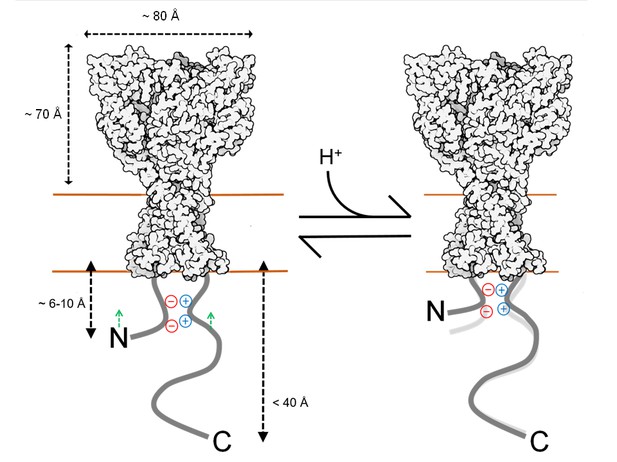
Topography and motion of acid-sensing ion channel intracellular domains
Figures
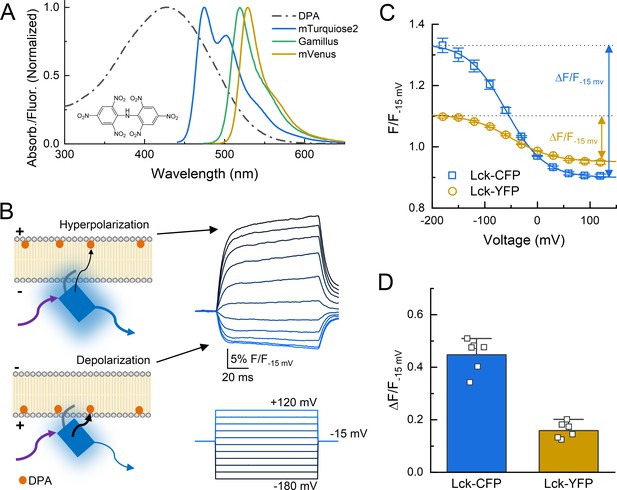
Schematic of dipicrylamine (DPA) quenching measurements.
(A) Absorption (dotted line) or fluorescence emission (solid lines) spectra of DPA (black), CFP variant mTurqoise2 (blue), GFP variant Gamillus (green) and YFP variant mVenus (yellow). Structure of DPA inset. (B, left) DPA (orange circle) localizes to the outer leaflet with hyperpolarization and moves to inner leaflet upon depolarization, resulting in stronger quenching. Arrows reflect excitation (purple), emission (blue), or resonance transfer (black). (B, right) Exemplar patch fluorometry data of Lck-CFP fluorescence changing during voltage steps. (C) Single cell fluorescence, normalized to fluorescence at −15 mV, over a range of voltages for Lck-CFP (blue) and Lck-YFP (yellow). Solid lines are fits to Boltzmann function which yield ΔF/F. (D) Summary of ΔF/Fs from Lck-CFP or Lck-YFP. Squares are single cells (N = 6 cells per construct) and error bars depict SEM.
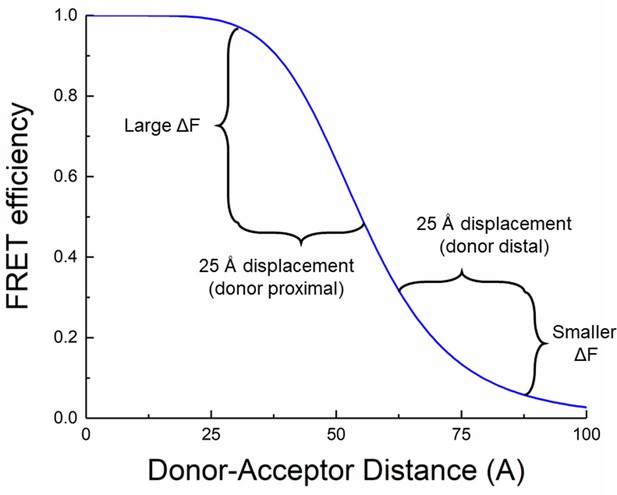
Non-linearity of fluorescence resonance energy transfer (FRET) provides for distance estimates.
Simulated FRET efficiency over distance with an R0 of 55 Å. Moving donor and acceptor apart by a fixed distance has a larger effect when the donor and acceptor are close (donor proximal case) compared to when they are further apart (donor distal case).
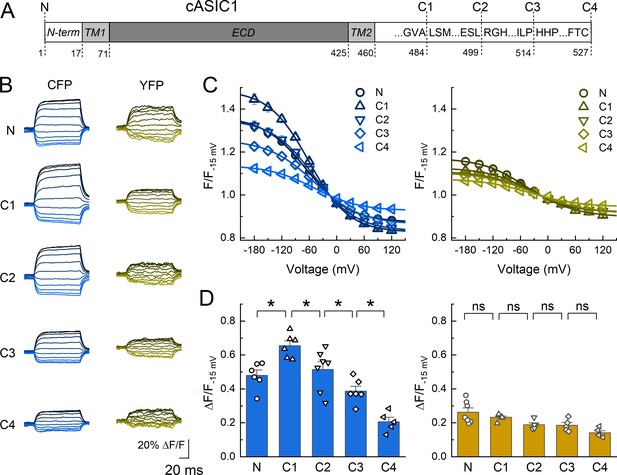
Dipicrylamine (DPA) quenching of CFP or YFP insertions in cASIC1 intracellular domains.
(A) Schematic of cASIC1 constructs used. The extracellular domain (ECD) and transmembrane helices (TM) are gray with amino and carboxy termini depicted in white. Letters within the carboxy tail indicate the amino acid sequence around the insertion site while the lower numbers give the position. (B) Example fluorescence traces for each of the CFP or YFP insertions. Voltage protocol and coloring is the same as in Figure 1B. (C and D) Quenching curves and summary plots for CFP (left) and YFP (right) insertions. Symbols denote individual cells (N = 5–7 cells) and error bars represent SEM. Asterisks mark significant differences with p values of 0.004 (N–C1), 0.0475 (C1–C2), 0.0483 (C2–C3), and 0.0163 (C3–C4) for CFP.
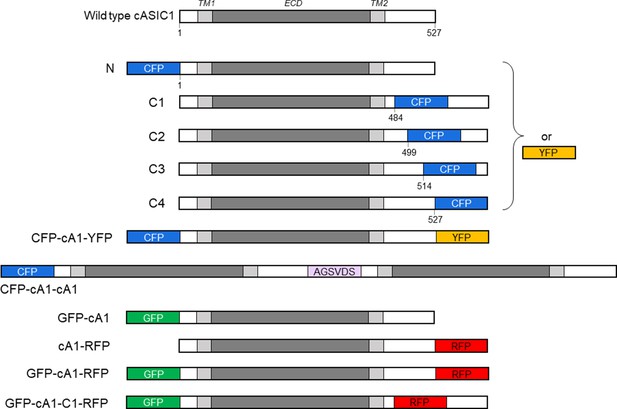
cASIC1 constructs used in this study.
The extracellular domain (ECD) and transmembrane helices (TM) are gray with amino and carboxy termini depicted in white. Numbers indicate the amino acid position for insertion. The violet bar shows the amino acid linker between the first and second subunit in the cASIC1 dimer. CFP, YFP, GFP, and RFP represent mTurq2, mVenus, Gamillus, and tagRFP, respectively. Sizes of fluorescent protein (FP) insertions are not drawn to scale. See Materials and methods for further details.
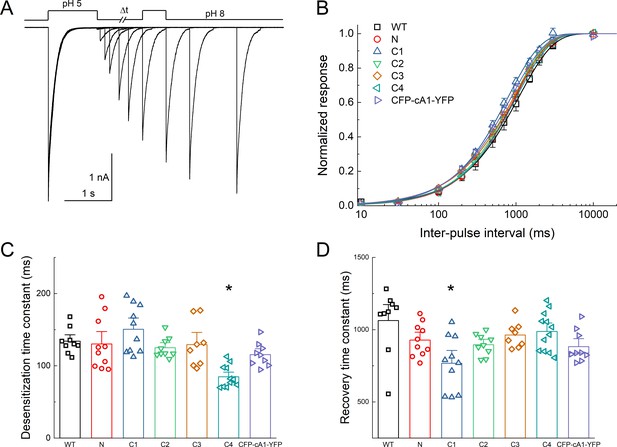
Fluorescent protein (FP) insertion does not substantially alter desensitization kinetics.
(A) Outside-out patch recording of cASIC1 CFP at C3 during paired pulse stimulation protocol using pH 8 and pH 5. (B) Recovery from desensitization curves over various interpulse intervals for the indicated constructs. The recovery slope, m, ranged from 0.94 ± 0.08 to 1.06 ± 0.02 (mean ± SEM). (C and D) Summary of desensitization entry (C) and recovery (D) time constants for the indicated constructs. Symbols denote individual excised patches (N = 9–12 patches per construct). Asterisks indicate significant difference compared to wild type using randomization test. For C4 desensitization, p = 0.00012. For C1 recovery, p = 0.046. Error bars represent SEM in all panels.
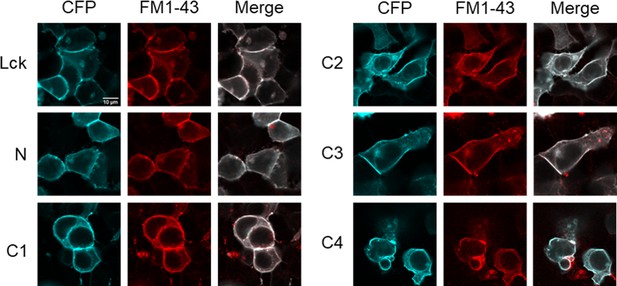
Lck-CFP and CFP-tagged cASIC1 constructs localize to the plasma membrane.
Confocal images of HEK293T cells transfected with the indicated constructs 2 days prior. Plasma membrane was stained with FM1–43 (5 µM) immediately prior to imaging.
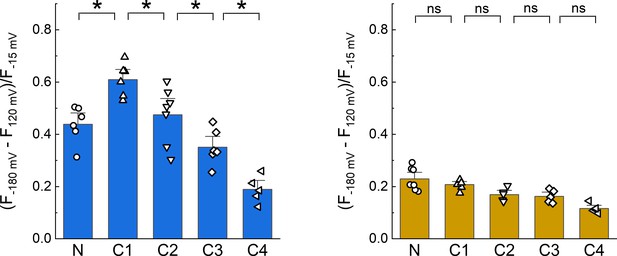
Dipicrylamine (DPA) quenching of CFP or YFP insertions in cASIC1 intracellular domains without Boltzmann fits.
Summary plots of DPA quenching for CFP (left) and YFP (right) insertions analyzed by subtraction of fluorescence values at +120 mV from −180 mV without fitting to a model. Symbols denote individual cells (N = 5–7 cells) and error bars represent SEM. Asterisks mark significant differences with p-values of 0.005 (N–C1), 0.0373 (C1–C2), 0.0396 (C2–C3), and 0.013 (C3–C4) for CFP.
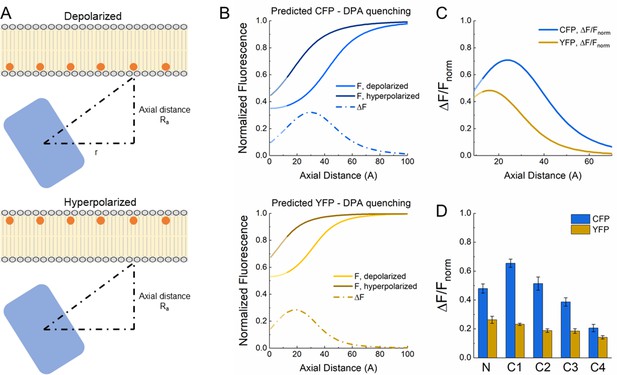
Theoretical analysis of fluorescent protein-dipicrylamine (FP-DPA) quenching.
(A) Schematic of DPA quenching of a fluorophore positioned away from the plasma membrane. (B) Predicted quenching curves for CFP (upper) and YFP (lower) at depolarized (light color) or hyperpolarized (darker color) voltages. Dotted line depicts the difference between hyperpolarized and depolarized curves or ΔF as a function of axial distance. (C) ΔF versus distance curves normalized to fluorescence at hyperpolarized voltage or ΔF/Fnorm as a function of distance. (D) Plot of observed quenching (i.e. ΔF/Fnorm) for each FP insertion. Same data as in Figure 2D. Note similarity with predicted ΔF/Fnorm curves in panel C.
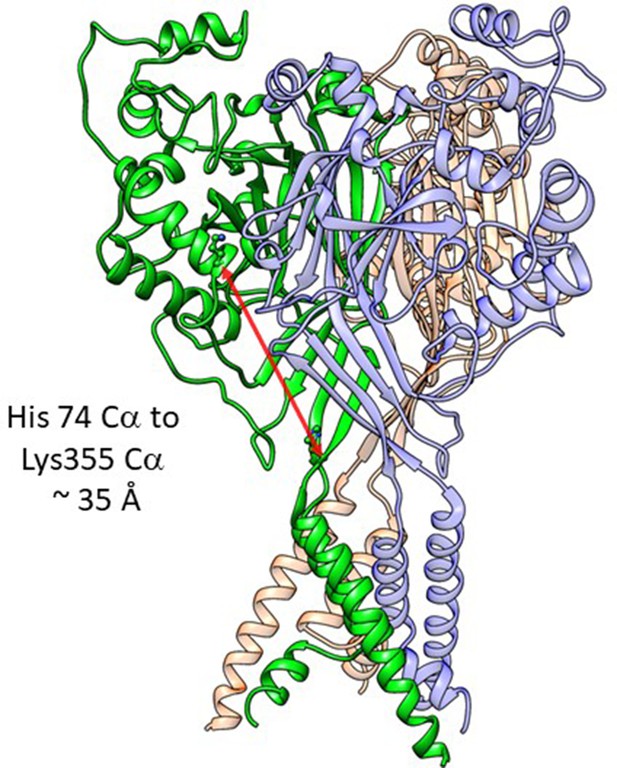
Distance in extracellular domain for reference.
Structure of cASIC1 in the resting stats (PDB: 6VTL) with individual subunits colored green, red, and blue. The distance between the critical His74 and the Lys355 at the base of the thumb domain is shown in a red arrow and is approximately 35 Å.
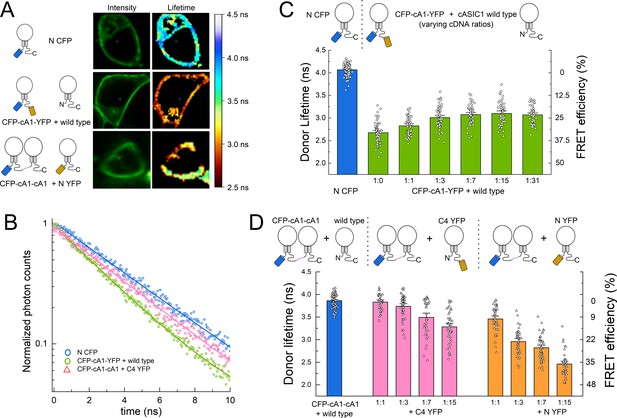
Fluorescence resonance energy transfer (FRET) is stronger within subunits than between subunits.
(A) Example cells transfected with the indicated constructs showing intensity (left) and lifetime (right) images. (B) Time-correlated single photon counting (TCSPC) histograms from the plasma membrane of cells in A and additional control. Solid line is a single exponential fit to the data. (C) Cartoon (upper) and lifetimes (lower) from the indicated constructs for measuring intra-subunit FRET. (D) Cartoon (upper) and lifetimes (lower) for measuring inter-subunit FRET. Symbols denote single cells (N = 35–76 cells) and error bars are SEM.
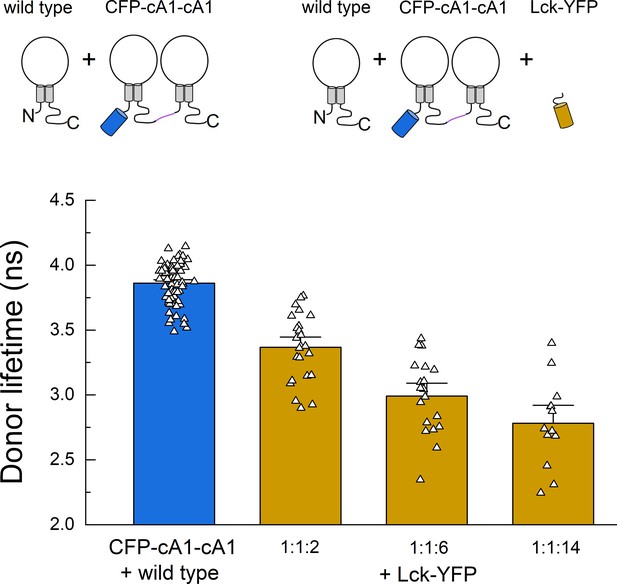
Fluorescence resonance energy transfer (FRET) due to membrane ‘crowding’.
Cartoon (upper) and lifetimes (lower) from the indicated constructs for measuring FRET between cASIC dimer and membrane tethered YFP (Lck-YFP) over varying cDNA ratios. Symbols denote single cells (N = 13–74 cells per construct) and error bars are SEM.
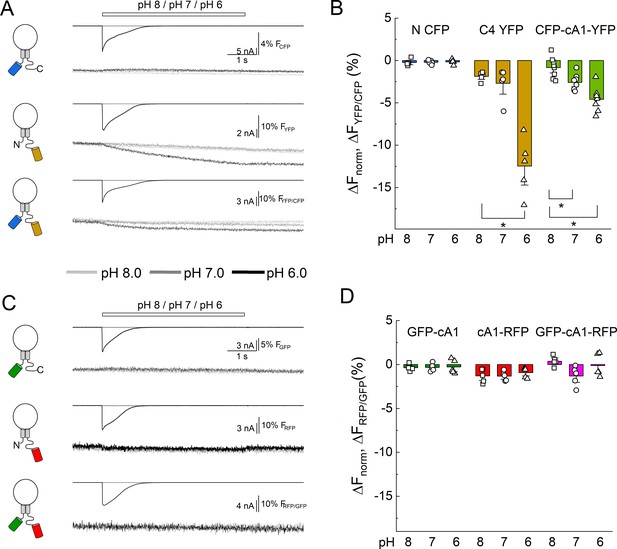
Extracellular acidification produces no change in fluorescence resonance energy transfer (FRET) when using pH-insensitive fluorescent proteins (FPs).
(A) Cartoons (left), electrophysiology (upper), and fluorescence traces (lower) from single cells transfected with the CFP, YFP, or both FPs attached to cASIC1 during pH jumps from eight into pH 8, pH 7, or pH 6. Fluorescence trace is the acceptor fluorescence signal divided by the donor. (B) Summary of change in acceptor/donor ratio during pH changes. (C) Same as in A but for pH-insensitive variants of GFP and RFP (see Materials and methods). (D) Summary of change in acceptor/donor ratio during pH changes. Symbols indicate single cells (N = 5–8 cells), error bars are SEM. Asterisks denote statistical significance with p-values of 0.028 for YFP pH 8 versus 6, 0.031 and 0.0025 for CFP-cA1-YFP pH 8 versus 7 and pH 8 versus 6, respectively.
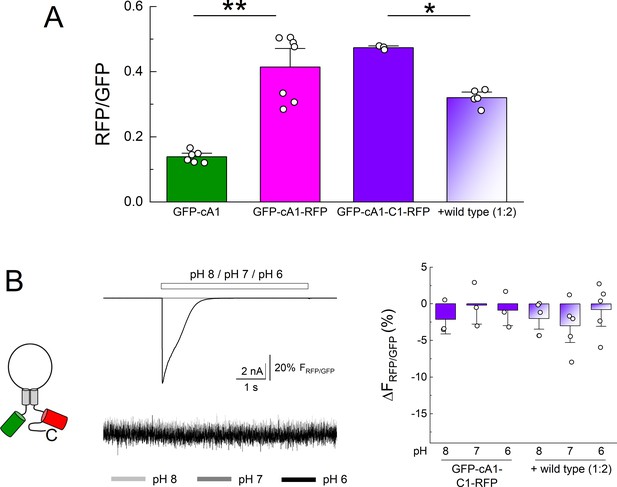
N and C terminal GFP and RFP do not change fluorescence resonance energy transfer (FRET) efficiency upon extracellular acidification.
(A) Summary of green fluorescence emission divided by red fluorescence emission upon GFP excitation for the indicated clones. ** denotes p = 0.0014 and * indicates p = 0.0117 by randomization test. (B) Cartoon (left), example traces of current and fluorescence ratio (middle) and summary changes to apparent FRET (right) from the indicated. Symbols denote single cells (N = 3–7 cells per construct) and error bars are SEM.
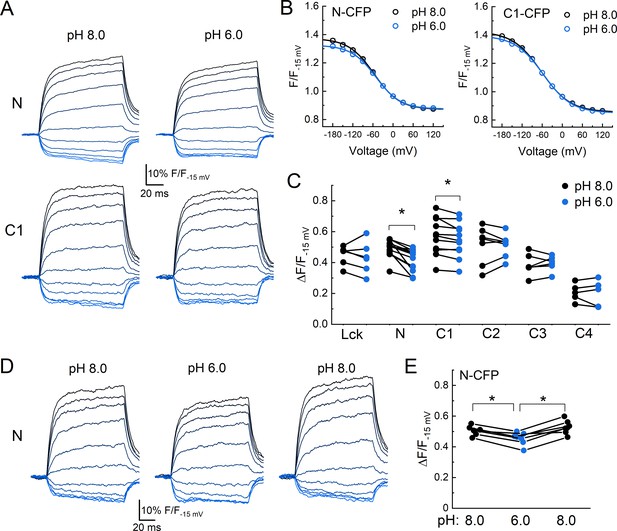
Acidification induces an axial motion of the membrane proximal region.
(A) Example CFP fluorescence quenching traces from single cells expressing N or C1 CFP-cASIC1 with extracellular pH 8 (left) or pH 6 (right). (B) Quenching voltage curves for CFP inserted in N (left) or C1 (right) position. Curves in extracellular pH 8.0 (resting state) shown in black and pH 6.0 (desensitized) in blue. (C) Summary of the extents of quenching at each position with extracellular pH 8 (black) and pH 6 (blue). Symbols denote individual cells (N = 5–13 cells per construct) and error bars represent SEM. Asterisks denote statistical significance by Wilcoxon signed-rank test after correction for multiple comparisons. The p-value for N and C1 are 0.0.0010 and 0.0.033, respectively. (D) Fluorescence-voltage traces from single cell expressing N CFP in the indicated extracellular buffer with 5 µM dipicrylamine (DPA). (E) Summary of the voltage-dependent extent of DPA quenching from N CFP in the indicated extracellular pH. Symbols denote individual cells (N = 7) and error bars represent SEM. * indicates p-value < 0.05 by paired Wilcoxon signed-rank test.
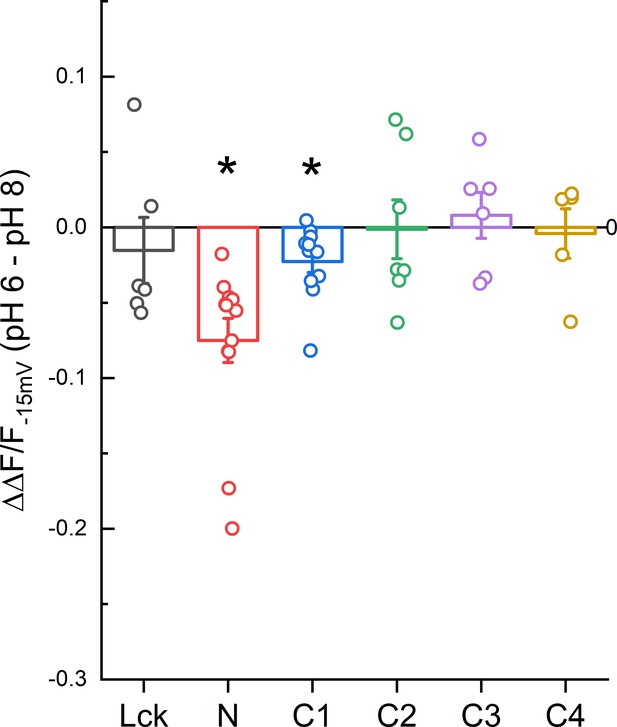
Magnitude of ΔF/F change between pH 8.0 and 6.0 for CFP insertions.
Data points represent the difference between the ΔF/F−15 mV at pH 6.0 and 8.0 of individual cells. Error bars represent SEM. Asterisk indicates that the median is significantly different from 0 by the one-sample Wilcoxon signed-rank test after correcting for multiple tests (p = 0.0015 and 0.015 for N and C1, respectively).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293T ASIC1 KO | Rook et al., 2020a | Clone 2C1 | |
| Recombinant DNA reagent | pcDNA3.1-cocASIC1 | Rook et al., 2020b | ||
| Recombinant DNA reagent | pmVenus(L68V)-mTurquiose2 | RRID:addgene_60493 | ||
| Recombinant DNA reagent | Gamillus/pcDNA3 | RRID:addgene_124837 | ||
| Recombinant DNA reagent | PSD-95-pTagRFP | RRID:addgene_52671 | ||
| Recombinant DNA reagent | Lck-mScarlet-I | RRID:addgene_98821 | ||
| Commercial assay or kit | Q5 Hot Start High-Fidelity 2X Master Mix | New England Biolabs, Inc | M0494L | PCR |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs, Inc | E2621L | Insertion of FP tags |
| Chemical compound, drug | Polyethylenimine 25 k | Polysciences, Inc | 23966–1 | Transfection reagent |
| Chemical compound, drug | Dipicrylamine | Biotium | 60037 | Quencher |
| Software, algorithm | Axograph | RRID:SCR_014284 | Patch clamp and fluorometry acquisition | |
| Software, algorithm | Clampfit | Molecular Devices | RRID:SCR_011323 (pClamp) | Patch clamp and fluorometry analysis |
| Software, algorithm | OriginPro 2020b | OriginLab Corp | 9.7.5.184 (Student Version) | Data fitting and figure preparation |
Additional files
-
Source data 1
Patch clamp and fluorescence experimental data.
- https://cdn.elifesciences.org/articles/68955/elife-68955-data1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68955/elife-68955-transrepform-v3.docx