Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells
Figures
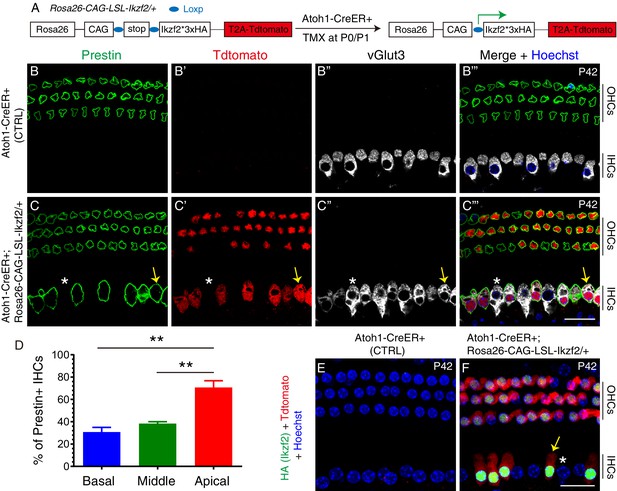
Prestin was expressed in IHCs upon ectopic Ikzf2 induction.
(A) Illustration of approach used to induce Ikzf2 expression in HCs (both IHCs and OHCs) using the Cre-LoxP system. The Atoh1-CreER+ line was used to efficiently drive Cre expression in HCs at neonatal ages when Tamoxifen (TMX) was given at P0 and P1. Tdtomato and Ikzf2 (tagged with HA) were placed in close proximity in the construct to ensure joint expression. (B–C’’’) Triple labeling against Prestin, Tdtomato, and vGlut3 in P42 cochlear samples: control Atoh1-CreER+ (B–B’’’) and experimental Atoh1-CreER+; Rosa26CAG-LSL-Ikzf2/+ (C–C’’’). In wild-type (B–B’’’) Prestin was only expressed in OHCs; in the experimental group (C–C’’’), expression was observed in both OHCs and some IHCs. Arrows in (C–C’’’): Tdtomato+/vGlut3+/Prestin+ IHC; asterisks in (C–C’’’): vGlut3+/Tdtomato-/Prestin- IHC. (D) Quantification of Prestin+ IHCs. More Prestin+ IHCs were present in the apical turn than in basal and middle turns of the cochleae. Data was presented as mean ± SEM, **p<0.01 (Student’s t test). (E, F) Co-staining against HA (Ikzf2) and Tdtomato in control (E) and experimental (F) mice at P42. HA+/Tdtomato + cells were present in experimental mice only. Arrow/asterisk in (F): IHC with/without HA (Ikzf2) expression. All Tdtomato+ cells were HA+ (Ikzf2 expressing) cells, and vice versa. Scale bar: 20 μm.
-
Figure 1—source data 1
It contained the numerical values of the data plotted in the graph of Figure 1D.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig1-data1-v2.xlsx
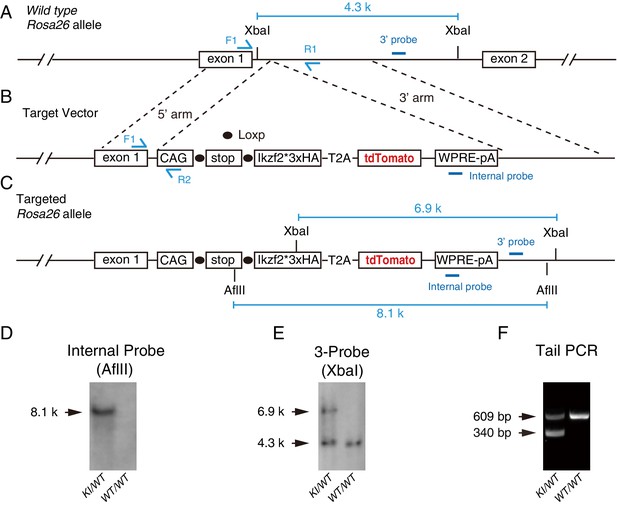
Generation of Rosa26CAG-LSL-Ikzf2/+ knock in mouse line using CRISPR/Cas9 technology.
(A) Wild-type Rosa26 allele. (B) Illustration of target vector. Ikzf2 was tagged with 3× HA fragments at its C-terminus; this was followed by T2A-Tdtomato. Ikzf2 and Tdtomato were transcribed and translated together as a fusion protein, and subsequently cleaved into Ikzf2 and Tdtomato through the 2A peptide. (C) Illustration of the Rosa26 locus after successful gene targeting. (D–E) Southern blotting assay testing for the presence of the internal probe (D) and 3ʹ-end probe (E). (F) Results of PCR genotyping tail DNA extracted from heterozygous (KI/WT) and wild-type (WT/WT) mice. WT mice showed a single 609 bp band, whereas heterozygous mice showed a 609- and a 340-bp band.
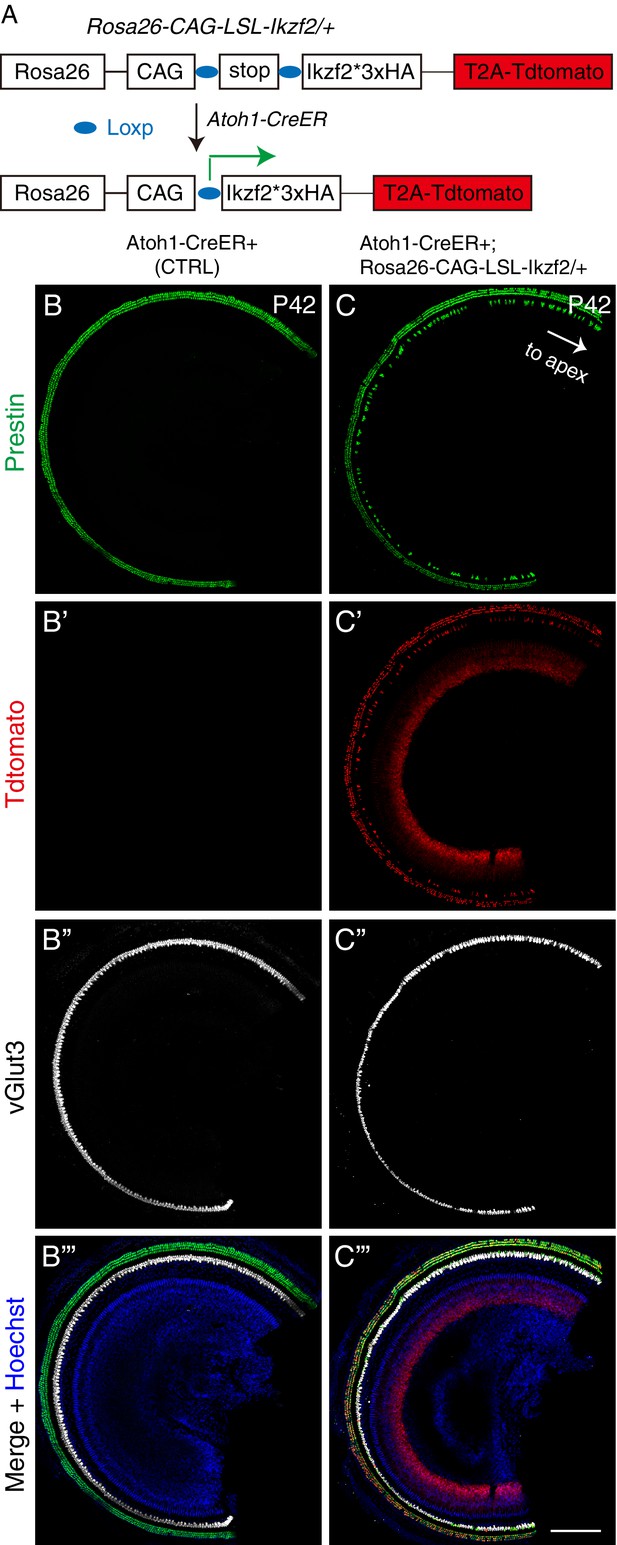
Prestin was expressed in IHCs upon ectopic Ikzf2 induction.
(A) Same illustration as in Figure 1A. (B–C’’’) Triple labeling against Prestin, Tdtomato, and vGlut3 in both control Atoh1-CreER+ mice (B–B’’’) and experimental Atoh1-CreER+; Rosa26CAG-LSL-Ikzf2/+ mice (C–C’’’) at P42. Same data as presented in Figure 1 but with larger field of view. Prestin was expressed in vGlut3+/Tdtomato+ IHCs of experimental but not control mice. Scale bar: 200 μm.
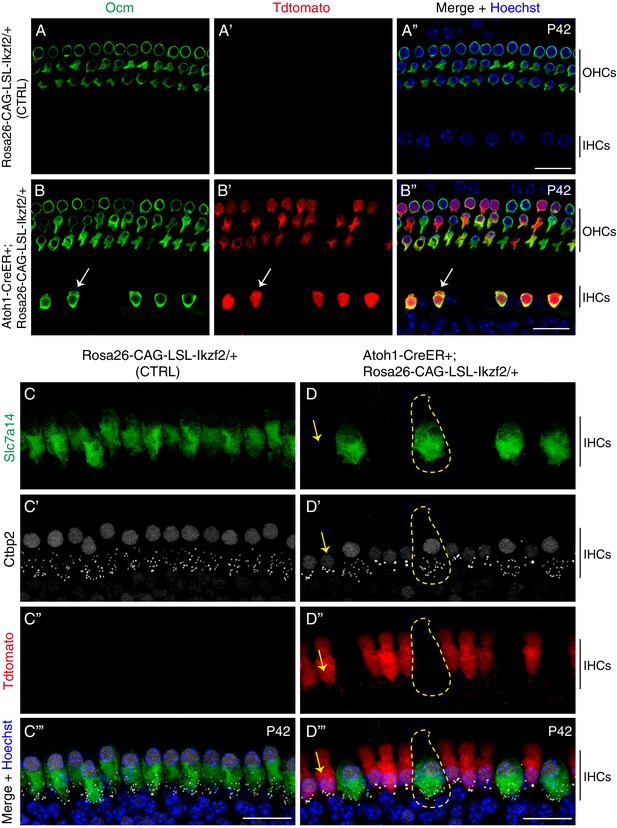
IHCs with ectopic Ikzf2 expression activated Ocm and suppressed Slc7a14 expression, as well as exhibited fewer ribbon synapses.
(A–B’’) Co-labeling against Ocm (OHC marker) and Tdtomato in both control Rosa26CAG-LSL-Ikzf2/+ mice (A–A’’) and experimental Atoh1-CreER+; Rosa26CAG-LSL-Ikzf2/+ mice (B–B’’) at P42. Arrows in (B–B’’) marked a Tdtomato+/Ocm+ IHC. (C–D’’’) Triple labeling against Slc7a14, Ctbp2 and Tdtomato in control (C–C’’’) and experimental mice (D–D’’’) at P42. Arrows in (D–D’’’) pointed to a Tdtomato + IHC (indicating ectopic Ikzf2) which suppressed Slc7a14 expression. Dotted circle in (D–D’’’) represented a Tdtomato- IHC (thus lacking ectopic Ikzf2) which maintained Slc7a14 expression. Notably, both nuclei size and numbers of Ctbp2+ ribbon synapse were decreased in Tdtomato+ IHCs, relative to the Tdtomato− IHCs. Scale bar: 20 μm.
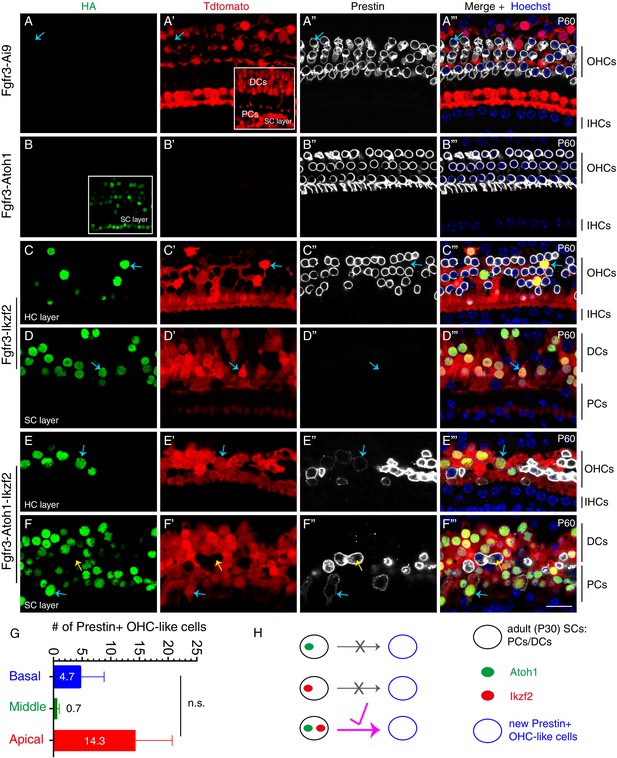
Joint expression of Atoh1 and Ikzf2 in adult SCs (PCs and DCs) induced transition to an OHC-like cell state at low frequency.
Triple labeling against HA, Tdtomato, and Prestin in four different mouse genetic models that were administered tamoxifen at P30 and P31 and sacrificed at P60. Confocal images scanned at HC or SC layers. (A–A’’’) In the control Fgfr3-iCreER+; Ai9/+ (Fgfr3-Ai9) mice, Tdtomato labelled SCs (primarily PCs and DCs) and no Prestin expression was observed in them. Inset in (A’): scan at SC layer. Arrows in (A–A’’’): Prestin+/Tdtomato- OHC. (B–B’’’) In mice expressing Atoh1 in adult SCs, Fgfr3-iCreER+; CAG-LSL-Atoh1+ (Fgfr3-Atoh1), no Tdtomato signal was detected and no HA+/Prestin+ cells were observed. Inset in (B): scan at SC layer. (C–D’’’) In mice expressing Ikzf2 in adult SCs, Fgfr3-iCreER+; Rosa26CAG-LSL-Ikzf2/+ (Fgfr3-Ikzf2), HA and Tdtomato were co-expressed in both the HC (C–C’’’) and SC (D–D’’’) layer. Arrows in both layers: two cells that were HA+/Tdtomato + but did not express Prestin. Alignment of endogenous Prestin+ OHCs was abnormal and some of them were lost. (E–F’’’) In mice, Fgfr3-iCreER+; CAG-LSL-Atoh1+; Rosa26CAG-LSL-Ikzf2/+ mice (Fgfr3-Atoh1-Ikzf2), some adult SCs were HA+/Tdtomato+/Prestin+, taking on an OHC-like phenotype. Arrows in (E–E’’’) indicated an OHC-like cell originating from an adult DC; in contrast, the blue arrows in (F–F’’’) indicated another OHC-like cell derived from adult PCs. Yellow arrows in (F–F’’’): Prestin+/Tdtomato- endogenous OHC appearing in the SC layer. Prestin expression in the new OHC-like cells was noted to be lower than that in wild-type endogenous OHCs. (G) Quantification of the number of OHC-like cells throughout the cochlear turns in the Fgfr3-Atoh1-Ikzf2 mice. Data was presented as mean ± SEM (n = 3); no statistical difference was detected between regions (Student’s t test). OHC-like cells were reliably observed in all animals, but cell numbers were low and showed large variations. (H) Summary of reprogramming outcomes in the three mice models. OHC-like cells were present only when both Atoh1 and Ikzf2 were concurrently reactivated in adult cochlear SCs. Scale bar: 20 μm.
-
Figure 2—source data 1
It contained the numerical values of the data plotted in the graph of Figure 2G.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig2-data1-v2.xlsx
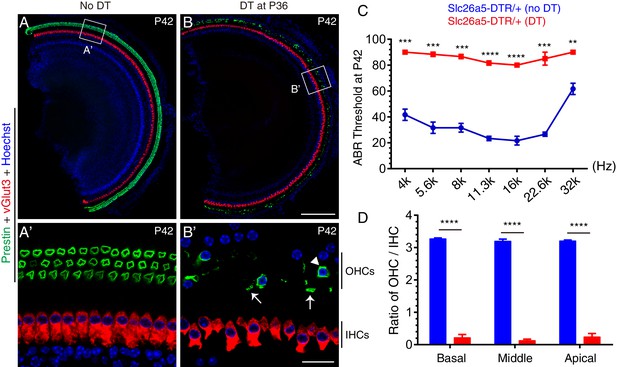
Targeted lesioning of adult endogenous OHCs via diphtheria toxin treatment.
(A–B’) Slc26a5DTR/+ mice were treated without (A–A’, control) or with (B-B’, experimental) diphtheria toxin (DT) at P36 and analyzed at P42. Samples were co-stained against Prestin and vGlut3. (A’) and (B’): magnified images of boxed region in (A) and (B). DT treatment led to rapid OHC death within 6 days; only a few OHCs remained (arrowhead in B’); debris of dying OHCs were frequently detected (arrows in B’). Much of the green signal in (B) came from the dying OHC debris. (C) Auditory brainstem response (ABR) threshold measurement of Slc26a5DTR/+ mice treated with (red line) or without (blue line) DT. After DT treatment ABR thresholds were significantly higher throughout the frequency ranges. (D) Ratios of OHCs to IHCs within a same scanning area in control mice (blue) and experimental mice (red); relative OHC numbers were significantly reduced in the experimental mice. Data was presented as mean ± SEM (n = 3). Student’s t-test were run for statistical testing. **p<0.01, ***p<0.001, ****P<0.0001. Scale bars: 200 μm (B), 20 μm (B’).
-
Figure 3—source data 1
It contained the numerical values of the ABR thresholds presented in Figure 3C.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig3-data1-v2.xlsx
-
Figure 3—source data 2
It contained the numerical value of the ratio of OHCs to IHCs described in Figure 3D.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig3-data2-v2.xlsx
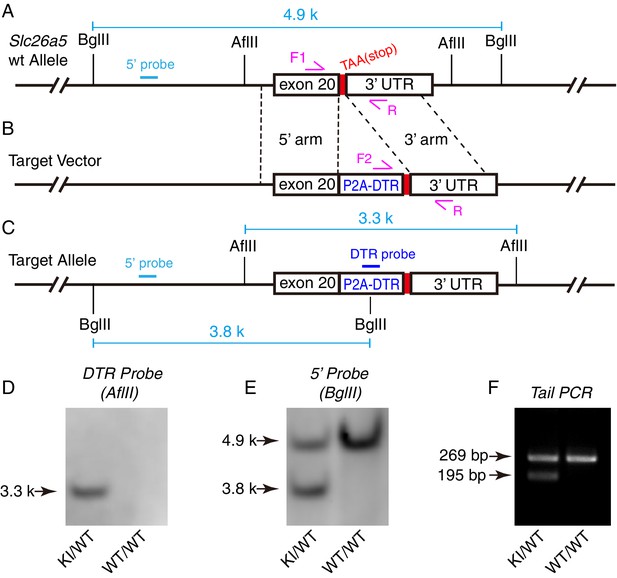
Generation of the Slc26a5DTR/+ knockin mouse model.
(A) Wild-type Slc26a5 (Prestin) allele. (B) Illustration of the target vector. P2A-DTR (diphtheria toxin receptor) was inserted immediately before the TAA stop codon. This construct allowed for DTR expression to be driven by the endogenous Slc26a5 enhancer/promoter. (C) Illustration of the Slc26a5 allele after successful gene targeting. (D–E) Southern blotting assay testing for the presence of the internal DTR probe (D) and 5ʹ-end probe (E). (F) Results of PCR genotyping tail DNA extracted from heterozygous (KI/WT) and wild-type (WT/WT) mice. WT mice displayed a single 269 bp band, whereas heterozygous mice showed a 195- and a 269bp band.
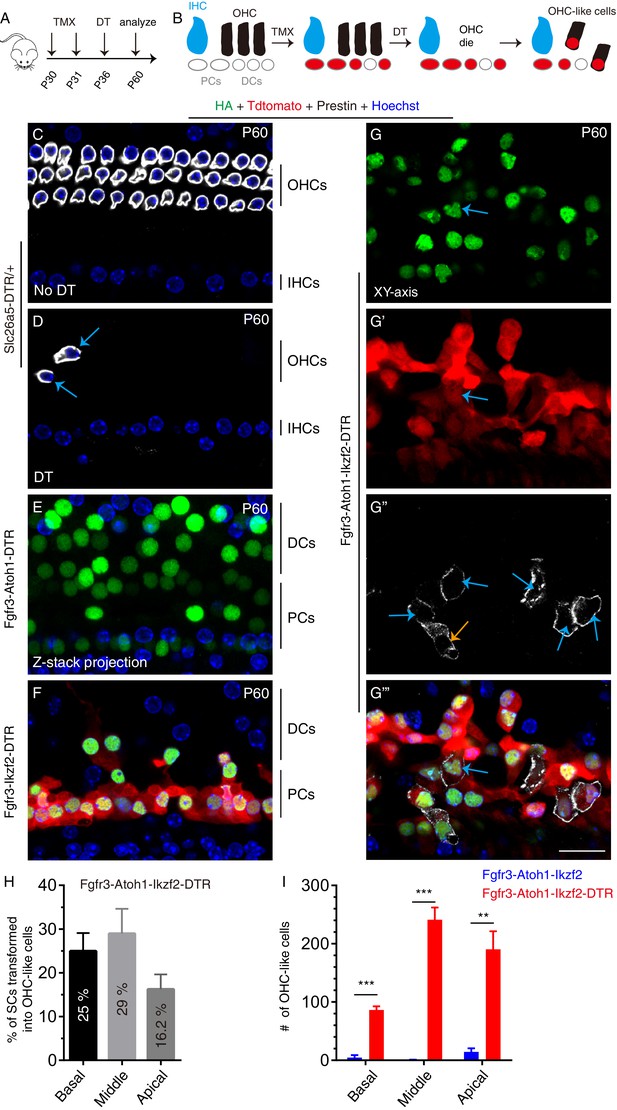
Lesion to endogenous OHCs enhances reprogramming efficiency of adult SCs by Atoh1 and Ikzf2.
(A) Three distinct mice models were subject to tamoxifen (TMX) treatment at P30 and P31, followed by DT treatment at P36 and analysis at P60. (B) Illustration of key cellular events occurring during the experiment: induction of Atoh1 and Ikzf2 in adult PCs and DCs, which were permanently labeled with Tdtomato for the subsequent fate-mapping analysis. (C–G’’’) Triple labeling against HA, Tdtomato, and Prestin in four models: (1) Slc26a5DTR/+ (C, D), (2) Fgfr3-iCreER+; CAG-LSL-Atoh1+; Slc26a5DTR/+ (Fgfr3-Atoh1-DTR; E), (3) Fgfr3-iCreER+; Rosa26CAG-LSL-Ikzf2/+; Slc26a5DTR/+ (Fgfr3-Ikzf2-DTR; F), and (4) Fgfr3-iCreER+; CAG-LSL-Atoh1+; Rosa26CAG-LSL-Ikzf2/+; Slc26a5DTR/+ (Fgfr3-Atoh1-Ikzf2-DTR; G–G’’’). Relative to Slc26a5DTR/+ mice not treated with DT (C), DT-treated Slc26a5DTR/+ mice harbored fewer normal Prestin+ OHCs at P60 (arrows in D). Debris of dying OHCs had disappeared. No OHC-like cells were observed in the first three models, but Tdtomato+/HA+/Prestin+ OHC-like cells (orange and blue arrows in G’’) were present in the Fgfr3-Atoh1-Ikzf2-DTR model (G-G’’’). (H) Percentages of SCs that were transformed into OHC-like cells at different cochlear turns in Fgfr3-Atoh1-Ikzf2-DTR mice. (I) Comparison of OHC-like cell numbers between Fgfr3-Atoh1-Ikzf2-DTR and Fgfr3-Atoh1-Ikzf2 models (without damaging adult wild-type OHCs). Fgfr3-Atoh1-Ikzf2-DTR mice harbored considerably more OHC-like cells than Fgfr3-Atoh1-Ikzf2 mice. Data was presented as mean ± SEM (n = 3). Student’s t test was used for statistical analysis. **p<0.01, ***p<0.001. Scale bar: 20 μm.
-
Figure 4—source data 1
It contained the numerical values of the data plotted in the graph of Figure 4H.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig4-data1-v2.xlsx
-
Figure 4—source data 2
It contained the numerical values of the data plotted in Figure 4I.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig4-data2-v2.xlsx
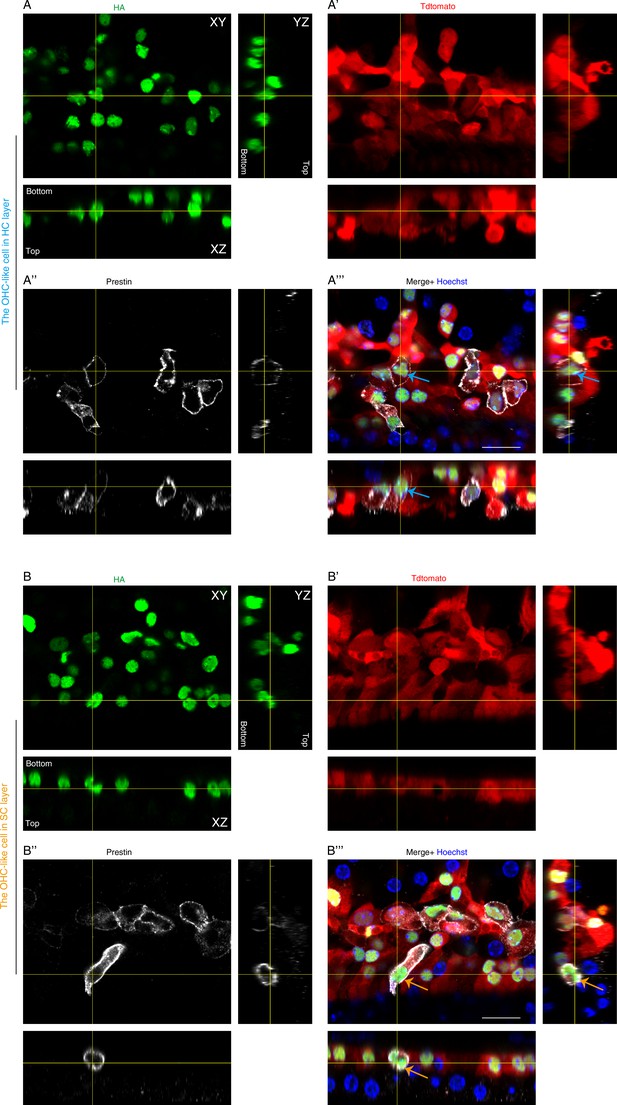
Visualization of OHC-like cells in orthogonal projections.
Triple staining against HA (A, B), Tdtomato (A’, B’), and Prestin (A’’, B’’) of cochlear samples from Fgfr3-iCreER+; CAG-LSL-Atoh1+; Rosa26CAG-LSL-Ikzf2/+; Slc26a5DTR/+ (Fgfr3-Atoh1-Ikzf2-DTR) mice. Blue arrows in the merged image (A’’’) marked the same OHC-like cell (blue arrow in Figure 4G’’’), which is located in the HC layer according to its nuclei position. Orange arrows in the merged image (B’’’) marked one OHC-like cell, which is located in the SC layer according to its nuclei position. Scale bar: 20 μm (A’’’, B’’’).
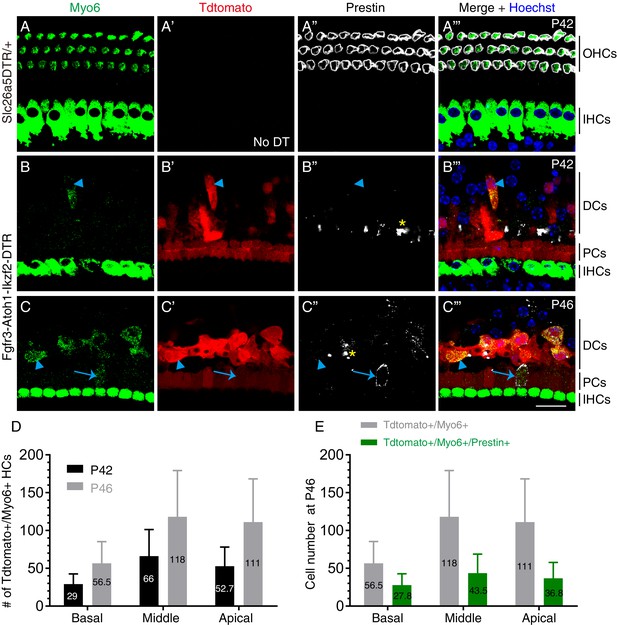
Nascent HCs emerged at P42 and OHC-like cells at P46.
(A–C’’’) Triple immunolabeling against early HC marker Myo6, Tdtomato, and Prestin in control P42 Slc26a5DTR/+ mice without DT treatment (A–A’’’) and in experimental P42 (B–B’’’) and P46 (C–C’’’) mice. Arrowheads: nascent HCs that were Tdtomato+/Myo6+/Prestin-; arrows: one Tdtomato+/Myo6+/Prestin+ OHC-like cell; asterisks: debris of dying endogenous OHCs. (D) Comparison between numbers of new HCs at P42 vs P46 in different cochlear turns in Fgfr3-Atoh1-Ikzf2-DTR mice. Data was presented as mean ± SEM. More new HCs tended to be present at P46 than P42, but statistical difference was not reached due to high variance. (E) Comparison between number of nascent HCs vs OHC-like cells within different cochlear turns in P46 Fgfr3-Atoh1-Ikzf2-DTR mice; Data was presented as mean ± SEM. Scale bar: 20 μm.
-
Figure 4—figure supplement 2—source data 1
It contained the number of new HCs at P42 and P46 presented in the graph of Figure 4—figure supplement 2D.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig4-figsupp2-data1-v2.xlsx
-
Figure 4—figure supplement 2—source data 2
It contained the numbers of new HCs at P46 presented in the graph of Figure 4—figure supplement 2E.
Nascent HCs and OHC-like cells are shown separately.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig4-figsupp2-data2-v2.xlsx
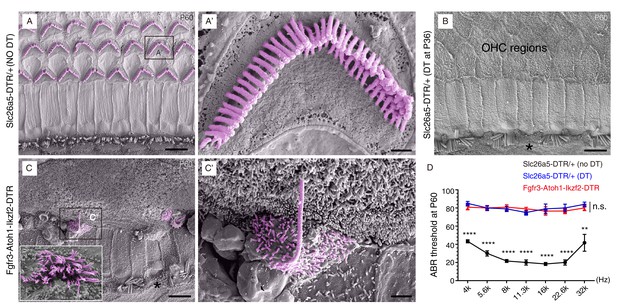
Hair bundles were present in OHC-like cells.
Scanning electron microscopy (SEM) analysis of samples from three different mouse models at P60. (A–A’) OHCs harbored V- or W-shaped hair bundles in Slc26a5DTR/+ mice not treated with DT. (A’): high-magnification view of boxed region in (A). (B) Majority of OHCs had disappeared by P60 in Slc26a5DTR/+ mice upon DT treatment at P36. Black asterisk: one IHC that was absent. (C) Immature hair bundles were frequently detected in the Fgfr3-iCreER+; CAG-LSL-Atoh1+; Rosa26CAG-LSL-Ikzf2/+; Slc26a5DTR/+ (Fgfr3-Atoh1-Ikzf2-DTR) model, but not in (A) and (B). These hair bundles were thought to originate from OHC-like cells. (C’): high-magnification view of boxed region in (C). Inset in C: instance of occasional hair bundles. Black asterisk: one IHC that was missing. (D) ABR measurements of the three mice models. Relative to the untreated Slc26a5DTR/+ control mice (black line, n = 3), the ABR thresholds of Slc26a5DTR/+ treated with DT (blue line, n = 5) and in Fgfr3-Atoh1-Ikzf2-DTR mice (red line, n = 6) were significantly higher. The blue and red lines showed no statistically significant difference at any frequency (n.s.). Data was presented as mean ± SEM. Student’s t-test was used for statistical testing. **p<0.01, ****p<0.0001. Scale bars: 5 μm (A–C), 1 μm (C’), and 500 nm (A’).
-
Figure 5—source data 1
It contained the numerical values of the ABR thresholds described in Figure 5D.
- https://cdn.elifesciences.org/articles/66547/elife-66547-fig5-data1-v2.xlsx
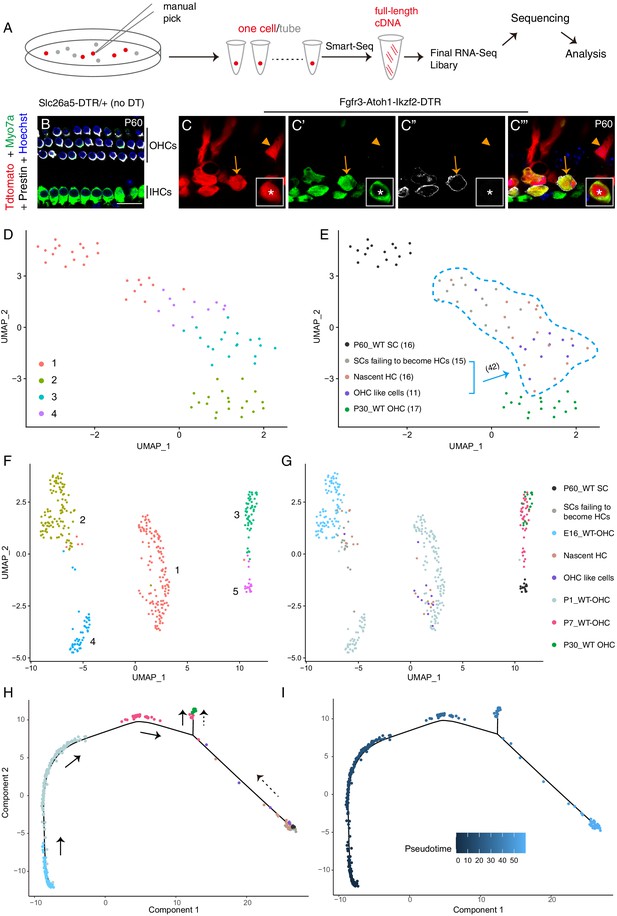
OHC-like cells were most akin to ~ P1 wild-type OHCs.
(A) Illustration of the single cell RNA-Seq experimental pipeline. (B–C’’’) Triple labeling against Myo7a (maker for HCs), Prestin, and Tdtomato in cochlear samples from control (B) and Fgfr3-Atoh1-Ikzf2-DTR (C) mice at P60. Arrows: Tdtomato+/Myo7a+/Prestin+ OHC-like cells; arrowheads: Tdtomato+/Myo7a-/Prestin− cell, defined as SCs failing to become HCs; asterisks: nascent HC that was Tdtomato+/Myo7a+/Prestin− (inset in C–C’’’). (D–E) UMAP embedding of RNA-seq data revealed four main clusters (D). The same plot was presented with the five different cell types included in the data labelled separately (E). The 42 Tdtomato+ cells within the light-blue dotted lines (E) originated from the P60 Fgfr3-Atoh1-Ikzf2-DTR mice. When unsupervised clustering was performed over all 42 Tdtomato+ cells, three clusters were identified. (F–G) UMAP analysis of cells in (D–E) as well as wild-type OHCs at E16, P1, and P7 (E16_WT OHCs, P1_WT OHCs, and P7_WT OHCs; further information was presented in Figure 6—figure supplement 5). OHC-like cells were overlapped with P1_WT OHCs. (H–I) Trajectory analysis of all cells in (F–G). Black arrows in (H) represented the developmental trajectory of endogenous OHCs between E16 and P30, whilst the black dotted arrows marked the reprogramming trajectory from endogenous adult SCs to endogenous adult OHCs. Scale bar: 20 μm.
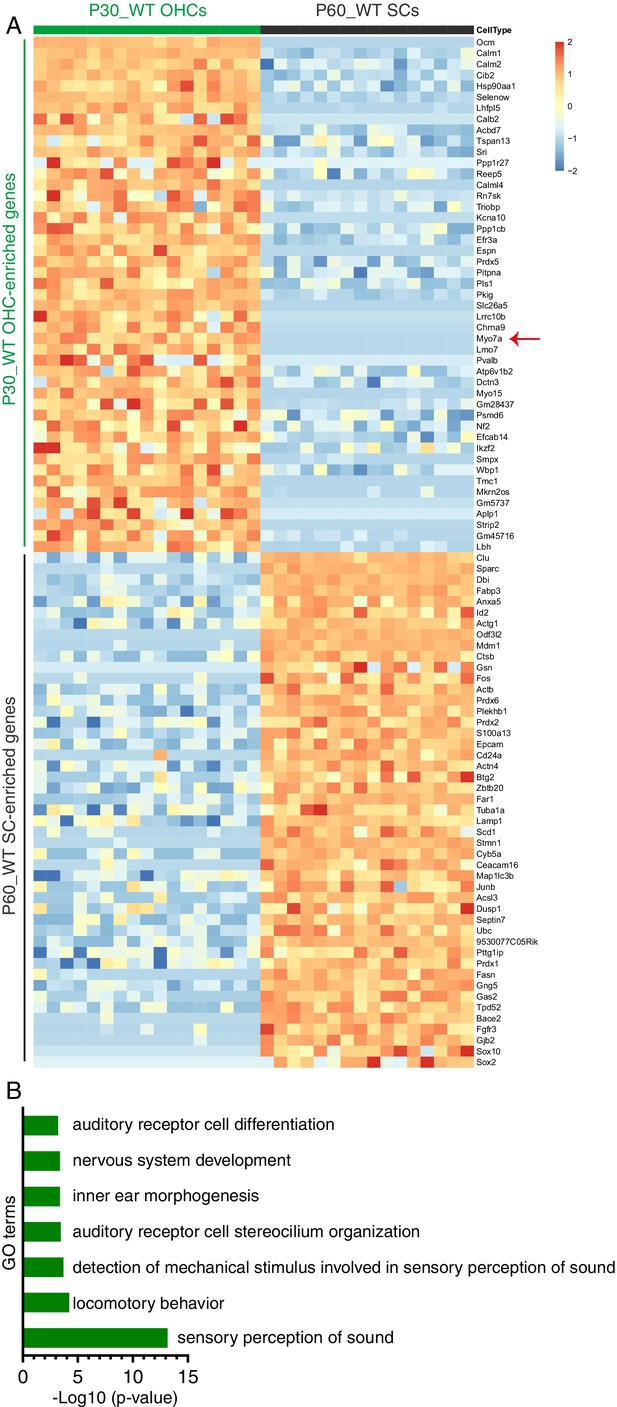
Transcriptomic comparison between adult wild-type OHCs and SCs.
17 wild-type OHCs were picked manually at P30 (P30_WT OHCs) and 16 SCs at P60 (P60_WT SCs) and single cell RNA-Seq was performed following SmartSeq methodology. (A) The top enriched genes in P30_WT OHCs and P60_WT SCs. The early pan-HC-specific gene, Myo7a (red arrow), OHC-specific genes, Ocm, Lbh, Slc26a5 (Prestin), and Ikzf2 were classified as OHC genes. Similarly, SC markers, Sox2, Sox10, Bace2, and Ceacam16 were defined as SC genes. The complete gene list can be found in Supplementary file 1. (B) Results of Gene Ontology (GO) enrichment analysis of genes enriched in OHCs. The gene lists associated with each GO category can be found in Supplementary file 2.
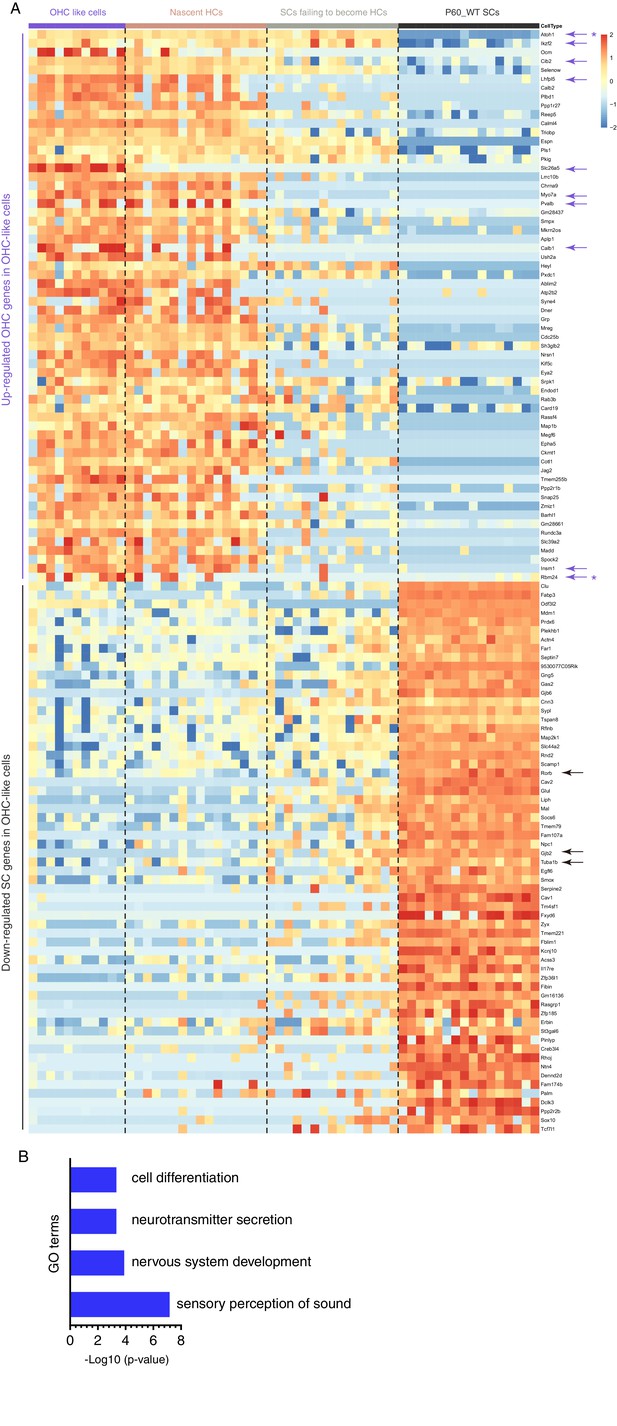
OHC-like cells upregulated 729 OHC genes and downregulated 331 SC genes.
Single cell RNA-Seq was applied to 42 Tdtomato+ cells from P60 Fgfr3-Atoh1-Ikzf2-DTR mice. Cells were categorized as OHC-like cells (Tdtomato+/Myo7a+/Prestin+), nascent HCs (Tdtomato+/Myo7a+/Prestin-) and SCs failing to become HCs (Tdtomato+/Myo7a-/Prestin-). We compared the transcriptomic profiles between OHC-like cells and P60_WT SCs. (A) Relative to P60_WT SCs, the OHC-or SC-genes that were significantly upregulated or down-regulated, respectively, in OHC-like cells were presented. The full list of those genes was presented in Supplementary file 4. Nascent HCs and SCs failing to become HCs were included as references only. Note that Rbm24 (purple arrow and asterisk) was also included to highlight its expression in OHC-like cells, although its p-value is between 0.01 and 0.001. In addition, Atoh1 (purple arrow and asterisk), which was not a gene enriched in adult OHCs, was also included to show its permanent expression in OHC-like cells. (B) GO analysis of all genes (without overlapping with OHC genes) that were significantly upregulated in OHC-like cells, relative to P60_WT SCs. The gene lists of each GO category were presented in Supplementary file 3.
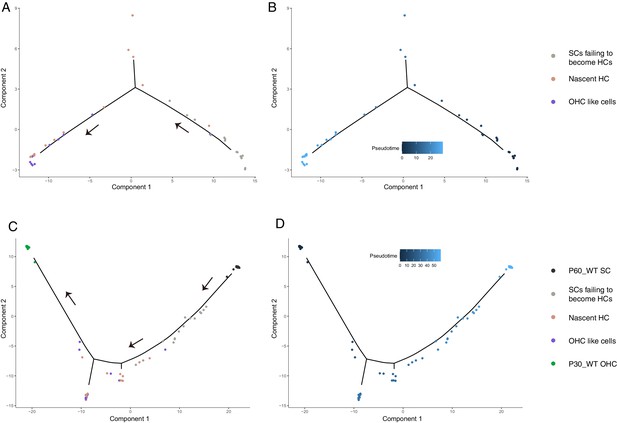
Two trajectory analyses by different cell pooling.
(A, B) Trajectory analysis of the 42 Tdtomato+ cells from P60 Fgfr3-Atoh1-Ikzf2-DTR mice. Nascent HCs and OHC-like cells were found to be closer to each other compared to SCs failing to become HCs. (C, D) Trajectory analysis which also included WT_P30 OHCs and WT_P60 SCs. Akin to the results above, nascent HCs and OHC-like cells were intermingled and neighbored WT_P30 OHCs, whereas SCs failing to become HCs were closer to WT_P30 SCs. Arrows in (A and C) represented reprogramming direction from SCs to OHCs. Trajectory analysis including embryonic and neonatal OHCs are presented in Figure 6H and I.
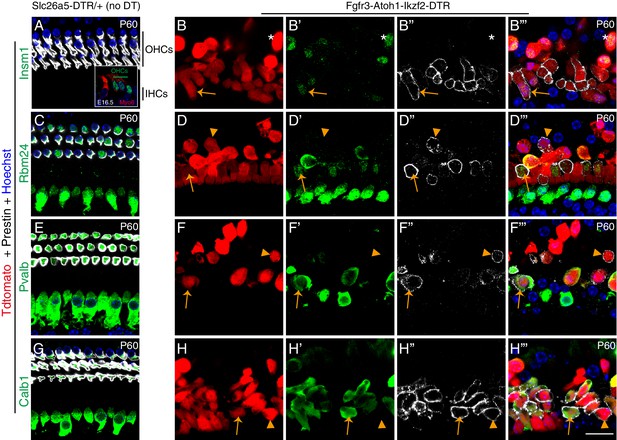
Heterogeneous expression of OHC genes across OHC-like cells.
Cochlear samples (P60) from control Slc26a5DTR/+ mice not treated with DT (A, C, E, G) and experimental Fgfr3-Atoh1-Ikzf2-DTR mice treated with DT (B–B’’’, D–D’’’, F–F’’’, H–H’’’). Samples were triple immunolabeled against Tdtomato (red), Prestin (white in images), and a third different marker (antibody staining, shown in green): Insm1 (A–B’’’), Rbm24 (C–D’’’), Pvalb (E–F’’’), or Calb1 (G–H’’’). Asterisks in (B–B’’’): cell that was Tdtomato+/Insm1+ but did not express Prestin; arrows in (B–B’’’): cell that was Tdtomato+/Insm1+/Prestin+. Inset in (A): cochlear cryosection sample at E16.5 stained for Myo6 and Insm1. Insm1 expression was detected in OHCs but not in IHCs, which was presented as another control to confirm the specificity of the Insm1 antibody. Arrows in (D–D’’’, F–F’’’, H–H’’’): triple-positive cells; arrowheads in (D–D’’’, F–F’’’, H–H’’’): cells that were Tdtomato+/Prestin+ but did not express Rbm24 (D–D’’’), Pvalb (F–F’’’), or Calb1 (H–H’’’). Prestin labelling in (B’’, D’’, F’’, H’’) was shown at a higher gain than in (A, C, E, G) for the sake of visualization clarity; Prestin level in OHC-like cells was considerably lower than that in wild-type endogenous OHCs. Scale bar: 20 μm.
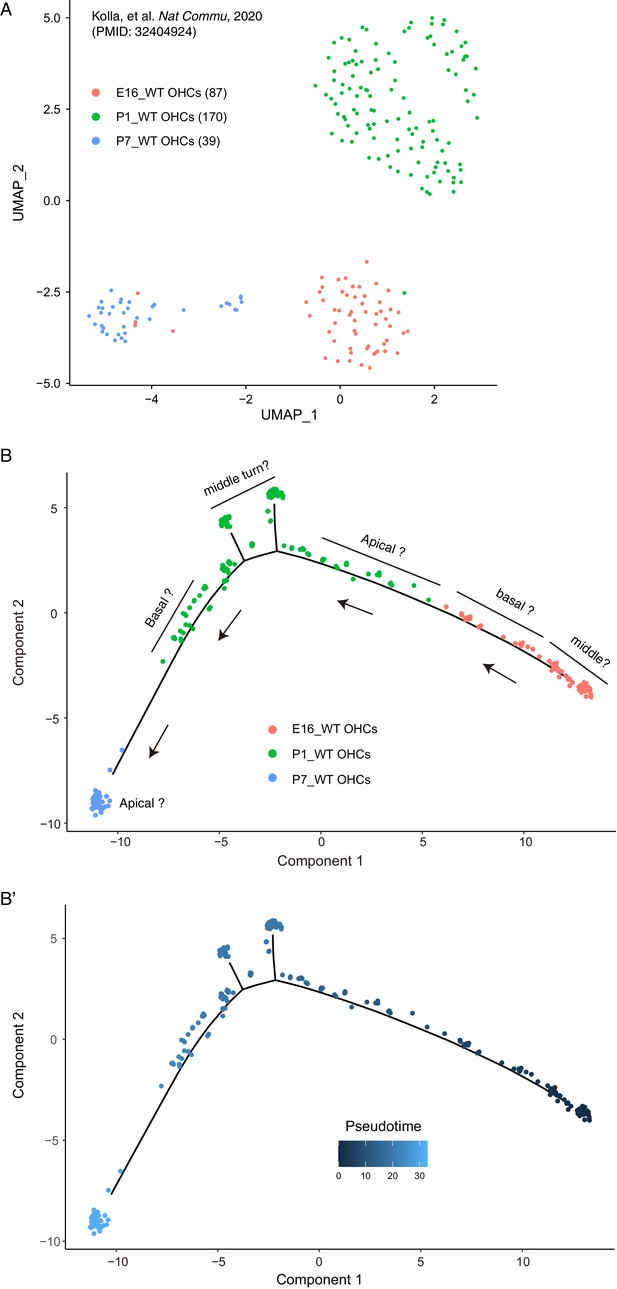
Trajectory analysis of wild type differentiating OHCs.
(A) UMAP analysis of wild-type OHCs at E16, P1, and P7. Those OHCs were clustered primarily according to age. (B–B’) Trajectory analysis of wild-type OHCs at three different ages by using Monocle. These data were included in the analyses presented in Figure 6.
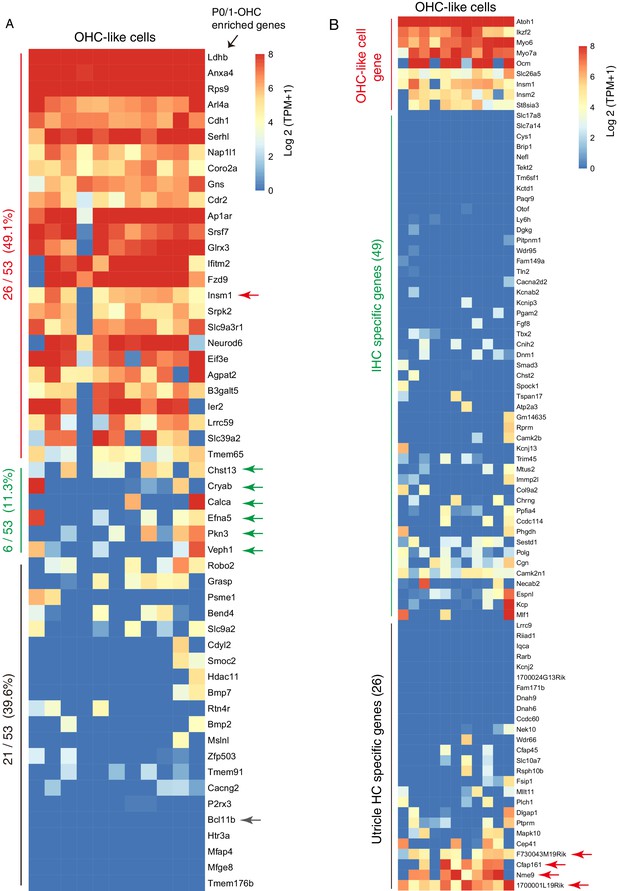
OHC-like cells primarily expressed neonatal OHC, but minimally expressed neonatal IHC or utricle HC genes.
(A) TPM values of 53 neonatal OHC genes. 26/53 were highly expressed in OHC-like cells, including Insm1 (red arrow). The remaining genes were expressed at low levels, including Bcl11b (gray arrow). Green arrows labelled six genes with average TPM >16, but that were nonetheless defined as low expressing genes as six or fewer OHC-like cells expressed them with TPM >16. (B) TPM values of 49 neonatal IHC and 26 neonatal utricle HC gene with red arrows highlighting four neonatal utricle HC genes that were highly expressed in OHC-like cells.
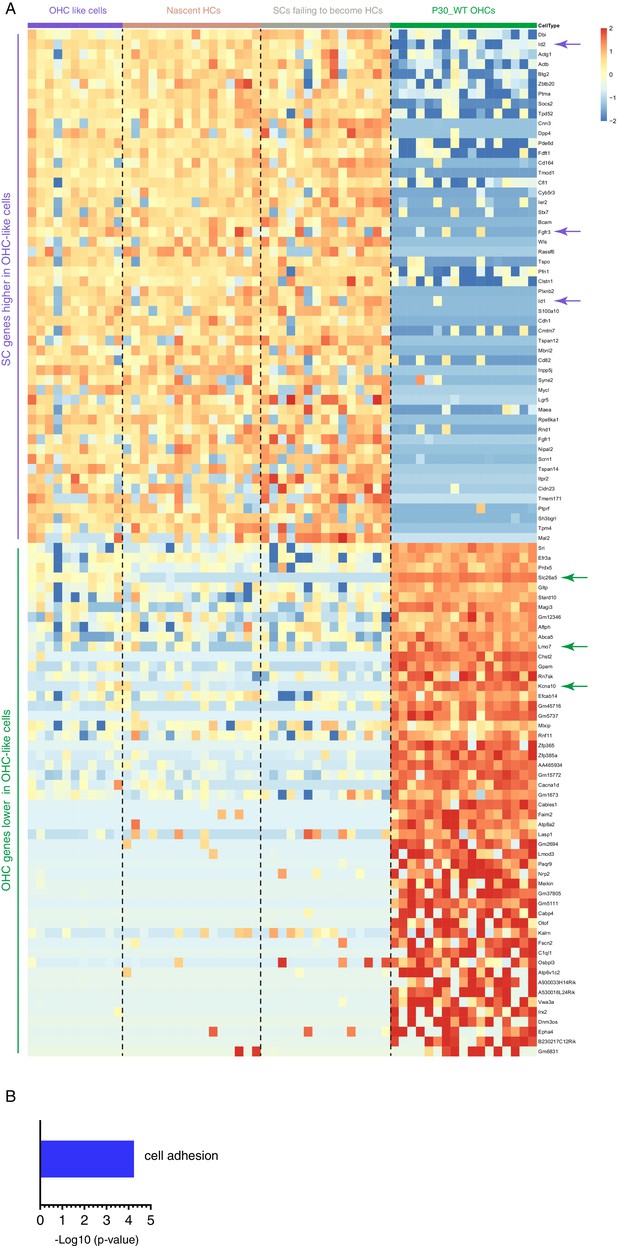
Transcriptomic differences between OHC-like cells and mature OHCs.
(A) Most differentially expressed genes (DEGs) between OHC-like cells and adult endogenous OHCs at P30. Nascent HCs and SCs failing to become HCs were included as references only. SC genes such as Id1, Id2, and Fgfr3 (purple arrows), were still higher in OHC-like cells than in adult endogenous OHCs. Conversely, OHC genes encoding functional proteins in mature OHCs such as Slc26a5 (Prestin), Lmo7, and Kcna10 (green arrows) were still lower in OHC-like cells than in adult endogenous OHCs. The complete gene lists were presented in Supplementary file 7. (B) GO analysis of genes higher in OHC-like cells than that in adult endogenous OHCs. The gene lists of the GO category were included in Supplementary file 8.
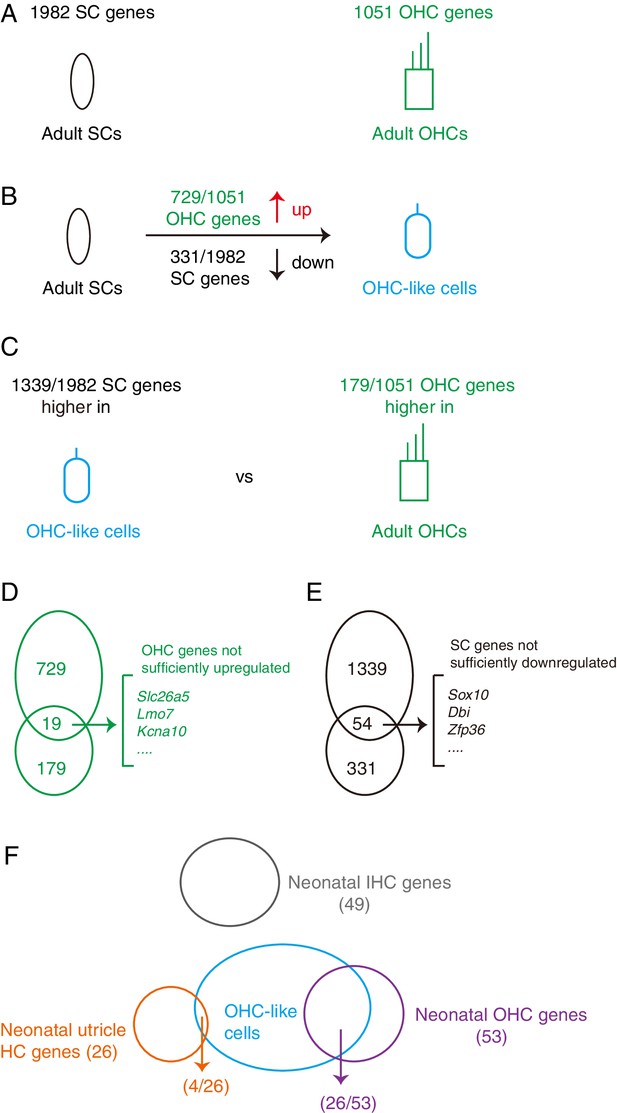
Summary of the differentiation status of the OHC-like cells.
(A) Comparison of transcriptomic profiles revealed 1982 SC and 1,051 OHC differentially expressed genes. (B) Compared to adult SCs, OHC-like cells upregulated 729 OHC genes and downregulated 331 SC genes, thus having become more OHC-like and less SC like. (C) Compared to adult OHCs, OHC-like cells expressed 1339 SC genes at higher levels, and 179 OHC genes at lower levels, showing that OHC status had not yet been achieved. (D) 19 OHC genes intersected between the upregulated 729 OHC genes (as compared to adult SCs) and the under-expressed 179 OHC genes (as compared to adult OHCs), suggesting that although these genes were up-regulated in OHC-like cells, their expression levels were still lower than that expected for adult OHCs. (E) Similar to (D), 54 SC genes overlapped between 1339 and 331 SC genes that were found to be at higher levels than expected for adult OHCs, but at lower levels than expected for adult SCs. These SC genes had thus successfully been downregulated, but not sufficiently so to reach the low levels in adult OHCs. (F) OHC-like cells expressed 26 of the 53 neonatal OHC genes, but only expressed 4 of the 26 neonatal utricle HC genes and did not express any neonatal IHC genes.
Videos
The confocal data presented in Figure 4G–G’’’ and Figure 4—figure supplement 1A–A''' was converted to Imaris format and loaded in the Imaris software to more readily visualize OHC-like cells using orthogonal projections of the data.
Red, green, and white colors represented Tdtomato signal and Prestin and HA labelling respectively. OHC-like cells were defined as triple-positive cells.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus. musculus) | FVB-Tg(Atoh1-cre/ERT)1Sbk/Mmnc | MMRRC | Stock#:029581-UNC | RRID:MMRRC_029581-UNC |
| Genetic reagent (Mus. musculus) | Tg(Fgfr3-icre/ERT2)4-2Wdr | Jackson Lab | Stock#:025809 | RRID:IMSR_JAX:025809 |
| Genetic reagent (Mus. musculus) | Ai9 | Jackson Lab | Stock#:007909 | RRID:IMSR_JAX:007909 |
| Genetic reagent (Mus. musculus) | Slc26a5CreER/+ | Fang et al., 2012 | PMID:21954035 | |
| Genetic reagent (Mus. musculus) | CAG-LSL-Atoh1+ | Liu et al., 2012a | PMID:22573682 | |
| Genetic reagent (Mus. musculus) | Rosa26CAG-LSL-Ikzf2/+ | Jackson Lab | Stock#:036272 | Knock-in mouse line that will be available soon |
| Genetic reagent (Mus. musculus) | Slc26a5DTR/+ | Jackson Lab | Stock#:036271 | Knock-in mouse line that will be available soon |
| Antibody | anti-HA (Rat monoclonal) | Roche | Cat.#:11867423001 | RRID:AB_390918 |
| Antibody | anti-Prestin (Goat polyclonal) | Santa Cruz | Cat.#:sc-22692 | RRID:AB_2302038 |
| Antibody | anti-Myosin-VI (Rabbit polyclonal) | Proteus Bioscience | Cat.#:25–6791 | RRID:AB_2314836 |
| Antibody | anti-Myosin-VIIa (Rabbit polyclonal) | Proteus Bioscience | Cat.#:25–6790 | RRID:AB_10015251 |
| Antibody | anti-Insm1 (Guinea pig) | A kind gift from Dr. Shiqi Jia and Dr. Carmen Birchmeier | Jia et al., 2015 | PMID:25828096 |
| Antibody | anti-Parvalbumin (Mouse monoclonal) | Sigma-Aldrich | Cat.#:P3088 | RRID:AB_477329 |
| Antibody | anti-Rbm24 (Rabbit polyclonal) | Proteintech | Cat.#:18178–1-AP | RRID:AB_2878513 |
| Antibody | anti-Calbindin (Rabbit monoclonal) | Sigma-Aldrich | Cat.#:C9848 | RRID:AB_2314067 |
| Antibody | anti-vGlut3 (Rabbit polyclonal) | Synaptic System | Cat.#:135,203 | RRID:AB_887886 |
| Antibody | anti-Ocm (Rabbit) | Swant | Cat.#:OMG4 | RRID:AB_10000346 |
| Antibody | anti-Slc7a14 (Rabbit polyclonal) | Sigma-Aldrich | Cat.#:HPA045929 | RRID:AB_2679501 |
| Antibody | anti-Ctbp2(Mouse monoclonal) | BD Biosciences | Cat.#:612,044 | RRID:AB_399431 |
| Commercial assay, kit | SMART-Seq HT Kit | Takara | Cat.#:634,437 | For reverse-transcription and cDNA amplification |
| Commercial assay, kit | TruePrep DNA Library Prep Kit V2 for Illumina | Vazyme | Cat.#:TD503-02 | For sequencing library construction |
| Commercial assay, kit | TruePrep Index Kit V2 for Illumina | Vazyme | Cat.#:TD202 | For sequencing library construction (index) |
Additional files
-
Supplementary file 1
List of all genes that were expressed at higher (P < 0.001) levels in WT_P30 OHCs as compared to WT_P60 SCs, and vice versa.
The most differentially expressed genes were listed in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp1-v2.xlsx
-
Supplementary file 2
GO analysis of genes that were significantly enriched in WT_P30 OHCs, relative to WT_P60 SCs.
Top GO terms were shown in Figure 6—figure supplement 1B.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp2-v2.xlsx
-
Supplementary file 3
GO analysis of genes that were significantly enriched in OHC-like cells, relative to WT_P60 SCs.
Top GO terms were presented in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp3-v2.xlsx
-
Supplementary file 4
List of genes expressed at higher level in OHC-like cells relative to WT_P60 SCs, and vice versa.
In addition, these genes which were overlapped with OHC or SC-enriched genes (Figure 6—figure supplement 2A) are also included in a separate tab in the file.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp4-v2.xlsx
-
Supplementary file 5
TPM values in OHC-like cells of 53 neonatal OHC genes, as presented in Figure 6—figure supplement 6A.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp5-v2.xlsx
-
Supplementary file 6
TPM values in OHC-like cells of 9 pan-HC or OHC-enriched genes, 49 neonatal IHC genes, and 26 neonatal utricle HC genes, as presented in Figure 6—figure supplement 6B.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp6-v2.xlsx
-
Supplementary file 7
List of all genes that were differently expressed between OHC-like cells and WT_P30 OHCs.
In addition, we also included those differently expressed genes that were overlapped with SC or OHC genes (presented in Figure 6—figure supplement 7A), as well as the 19 OHC genes (not sufficiently upregulated) or 54 SC genes (not sufficiently downregulated) in OHC-like cells.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp7-v2.xlsx
-
Supplementary file 8
GO analysis of genes that were significantly enriched in OHC-like cells relative to WT_P30 OHCs.
The results were presented in Figure 6—figure supplement 7B.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp8-v2.xlsx
-
Supplementary file 9
Genotyping primers, and sizes of PCR amplicons for the various knock-in and transgenic mouse strains used in this study.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp9-v2.xlsx
-
Supplementary file 10
Detailed information of all the 75 single cells that were subject to smartseq analysis.
Notably, the OHC-like cells, nascent HCs and SCs failing to become HCs were picked from the same model and labelled exp-1 to exp-42 in the file. Their cell identity was defined after bioinformatic analysis.
- https://cdn.elifesciences.org/articles/66547/elife-66547-supp10-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66547/elife-66547-transrepform1-v2.docx





