Structural intermediates observed only in intact Escherichia coli indicate a mechanism for TonB-dependent transport
Figures
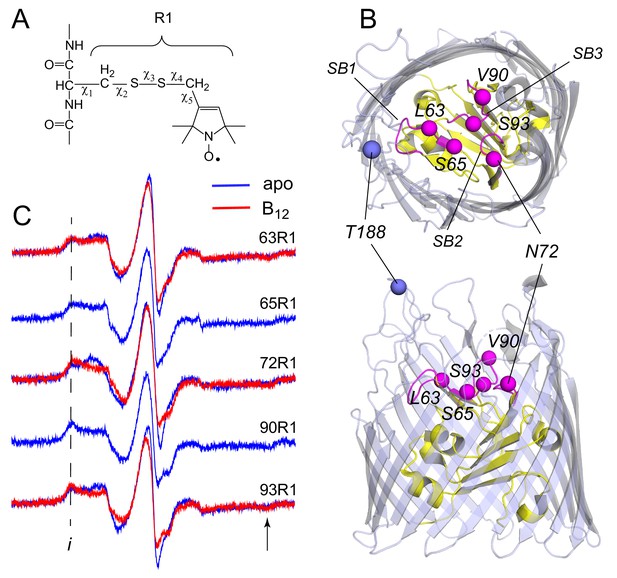
Electron paramagnetic resonance (EPR) spectra obtained in vivo from spin labeled core sites on extracellular face of the BtuB.
The spin labeled side chain (A) R1 was attached to sites on the extracelluar core of BtuB shown in (B). BtuB is shown in both extracellular (top) and side views (PDB ID: 1NQH), with the core in yellow and barrel in light blue. The Cα atom on site 188 on the second extracellular loop is shown, which is used as a reference point for measurements to the core. Labeled Cα sites in the core are shown in magenta, along with substrate binding loops SB1, SB2, and SB3. In (C) EPR spectra are shown in the absence (apo state in blue) and presence of substrate (vitamin B12 bound state in red). No change in the spectrum is observed at sites 65 and 90 upon the addition of substrate. The spectra are characterized by well-defined hyperfine extrema ((i) and location of arrow), where the difference between these points is approximately 69 Gauss. This value is close to 2Azz, twice the value of the hyperfine tensor, and indicates that the label is immobilized on the ns time scale. Spectra are the sum of 10–100 Gauss scans.
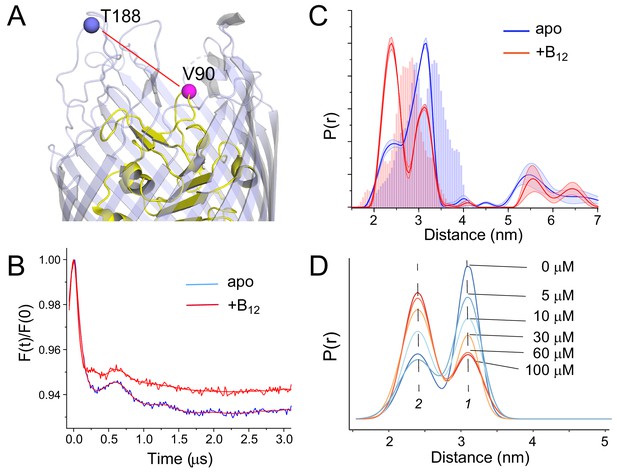
Substrate-dependent shifts are detected at the apex of substrate binding loop 3 (SB3) in whole cells.
A side view of BtuB (A) with the locations of site 90 in SB3 of the core domain (yellow) and site 188 near the apex of the 3/4 extracellular loop (light blue). (B) Background corrected double electron-electron resonance (DEER) data for the apo (blue) and substrate bound states (red) of the V90R1-T188R1 spin pair, where the red traces represent the fits to the data. The resulting distance distributions are shown in (C) where the histograms represent predicted distance ranges obtained from the in surfo crystal structures 1NQG (blue) and 1NQH (red) using the software package MMM (Jeschke, 2018). (D) This structural change titrates between two states (labeled 1 and 2) with the addition of vitamin B12. The conversion between states saturates at concentrations above 60 µM. For the distributions shown in (C), data were analyzed using LongDistances v932 using the model-free fitting mode, whereas for distributions in (D), data were fit to a two-Gaussian model where position, width, and amplitude were variables in the fit. Errors in the fit to the distributions in (D) are shown in Supplementary file 1.
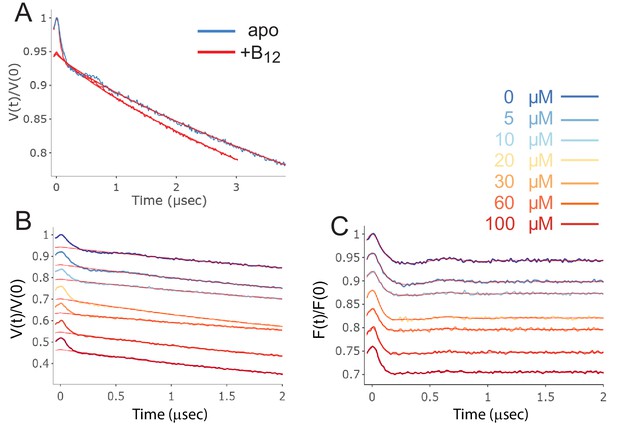
Raw and background corrected double electron-electron resonance (DEER) data for V90R1-T188R1.
In (A) raw DEER data, V(t)/V(0), along with the background form factor that was subtracted (straight red lines). In (B) raw DEER data as a function of substrate (vitamin B12) concentration (see inset). In (C) background corrected DEER data, (F(t)/F(0)), as a function of vitamin B12 concentration (see inset).
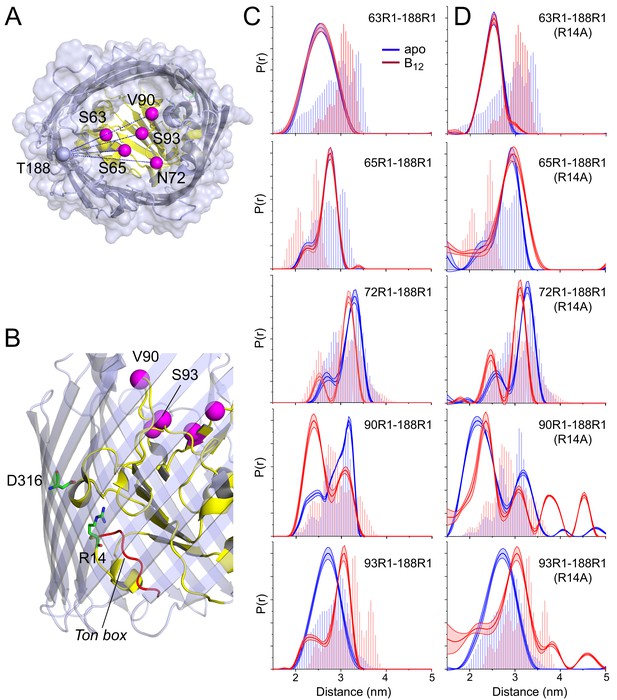
Substrate-dependent conformational shifts are limited to substrate binding loop 3 (SB3) and are altered by mutation of the R14-D316 ionic lock.
(A) Top view of BtuB (PDB ID: 1NQH) showing the locations of the hatch sites relative to the reference site, 188, in the 3/4 extracellular loop. In (B) the location of the R14-D316 ionic interaction between the core and the barrel is also shown along with the Ton box (red). (C) Distance distributions obtained for hatch: barrel pairs in the apo state (blue) and with substrate (red). (D) Distance distributions obtained for hatch: barrel pairs in the apo state (blue) and with substrate (red) in the presence of the R14A mutation. Both V90R1-T188R1 and S93R1-T188R1 spin pairs show additional distances at 3.8 and 4.5 nm in the presence of R14A. Data were analyzed using LongDistances v932 and the model-free fitting regime. The shaded error bands in (C) and (D) represent variation due to background noise, start time, dimensionality, and regularization. Histograms are predicted distances generated from the in surfo crystal structures PDB ID: 1NQG (light blue) and PDB ID: 1NQH (light red) using the software package MMM (Jeschke, 2018).
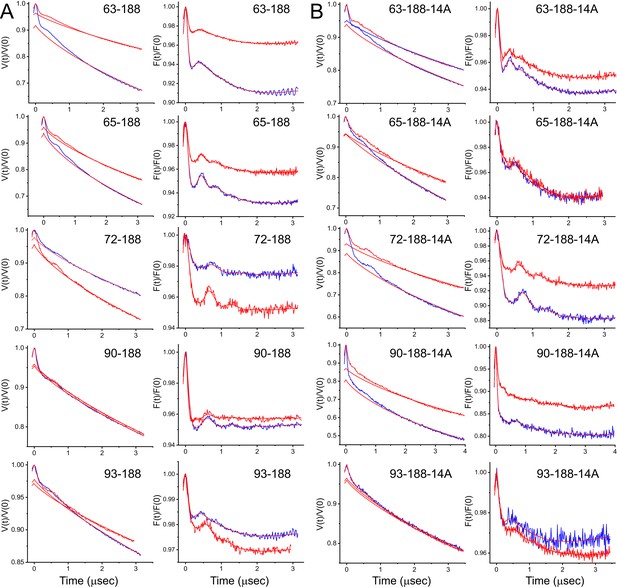
Raw and background corrected double electron-electron resonance (DEER) data for loop-core spin pairs.
In (A) data obtained without the R14A mutation and (B) data with the R14A mutation. Data for the apo state is shown in blue and the data for the vitamin B12 bound state is shown in red. For each of the 20 sets of distributions in Figure 3 are shown the raw DEER data, V(t)/V(0) (left panels), along with the background form factor that was subtracted (straight red lines). The background corrected DEER data, (F(t)/F(0)) (right panels), are shown along with the fits to the data (straight red lines) used to generate the distributions in Figure 3.
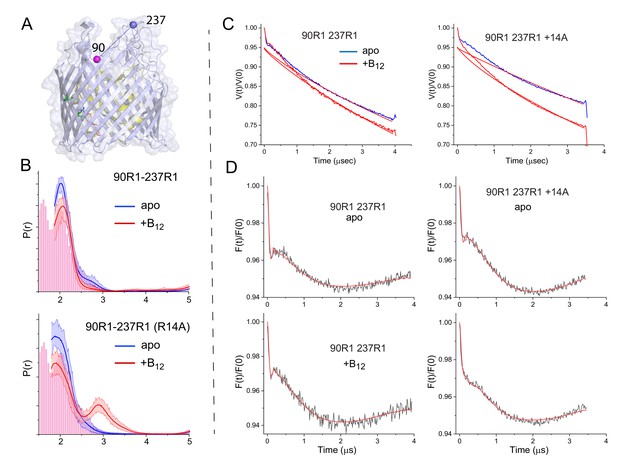
The substrate-dependent movement in substrate binding loop 3 (SB3) promoted by the R14A mutation is observed at site 237.
Shown in (A) is a side view of sites 90 and 237 (PDB ID:1NQH). In (B) is shown the distribution for the V90R1-S237R1 spin pair in the absence and presence of R14A with and without vitamin B12, where the histogram represents prediction from the vitamin B12 bound structure (PDB ID: 1NHQ). A significant portion of the predicted distance distribution is below the minimum range observable using double electron-electron resonance (DEER) (1.5–2 nm). Shown in (C) are the raw data and background for the V90R1-S237R1 spin pair without and with the R14A mutation, and in (D) are the background correct DEER data. These data were analyzed using the DeerNet routine (Worswick et al., 2018) in DEERAnalysis (Jeschke et al., 2006). Histograms are predicted distances generated from the in surfo crystal structures (PDB ID: 1NQG (blue) and PDB ID: 1NQH (red)) using MMM (Jeschke, 2018).
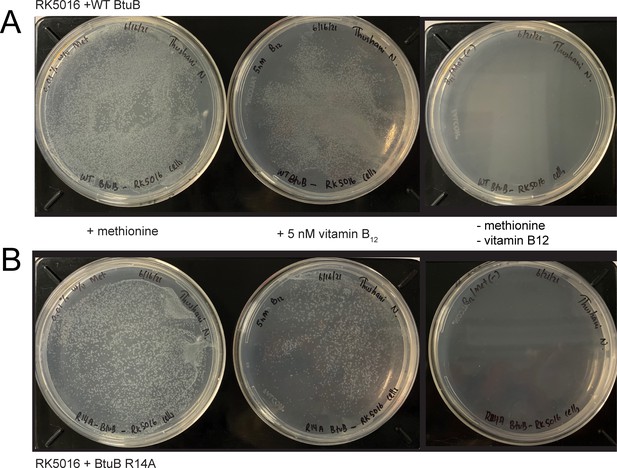
Both wild-type (WT) BtuB and BtuB mutant R14A support growth in minimal media containing vitamin B12.
Using an approach described previously (Lathrop et al., 1995), RK5016 cells (-argH, - btuB, -metE) expressing (A) WT BtuB or (B) BtuB mutant R14A were grown in minimal media plates supplemented with 200 µg/ml ampicillin, 0.2 % w/v glucose, 150 µM thiamine, 3 mM MgSO4, 300 µM CaCl2, 0.01 % w/v arginine and either 0.01% w/v methionine (left), 5 nM vitamin B12 (middle), or in the absence of methionine and vitamin B12 (right). Plates were incubated overnight at 37°C. WT BtuB and R14A BtuB in RK5016 both produce similar size colonies. As expected for the RK5016 phenotype, growth is not supported in the absence of methionine or vitamin B12.
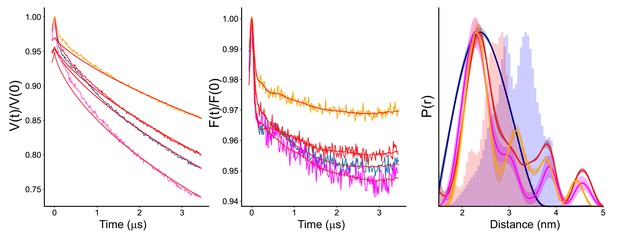
Time domain (left), dipolar (middle), and distance (right) data obtained for the V90R1-T188R1 pair with the R14A mutation at various time points show relative stability of these distance components.
The initial apo (blue) and vitamin B12 (red) traces were processed and frozen more quickly than in other figures, and it can be seen that the starting distribution, particularly for the apo condition, is broader with a single peak around 2.6 nm. From 0 to 30 min (magenta) after addition of substrate, there is a narrowing of the distance elements, which continues to 60 min after addition (orange). Despite this, and the loss of signal by 60 min seen in the decrease in modulation depth, the locations of the peaks are not altered, indicating that either these conformations, or their percentage abundance, are stable on the 60 min time scale. Data were analyzed using LongDistances v932 and the model-free fitting regime. Histograms are predicted distances generated from the in surfo crystal structures 1NQG and 1NQH using the software package MMM.
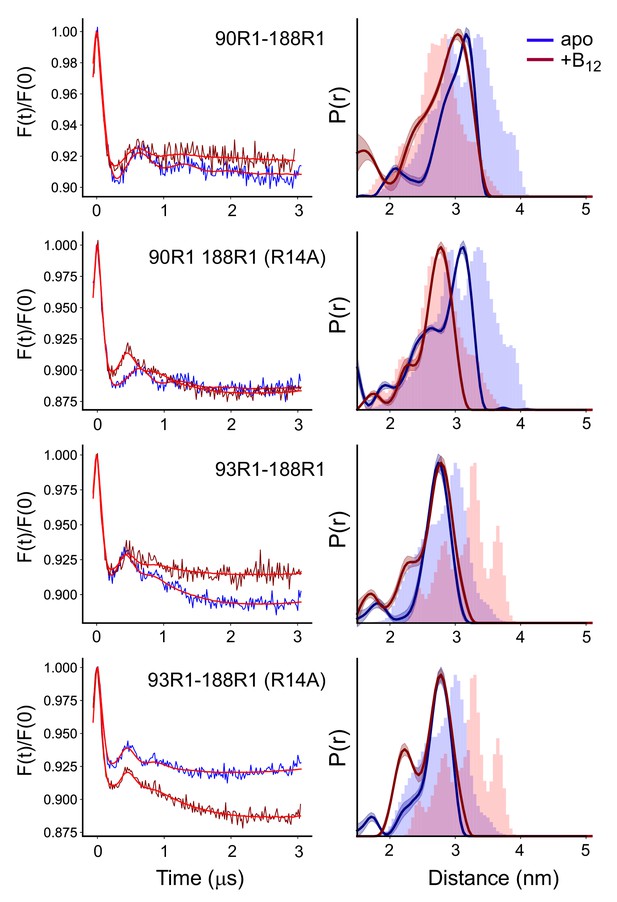
The substrate-induced changes in substrate binding loop 3 (SB3) are altered or absent in proteoliposomes.
Background corrected double electron-electron resonance (DEER) signals (left) and distance distributions (right) for the V90R1-T188R1 and S93R1-T188R1 pairs involving SB3 in the absence and presence of the R14A mutation, where the labeled BtuB have been reconstituted into 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) vesicles. Data were analyzed using LongDistances v932 and the model-free fitting mode. The shaded error bands represent variation due to background noise, start time, dimensionality, and regularization. Histograms are predicted distances generated from the in surfo crystal structures 1NQG (blue) and 1NQH (pink) using the software package MMM (Jeschke, 2018).
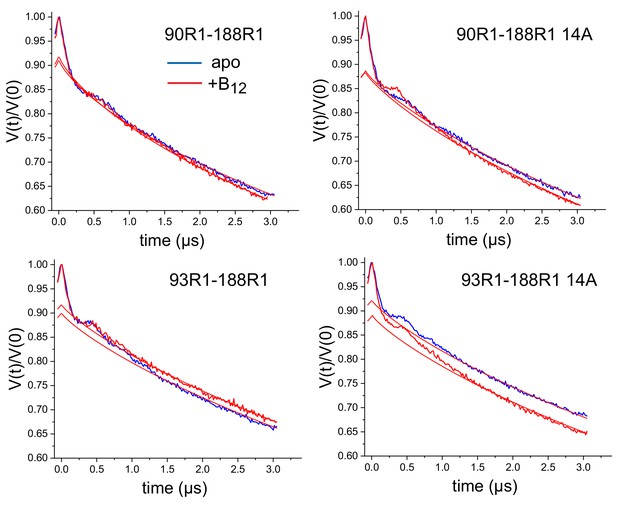
Raw double electron-electron resonance (DEER) data for the V90R1-T188R1 and S93R1-T188R1 spin pairs purified and reconstituted into 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC).
Solid red lines indicate the background form factors that were used to obtain the background corrected data shown in Figure 4.
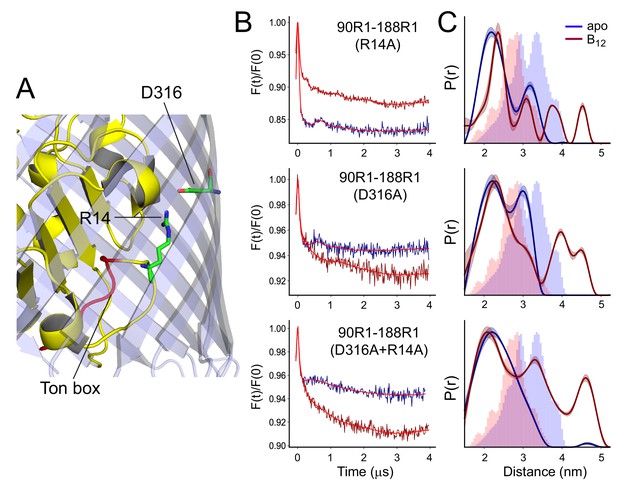
R14A, D316A, or D316A-R14A have similar effects on the conformation of SB3.
In (A) is shown the structure of BtuB highlighting the positions of the R14 and D316 side chains and the location of the Ton box (from PDB ID: 1NQG). Background corrected double electron-electron resonance (DEER) data are shown in (B) and distance distributions in (C) for the V90R1-T188R1 spin pair in whole cells in the presence of the R14A, D316A, or the combined R14A-D316A mutants. Data are shown for both apo (blue) and vitamin B12 bound (red) states. Lines through the DEER data represent the fits for the distributions shown on the right. These data were analyzed using LongDistances v932 and the model-free fitting mode. The shaded error bands in (C) represent variation due to background noise, start time, dimensionality, and regularization. Histograms represent predicted distances generated from the in surfo crystal structures for PDB ID: 1NQG (light blue) and PDB ID: 1NQH (pink) using the software package MMM (Jeschke, 2018).
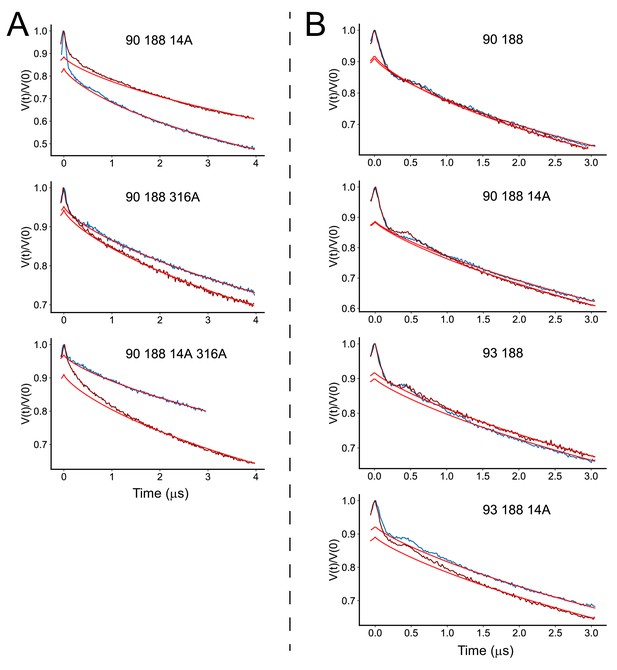
Raw double electron-electron resonance (DEER) data for the V90R1-T188R1 spin pair in the presence of the R14A, D316A, and R14A/D316A mutations.
Solid red lines indicate the background form factors that were used to obtain the background corrected data shown in Figure 5.
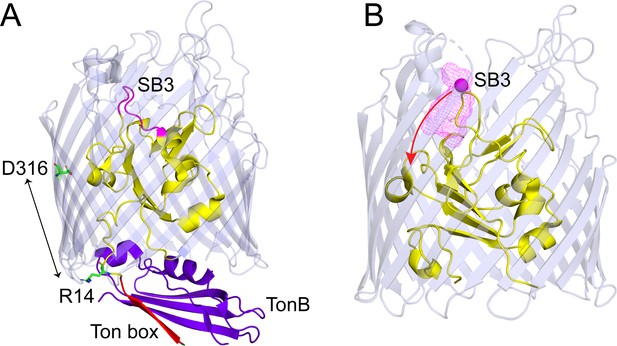
Conformational shifts in substrate binding loop 3 (SB3) with release of R14-D316 ionic lock.
(A) View of BtuB (PDB ID: 2GSK) in complex with the C-terminal domain of TonB (purple) showing the core (yellow) and barrel (light blue) with the substrate binding loop SB3 (magenta), and the Ton box (red), where the R14-D316 ion pair has been broken. (B) An Xplor-NIH simulation showing positions of the Cβ carbon (magenta mesh) on site 90 in SB3 for 300 structures that are consistent with the distributions obtained by double electron-electron resonance (DEER) for V90R1-T188R1-R14A and V90R1-S237R1-R14A in the presence of vitamin B12 (see Materials and methods). TonB binding extracts the Ton box from the core to create an edge-to-edge interaction with TonB, thereby breaking the R14-D316 ionic interaction. Breaking this interaction promotes the movement of SB3 toward the periplasmic interface of the transporter (red arrow) and may facilitate passage of vitamin B12.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | RK5016 (A derivative of MC4100 with the genotype araD139 Δ(argF-lac)169 flbB5301 ptsF25 relA1 rpsL150 rbsR22 deoC1 gyrA219 non-9 metE70 argH1 btuB461 recA56) | Robert Kadner (University of Virginia) | PMID:2982793 | E. coli strain lacking chromosomal gene for BtuB This strain was authenticated using phenotype assays |
| Strain, strain background (Escherichia coli) | RI90 (araD139 Δ(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi phoR Δara714 leu+, dsbA:: Kanr) | Coli Genetic Stock Center (Yale University, New Haven, CT) | PMID:8917542 | E. coli DsbA null strain This strain was authenticated using phenotype assays |
| Recombinant DNA reagent | pAG1 (plasmid) | Robert Kadner (University of Virginia) | pUC8 with btuB ORF (2.4 kb) and regulatory region | Plasmid containing WT BtuB gene |
| Recombinant DNA reagent | pAG1 with single point mutations in BtuB (L63C, S65C, N72C, S93C and S237C) | Applied Biological Materials (Richmond, BC, Canada) | Plasmids used to construct and express BtuB with single mutations | |
| Sequence-based reagent | BtuB D316A-FP | This paper | PCR primers | (5’ – 3’) GGTGCGGGTGTCGCCTGGCAGAAACAGACTAC |
| Sequence-based reagent | BtuB D316A-RP | This paper | PCR primers | (5’ – 3’) GTAGTCTGTTTCTGCCAGGCGACACCCGCACC |
| Sequence-based reagent | BtuB R14A-FP | This paper | PCR primers | (5’ – 3’) GTTACTGCTAACGCTTTTGAACAGCCGCGCA |
| Sequence-based reagent | BtuB R14A-RP | This paper | PCR primers | (5’ – 3’) TGCGCGGCTGTTCAAAAGCGTTAGCAGTAAC |
| Sequence-based reagent | BtuB V90C-FP | Nilaweera et al., Biophys. J. 117, 1476–1484. PMID:31582182 | PCR primers | (5’ – 3’) GAATCTGGCGGGGTGTAGTGGTTCTGCCG |
| Sequence-based reagent | BtuB V90C-RP | Nilaweera et al., Biophys. J. 117, 1476–1484. PMID:31582182 | PCR primers | (5’ – 3’) CGGCAGAACCACTACACCCCGCCAGATTC |
| Sequence-based reagent | BtuB T188C-FP | Nilaweera et al., Biophys. J. 117, 1476–1484. PMID:31582182 | PCR primers | (5’ – 3’) ACCGGATGCCAAGCGCAGACAGATAACGATGG |
| Sequence-based reagent | BtuB T188C-RP | Nilaweera et al., Biophys. J. 117, 1476–1484. PMID:31582182 | PCR primers | (5’ – 3’) GCGCTTGGCATCCGGTATTACCATAGGCAACAAC |
| Chemical compound, drug | OG (octylglucoside or n-octyl-β-D-glucopyranoside) | Chem-Impex, international (Wood Dale, IL) | Cat# 00234 | Detergent for BtuB reconstitution |
| Chemical compound, drug | Vitamin B12(CN-Cbl, Cyanocobalamin) | Sigma Aldrich | Cat# V2876 | Substrate for BtuB |
| Chemical compound, drug | 1-Palmitoyl-2-oleoyl-glycero-3-phosphocholine | Avanti Polar Lipids, (Alabaster, AL) | POPC Cat#8 50457 | Lipid used for membrane reconstitution of BtuB |
| Chemical compound, drug | (1-Oxy-2,2,5,5-tetramethylpyrrolinyl-3-methyl)methanethiosulfonate | Cayman Chemical, Ann Arbor Michigan | MTSSL Cat# 16463 | Reagent for spin labeling protein cysteine residues |
| Software, algorithm | LongDistances (v. 932) | Christian Altenbach (UCLA) | LabVIEW software routine for the analysis of pulse EPR data | Used to examine DEER data |
| Software, algorithm | DeerAnalysis (v. 2019) | Gunnar Jeschke (ETH Zürich) | MATLAB routine for the analysis of pulse EPR data | Used to examine DEER data |
| Software, algorithm | MMM (v. 2018.2) | Gunnar Jeschke (ETH Zürich) | MATLAB routine for the determination of spin label rotamers and predicted label-label distances | Used in this study to predict distance distributions from crystal structures and in silico BtuB structures for simulated annealing |
| Software, algorithm | Xplor-NIH (v. 3.2) | Charles Schwieters, Marius Clore (NIH, NIDDK) | Used for simulated annealing to generate structures consistent with DEER data | |
| Software, algorithm | MATLAB (v. 2020a) | MathWorks, Inc (Natick, MA) | Program needed to execute DeerAnalysis and MMM | |
| Software, algorithm | Pymol (v. 2.4.0a0) | Schrödinger, LLC (New York, NY) | Program for molecular graphics |
Additional files
-
Source data 1
Unprocessed EPR data.
- https://cdn.elifesciences.org/articles/68548/elife-68548-data1-v4.zip
-
Supplementary file 1
Table showing the results of fitting double electron-electron resonance (DEER) data for V90R1-T188R1 taken at increasing concentrations of substrate to a two-component model.
- https://cdn.elifesciences.org/articles/68548/elife-68548-supp1-v4.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68548/elife-68548-transrepform-v4.docx





