 1 μm line in pHRodo-positive vesicles; WT (n = 15); MZrab25a (n = 46); MZrab25b (n = 54). Mean surface area of pHRodo vesicles; WT(n = 6); MZrab25a and MZrab25b (n = 23,13). Means: SEM; significance using Mann-Whitney test. Scale bars, (A–C) 20 μm.

Co-localization of N-terminally fluorescently tagged Rab11a, Rab25a, and Rab25b constructs and Rab11b endosome quantification.
(A) Confocal z-projections of N-terminally fluorescently tagged Rab constructs at 30% epiboly in WT embryos. Constructs co-localize at the plasma membrane, cytosol, centrosomes (black arrows), and tricellular vertices (red arrows); Scale bar 20 μm.
Videos
N-terminally tagged-Rab25 constructs dynamics during cell proliferation and early epiboly.
WT embryo expressing Venus-Rab25a (left panel) and eGfp-Rab25b (right panel) beginning at 30% epiboly; Scale bar 20 μm.
eGfp-Rab25b live distribution at the plasma membrane and cell vertices during late epiboly.
WT embryo expressing eGfp-Rab25b beginning at 6hpf; Scale bar 20 μm.
Venus-Rab25a is dynamically recruited to vertices of rearranging marginal cells.
WT embryo expressing Venus-Rab25a beginning at 7hpf; Scale bar 20 μm.
WT EVL Mitosis.
WT embryo injected with mrfp (magenta) and gal4 mRNA and H2A-rfp(magenta)-dUAS-dcx-gfp (green) plasmid imaged beginning at dome stage. Scale bar 20 μm.
Cytokinetic bridge regression in Mrab25b mutant.
Mrab25b embryo injected with mrfp (magenta) and gal4 mRNA and H2A-rfp(magenta)-dUAS-dcx-gfp (green) plasmid imaged beginning at dome stage. Scale bar 20 μm.
Failed formation of the cytokinetic midbody in MZrab25b embryos.
Left panel: Tg:(Dcx-Gfp) (magenta) expressing mCherry-Mklp1 (green) imaged beginning at dome stage showing formation of midbody in wild-type embryo. Right panel: Apical z-projection of Mrab25b embryo expressing mRfp (magenta) and mCherry-Mklp1 (green); coalescence of the cytokinetic midbody which precedes bridge regression; Scale bar 20 μm.
Torn open intercellular bridge during multipolar mitosis.
Mrab25b embryo injected with mrfp (magenta) and gal4 mRNA and H2A-rfp (magenta)-dUAS-dcx-gfp (green) plasmids imaged beginning at dome stage. Scale bar 20 μm.
EVL cells interconnected by long cytokinetic bridges in Mrab25b embryos.
Mrab25b embryo injected with mrfp (magenta) and gal4 mRNA and H2A-rfp(magenta)-dUAS-dcx-gfp (green) plasmid imaged beginning at dome stage. Scale bar 20 μm.
Heterogenous distribution of F-actin in MZrab25b EVL cells during late epiboly.
Left Panel: WT embryo expressing Gfp-Utrophin (Fire-LUT) imaged beginning at 7hpf; Enrichment of cortical F-actin during marginal EVL cell rearrangements. Right panel: MZrab25b embryo expressing Gfp-Utrophin (Fire-LUT) imaged beginning at 7hpf. Cells with reduced and large apices shorten their EVL-YC contact; Scale bar 20 μm.
Myosin-Gfp distribution in MZrab25b embryo during epiboly progression.
Left Panel: WT Tg(Myl1.1-Gfp) (Fire-LUT) embryo imaged starting at 7hpf. Right Panel: MZrab25b Tg(Myl1.1-Gfp) (Fire-LUT) embryo imaged starting at 7hpf; Myosin-Gfp becomes heterogeneously distributed in EVL marginal regions Scale bar 20 μm.
mCherry-Rab11a spatiotemporally overlaps with Venus-Rab25a in the cytosol, plasma membrane, and at centrosomes.
Left Panel: Confocal time-lapse of a WT embryo expressing mCherry-Rab11a (magenta) and Venus-Rab25a (green) beginning at 30% epiboly. Middle Panel: Venus-Rab25a. Right Panel: mCherry-Rab11a. Scale Bar 20 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound | rhodamine-phalloidin | Invitrogen | Cat #R415 | (1:200) |
| Chemical compound | sytox green | Invitrogen | Cat #S7020 | (1:1000) |
| Chemical compound | pHrodo Red Dextran | Invitrogen | Cat# P10361 | (1mg/ml) |
| Antibody | anti-phospho-myosin-light chain 2 Ser 19 (Rabbit polyclonal) | Cell Signalling | Cat #3671 | IF(1:100) |
| Antibody | anti-Cdh1 (Rabbit polyclonal) | AnaSpec | Cat# 55527s | IF (1:1000) |
| Commercial assay or kit | MEGAscript T7-Transcription Kit | Ambion | Cat#AMB1334 | |
| Commercial assay or kit | MegaClear Clean Up Kit | Ambion | Cat#AM1908 | |
| Commercial assay or kit | mMessage mMachine T3 Transcription Kit | ThermoFisher Scientific | Cat#AM1348 | |
| Commercial assay or kit | NucAway Spin Column | Ambion | Cat#AM10070 | |
| Strain (D. rerio) | AB wildtype | Zebrafish International Resource Centre | RRID:BDSC_5138 | |
| Strain (D. rerio) | Tg:(XIEef1a1:dclk2DeltaK-GFP) | Sepich et al., 2011 | ||
| Strain (D. rerio) | Tg:(XIEef1a:eGFP-tubα8I) | Fei et al., 2019 | ||
| Strain (D. rerio) | Tg:(actb1:myl12.1-eGFP) | Maître et al., 2012 | ||
| Strain (D. rerio) | MZrab25a4 | This study | https://zfin.org/ZDB-ALT-201221-11 | |
| Strain (D. rerio) | MZrab25a2.3 | This study | https://zfin.org/ZDB-ALT-201221-10 | |
| Strain (D. rerio) | MZrab25b | This study | https://zfin.org/ZDB-ALT-201221-12 | |
| Strain (D. rerio) | MZrab25b Tg:(actb1:rab25b) | This study | ||
| Strain (D. rerio) | MZrab25a4 Tg:(actb1:myl12.1-eGFP) | This study | ||
| Strain (D. rerio) | MZrab25b Tg:(actb1:myl12.1-eGFP) | This study | ||
| Sequence-based reagent | rab25a_F | This study | PCR Primers | Forward: TATTTATTCACCAAGCGGTTG (for genotyping) |
| Sequence-based reagent | rab25a_R | This study | PCR Primers | Reverse: GAGTGGTTCTGGGTGTGAGTC (for genotyping) |
| Sequence-based reagent | rab25b_F | This study | PCR Primers | Forward: TGTTTGCAGTGGTTCTTATTGGAG (for genotyping) |
| Sequence-based reagent | rab25b_R | This study | PCR Primers | Reverse: ATTACGTTCGCTTGCAGAATTT (for genotyping) |
| Sequence-based reagent | rab25a_F | This study | PCR Primers | Forward: ATGGGGACAGATTTAGCCTACAAC (for cDNA) |
| Sequence-based reagent | rab25a_R | This study | PCR Primers | Reverse: CGAAGCTGCTGCAAAAACTCCTGA (for cDNA) |
| Sequence-based reagent | rab25a_F | This study | PCR primers | Forward: GGATCCATGGGGACAGATTTAGCCTACAAC (ligation into pCS2+ via restriction digest via BamH1, Xho1) |
| Sequence-based reagent | rab25a_R | This study | PCR primers | Reverse: CTGGAGCGAAGCTGCTGCAAAAACTCCTGA (ligation into pCS2+ via restriction digest via BamH1, Xho1) |
| Sequence-based reagent | rab25a_F | This study | PCR Primers | Forward: CTCGAGGGCGCCACCAT GGGGACAGATTTAGCCTACAAC (ligation with into pCS2+ venus via Xho1 restriction digest) |
| Sequence-based reagent | rab25a_R | This study | PCR Primers | Reverse: CTCGAGCGAAGCTGCTGC AAAAACTCCTGA (ligation with into pCS2+ venus via Xho1 restriction digest) |
| Sequence-based reagent | rab25b_F | This study | PCR Primers | Forward: GGGGACAAGTTTGTACAAAAA AGCAGGCT TCATGGGGTCTGATGAGGCCTA (rab25b with attb1 for recombination into pDONR221) |
| Sequence-based reagent | rab25b_R | This study | PCR Primers | Reverse: GGGGACCACTTTGTACAAGAAAG CTGGGT GTCACAAGTTTTTACAGCAGG (rab25b with attb1 for recombination into pDONR221) |
| Sequence-based reagent | rab11a_F | This study | PCR Primers | Forward: AGAAAAACGGTCTGTCCTTC (qPCR) |
| Sequence-based reagent | rab11a_R | This study | PCR Primers | Reverse: TCAGGATGGTCTGAAAAGCA (qPCR) |
| Sequence-based reagent | rab25a_F | This study | PCR Primers | Forward: GAAGTGACCAGAGGCTCGAT (qPCR) |
| Sequence-based reagent | rab25a_R | This study | PCR Primers | Reverse: GGAGTTTTTGCAGCAGCTT (qPCR) |
| Sequence-based reagent | rab25b_F | This study | PCR Primers | Forward: TCGGAGCTCTGCTGGTTTAT (qPCR) |
| Sequence-based reagent | rab25b_R | This study | PCR Primers | Reverse: GCGTGATCGTAGAGCTCCTT (qPCR) |
| Sequence-based reagent | Lsm12_F | This study | PCR Primers | Forward: AGTTGTCCCAAGCCTATGCAATCAG (qPCR) |
| Sequence-based reagent | Lsm12_R | This study | PCR Primers | Reverse: CCACTCAGGAGGATAAAGACGAGTC (qPCR) |
| Recombinant DNA reagent | pCS2+ (plasmid) | Rupp et al., 1994 | SP6/T7 based backbone | |
| Recombinant DNA reagent | pCS2+egfp-rab25b (plasmid) | This study | egfp-Rab25b version of pCS2+ | |
| Recombinant DNA reagent | pCS2+mcherry-rab25b (plasmid) | This study | mcherry-rab25b version of pCS2+ | |
| Recombinant DNA reagent | pCS2+venus-rab25a (plasmid) | This study | venus-rab25a version of pCS2+ | |
| Recombinant DNA reagent | pCS2+-mcherry-Rab11a (plasmid) | Rathbun et al., 2020 | mcherry-Rab11a version of pCS2+ | |
| Recombinant DNA reagent | pCS2+-mcherry-Mklp1 (plasmid) | Rathbun et al., 2020 | mCherry-Mklp1 version of pCS2+ | |
| Recombinant DNA reagent | pCS2+ lyn-eGfp | A gift from Brian Ciruna | lyn-eGfp version of pCS2+ | |
| Recombinant DNA reagent | pTol2-actb1 | A gift from Brian Ciruna | Tol2 transgenics | |
| Recombinant DNA reagent | pTol2-actb1:rab25b | This study | rab25b version of pTol2-actb1 | |
| Recombinant DNA reagent | pCS2+- Gfp-Utrophin | Addgene | RRID: Addgene_26737 | Gfp-Utrophin version of pCS2+ |
| Recombinant DNA reagent | FP2 | Narayanan and Lekven, 2012 | Wnt8a enhancer based backbone | |
| Recombinant DNA reagent | FP2-GS1 | Fei et al., 2019 | GS1 version of FP2 | |
| Recombinant DNA reagent | FP2-SPVB | Fei et al., 2019 | SPVB version of FP2 | |
| Recombinant DNA reagent | FP2-DN-RhoA | This study | DN-RhoA version of FP2 | |
| Recombinant DNA reagent | PT3TS-nCas9 | Jao et al., 2013 | RRID: Addgene_46757 | T3 based backbone |