Tbx5 drives Aldh1a2 expression to regulate a RA-Hedgehog-Wnt gene regulatory network coordinating cardiopulmonary development
Figures
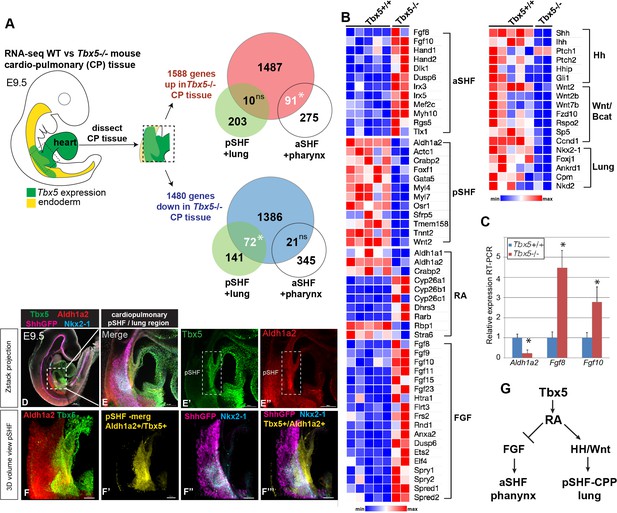
Tbx5 is required for posterior cardiopulmonary (CP) development.
(A) Schematic of an E9.5 mouse embryo highlighting the dissected CP tissue (containing foregut mesoderm and endoderm) profiled by bulk RNA-seq. Venn diagrams show genes differentially expressed in wild-type (WT) versus Tbx5−/− CP tissue (>1.5 fold change, 5% FDR; Steimle et al., 2018, GSE75077) intersected with gene sets from single-cell RNA-seq studies defining aSHF+ pharynx cells versus pSHF/CPP+ lung progenitor cells (de Soysa et al., 2019, GSE126128; Han et al., 2020, GSE136689) (Supplementary files 2 and 3). Statistically significant intersection based on hypergeometric tests. *p<0.0001. (B) Transcriptome analysis of Tbx5−/− CP tissue suggests disrupted SHF pattering and failed pulmonary development with reduced RA and increased FGF signaling. Heat map of selected differentially expressed genes in WT Tbx5+/+ (n=5) and Tbx5−/− mutant (n=2) CP tissue grouped by domain of expression or pathway. (C) RT-qPCR validation of decreased Aldh1a2 and increased Fgf8, Fgf10 expression in E9.5 WT and Tbx5−/− CP tissue. Relative mean expression+S.D. *p<0.05 Student’s t-test relative to WT littermates. (D–F) Whole-mount immunostaining of E9.5 Shh:GFP mouse embryos show that Tbx5 (green) and Ald1a2 (red) expression overlaps in a subset of the pSHF (yellow in (F’) and (F”’)) adjacent to the Nkx2-1+/Shh:GFP+ pulmonary domain (F”, F”’). Scale bar in (D) = 200 μM, (E–E”) = 100 μM, and (F–F”’) = 50 μM. (G) Model of the proposed Tbx5-RA signaling networks in the cardiopulmonary tissue. Also see Figure 1—figure supplement 1, Figure 1—figure supplement 1– tables 1-3. aSHF, anterior second heart field; fg endo, foregut endoderm; ns, not significant; oft, outflow tract; pSHF, posterior SHF; RA, retinoic acid; ventr, ventricle.
-
Figure 1—source data 1
Differentially expressed genes in mouse E9.5 micro-dissected cardiopulmonary progenitor (CPP) tissue based on bulk RNA-seq (Steimle et al., 2018, GSE GSE75077).
1588 upregulated genes and 1480 downregulated genes were observed in Tbx5 knockout CPP tissue (≥1.5 fold change and 5% FDR).
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig1-data1-v2.xlsx
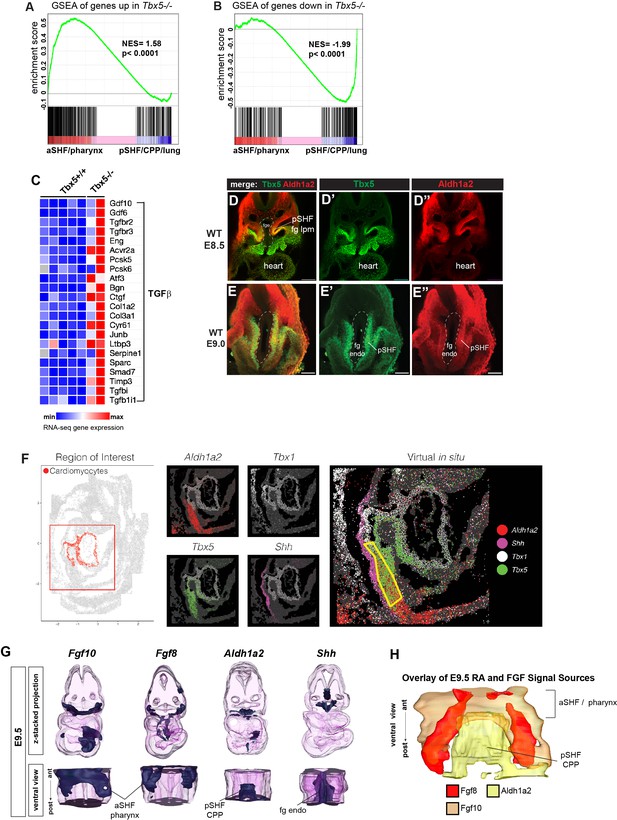
Analysis of the Tbx5 regulated transcriptome in mouse cardiopulmonary tissue and co-expression of Tbx5 with Aldh1a2 in the pSHF.
(A, B) Gene set enrichment analysis (GSEA) comparing Tbx5-regulated transcriptome in CP tissue (|>1.5| fold change, 5% FDR; Steimle et al., 2018, GSE75077) with gene sets from single-cell RNA-seq studies defining aSHF versus pSHF (de Soysa et al., 2019, GSE126128) and pharynx versus CPP+ lung progenitor cells (Han et al., 2020, GSE136689) (Supplementary files 2 and 3). Genes upregulated in Tbx5−/− CP tissue are enriched in aSHF and pharynx and depleted in pSHF/CPP/lung enriched markers (A). In contrast transcripts downregulated in Tbx5−/− CP tissue were significantly enrichened pSHF/CPP/lung marks and depleted for aSHF/pharynx markers (B). (C) Heat map of RNA-seq gene expression showing TGFβ pathway genes is upregulated in Tbx5−/− CP tissue. (D, E) Immunostaining of E8.5 (D) or E9 (E) mouse embryo foregut region shows co-expression of Tbx5 (green) and Aldh1a2 (red) protein in the foregut lateral plate mesoderm (fg lpm)/posterior second heart field (pSHF). (F) Digital in-situ hybridization using as online Spatial Mouse Atlas of single cell gene expression (Lohoff et al., 2021). E9.5 digital in-situ hybridizations show the domains of Aldh1a2, Tbx5, Tbx1, and Shh expression; yellow boxed region indicates Tbx5/Aldh1a2 co-expression in the pSHF lpm. (G, H) 3-D reconstructions from actual serial section in-situ hybridizations of E9.5 mouse CP foregut region showing expression domains of Fgf10, Fgf8, Aldh1a2, and Shh transcripts. Top row is a z-stack 3-D projection; bottom row is a rotated ventral view; (H) shows a ventral view, merged projection of the expression domains of Fg10, Fgf8, and Shh. aSHF, anterior second heart field; CPP, cardiopulmonary progenitor; NES, normalized enrichment score.
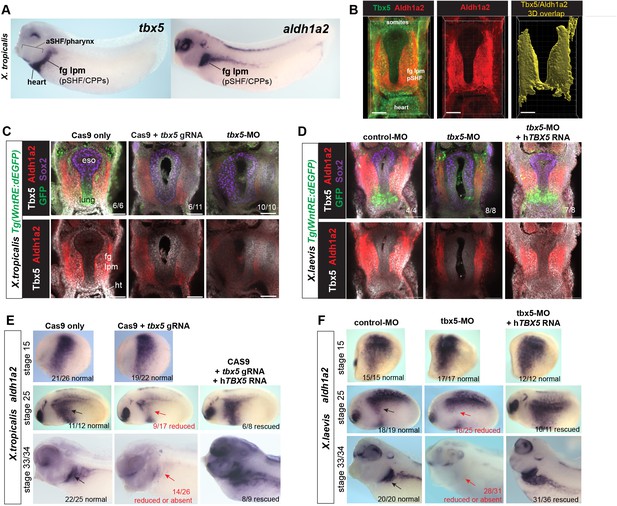
Tbx5 maintains aldh1a2 expression in Xenopus foregut lpm.
-
Figure 2—source data 1
Summary of in-situ hybridization results from Xenopus CAS9/CRISPR injection experiments in F0 embryos.
The tbx5 exon5 sgRNA (targeting the DNA binding domain; Steimle et al., 2018) causes approximately 40% of injected embryos to have a molecular phenotype as revealed by in-situ hybridization. 2 nl of a mixture containing 50 pg/nl sgRNA with 0.5 ng/nl Cas9 protein (PNA Bio CP01-20) was injected on either side of the sperm entry point at the 1 cell stage (total of 200 pg sgRNA and 2 ng Cas9 protein per embryo), embryos were cultured to NF34 and assayed by in-situ hybridization for the indicated genes.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig2-data1-v2.xlsx
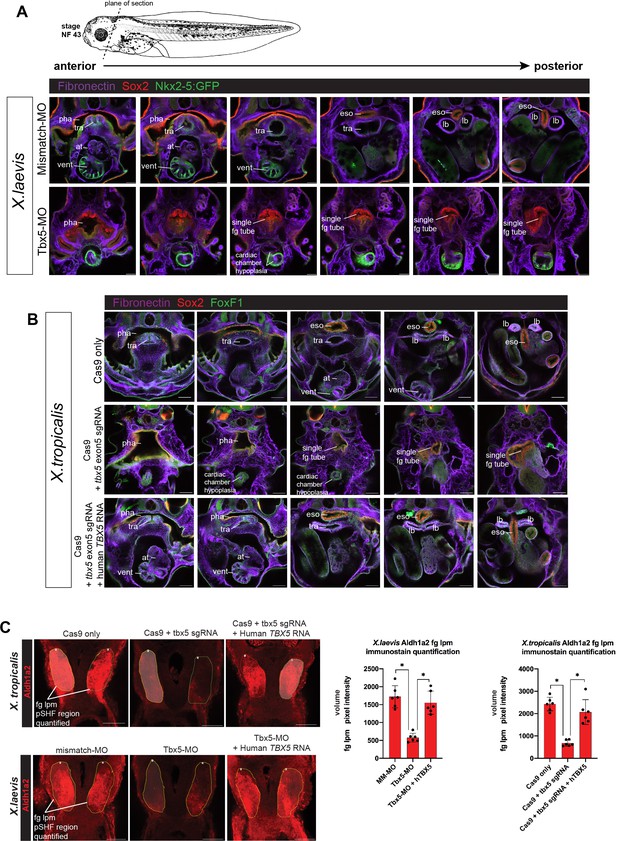
Tbx5 LOF phenotype at stage NF43/44 and quantification of reduced Aldh1a2 immunostaining.
(A) Immunostaining of stage NF43 transgenic Xenopus laevis Tg(nkx2-5:GFP) reporter embryos injected with either mismatch-MO (control) or Tbx5-MO. Confocal optical sections through the cardiopulmonary-foregut region showing Fibronectin+ tissue boundaries (purple), GFP in the ventral pharynx, trachea, and ventricle; and Sox2 (red) in the pharyngeal/esophageal endoderm. Tbx5-depleted embryos exhibit severe cardiac hypoplasia, a lack of ventricular trabeculae, and a single, undivided foregut tube lacking Nkx2-1 respiratory progenitors. (B) Immunostaining of NF44 X. tropicalis embryos for Fibronectin (purple), Sox2 (red), and FoxF1 (green) shows that tbx5 mutants, have a single Sox2+ foregut tube and severe cardiac hypoplasia phenocopying X. laevis Tbx5 morphants. Injection of human TBX5 RNA rescues trachea-esophageal morphogenesis and cardiac chamber development. (C) Representative images for quantitation of Aldh1a2 immunofluorescence signal in Tbx5 LOF embryos at NF34. Nikon Elements Analysis AR software was used to determine the average volume pixel intensity of the fg lpm region (dotted yellow lines). Each dot in the graphs represents a distinct fg lpm/pSHF region from N=3 total embryos, all imaged with identical confocal laser settings. *p<0.05, parametric two-tailed paired t-test. at, atrium; eso, esophagus; lb, lung bud; LOF, loss-of- function; lpm, lateral plate mesoderm; MO, morpholino; pha, pharynx; tra, trachea; vent, ventricle.
-
Figure 2—figure supplement 1—source data 1
Quantitation of Xenopus NF34 fg lpm/pSHF Aldh1a2 immunostaining volume pixel intensity.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig2-figsupp1-data1-v2.xlsx
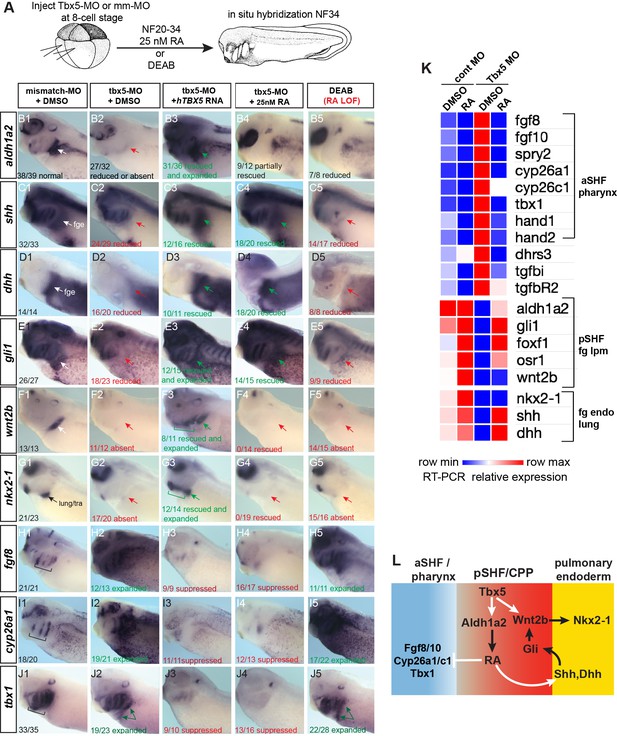
Tbx5 regulates Xenopus cardiopulmonary development in part via RA.
(A) Schematic of the experimental design. (B–J) Exogenous RA rescues Tbx5 LOF, while inhibition of RA phenocopies Tbx5 LOF. Whole-mount in-situ hybridization of NF34 X. laevis embryos after the indicated experimental treatments: injection of negative control 3 bp mismatch-MO (10 ng), Tbx5-MO (10 ng), Human TBX5 RNA (hTBX5; 100 pg), and/or 25 nM RA, 10 µM DEAB, DMSO vehicle control from NF20-34. The numbers of embryos with the observed expression pattern are indicated. Arrows indicate the relevant expression domain in the cardiopulmonar (CP) tissue. Brackets indicate the aSHF/pharyngeal domain. (K) Heat map showing relative expression from RT-PCR analysis of NF34 CP-foregut (fg) tissue dissected from control or Tbx5-MO injected embryos and treated with or without RA from NF20 to NF34. Each row is the average from the three biological replicates (n=4 explants per replicate). (L) Diagram of the proposed GRN model at NF25–35 showing the key role of Aldh1a2-dependent RA signaling downstream of Tbx5. White arrows indicate relationships tested in the above experiments and black arrows are demonstrated from the previous publications. Also see Figure 3—figure supplement 1, and related source data files. GRN, gene regulatory network; LOF, loss-of-function; MO, morpholino; RA, retinoic acid.
-
Figure 3—source data 1
Xenopus explant RT-qPCR source data. Explants were dissected at NF20, cultured 48 hr±DMSO or 25 nM all-trans retinoic acid (RA); harvested at NF34; three biological replicates for each condition; n=4 pooled explants in each replicate; pooled explants came from 2 to 3 separate fertilization/injection experiments.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig3-data1-v2.xlsx
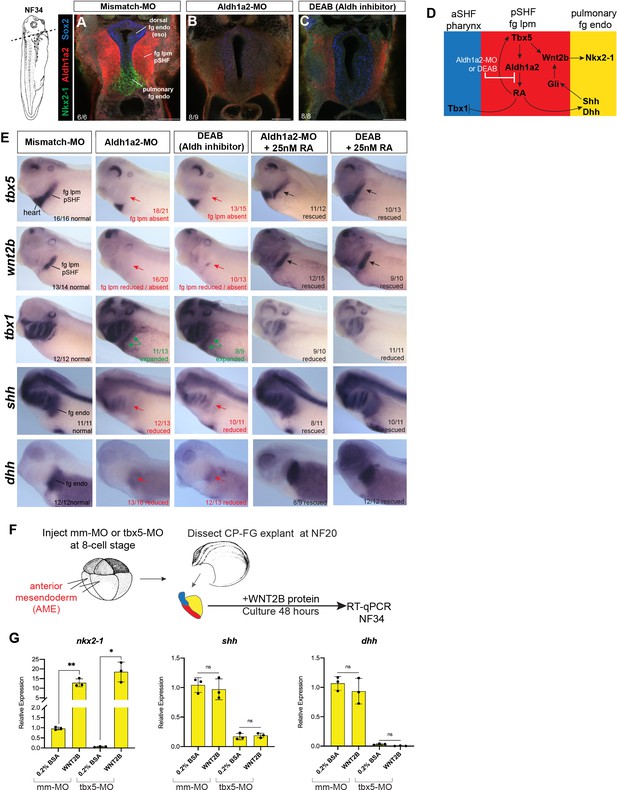
Aldh1a2 morpholino (MO) knockdown phenocopies DEAB treatment and WNT2B protein rescues Nkx2-1+ pulmonary fate.
(A–E) Aldh1a2 MO knockdown phenocopies the pharmacological inhibition by DEAB treatment. Embryos were injected at the 8-cell stage into dorsal mesendoderm targeting the CP domain or treated from NF20 to NF34 with 10 µM DEAB or 25 nM RA. (A–C) Immunostaining of NF34 embryos for Aldh1a2 (red), Nkx2-1 (green), and Sox2 (blue) confirms that Aldh1a2-MO injection or DEAB treatment disrupts the RA>Tbx5>Aldh1a2>RA positive feedback loop (diagrammed in (D)) and results in failed induction of Nkx2-1+ pulmonary progenitors. (E) Whole-mount in-situ hybridization of NF34 control, Aldh1a2-MO injection, or DEAB treatment (NF20–34) showing similar effects. Treatment with 25 nM RA (NF20–34) rescued the changes in gene expression. The numbers of embryos with the observed expression pattern are indicated. Arrows indicate the relevant expression domain in the foregut – cardiopulmonary region. (F) Experimental schematic. (G) WNT2B protein 100 ng/ml rescues nkx2-1 but not Hh ligand expression in Tbx5-depleted fg explants. Graphs show mean relative expression ± standard deviation from N=3 biological replicates (four explants/replicate). Each black dot in the graphs represents a biological replicate (pool of n=4 explants). *p<0.05, **p<0.01, parametric two-tailed paired t-test relative to uninjected, untreated explants. CP, cardiopulmonary; ns, not significant; RA, retinoic acid.
-
Figure 3—figure supplement 1—source data 1
Xenopus explant RT-qPCR source data.
Exogenous WNT2B protein treatment rescues nkx2-1 expression in Tbx5-depleted Xenopus cardiopulmonary foregut explants. RT-qPCR analysis of Xenopus foregut cardiopulmonary explants treated ± recombinant Human WNT2B protein (100 ng/ml) 48 hr NF20–34.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig3-figsupp1-data1-v2.xlsx
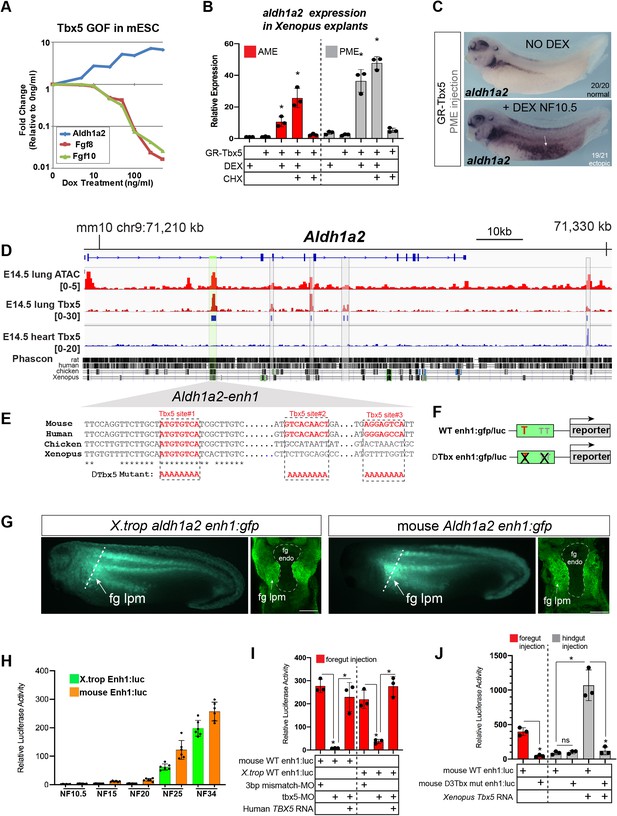
Tbx5 directly activates aldh1a2 transcription via an evolutionarily conserved first intron enhancer.
(A) Dox-inducible Tbx5 activated Aldh1a2 expression and repressed Fgf8 and Fgf10 during the directed differentiation of mouse embryonic stem cells (mESCs) into cardiac progenitors in a dose-dependent manner. (B) Tbx5 directly activated aldh1a2 expression in Xenopus anterior or posterior mesendoderm (AME, PME) explants. RT-qPCR shows that aldh1a2 transcription was induced by DEX activated GR-Tbx5 in the presence of translation inhibitor CHX. Graphs show mean relative expression ± standard deviation from N=3 biological replicates, four explants/replicate. *p<0.05, pair-wise Student’s t-test relative to uninjected, untreated explants. (C) Whole-mount in-situ hybridization of aldh1a2 expression of X. laevis NF34 embryos injected with GR-Tbx5 (100 pg) into the PME with or without DEX. (D) Genome browser of the mouse Aldh1a2 locus showing Tbx5 ChIP-seq tracks from E14.5 mouse lung (GSE167207) and E14.5 mouse heart (Burnicka-Turek et al., 2020, GSE139803) as well as ATAC-seq track from the ENCODE project (Castillo et al., 2010; Davis et al., 2018; ENCSR335VJW). Tbx5 ChIP-seq peaks in the E14.5 lung are indicated in blue. Phascon multiple species conservation track shows that the prominent Tbx5-bound first intron enhancer (enh1) is evolutionarily conserved from mammals to Xenopus. (E) Multiple species sequence alignment of enh1 reveals one Tbx5 DNA-binding site conserved from mammals to Xenopus and two additional mammalian-specific Tbx5 sites, which were mutated in reporter constructs. (F) Schematic of the Wild-type (WT) and mutant (ΔTbx) enh1:gfp and enh1:luciferase reporter constructs. (G) Both the Xenopus and mouse intronic enh1 enhancer are sufficient to drive GFP expression in the foregut lpm in Xenopus transgenic assays. (H) Time course of Xenopus and mouse enh1:luc reporter activity injected into X. laevis CP-foregut tissue, reflects endogenous Tbx5-dependent aldh1a2 expression between NF25 and NF34. Graphs show mean relative luciferase activity ± standard deviation. N=5 biological replicates/time point with five embryos/replicate. *p<0.05, parametric two-tailed paired t-test. (I) The Xenopus and mouse aldh1a2 enh1 reporter constructs are regulated by Tbx5. Graphs show relative mean luciferase activity ± standard deviation of reporters injected into CP-foregut tissue with control mm-MO, Tbx5-MO, and/or human TBX5 RNA. N=3 biological replicates/time point with five embryos/replicate. *p<0.05, parametric two-tailed paired t-test. (J) The three putative Tbx5 motifs in the mouse aldh1a2-enh1 enhancer are required for reporter activity in the CP-foregut tissue and Tbx5-dependent activation in the hindgut. Graphs show mean relative luciferase activity ± standard deviation. N=5 biological replicates/time point with five embryos/replicate. *p<0.05, parametric two-tailed paired t-test. Also see Figure 4—figure supplement 1, Figure 4—figure supplement 2 and related source data files. CP, cardiopulmonary.
-
Figure 4—source data 1
Xenopus explant RT-qPCR source data. GR-Tbx5 directly activated aldh1a2 in Xenopus anterior or posterior mesendoderm explants. NF10.5 anterior or posterior mesendoderm explants were treated for 2 hr in CHX prior to 6 hr in CHX+DEX.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Figure 4H Luciferase source data.
Temporal analyses of mouse and Xenopus tropicalis enh1-driven luciferase activity in the foregut.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Luciferase source data. NF34 analysis of mouse and Xenopus tropicalis enh1-driven luciferase activity in the foregut±mismatch MO, tbx5-MO,±Human TBX5 RNA.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Luciferase source data. NF34 analysis of mouse WT Enh1 or Tbx motif mutant Enh1-driven luciferase activity in fg or in hg±Tbx5 RNA.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-data4-v2.xlsx
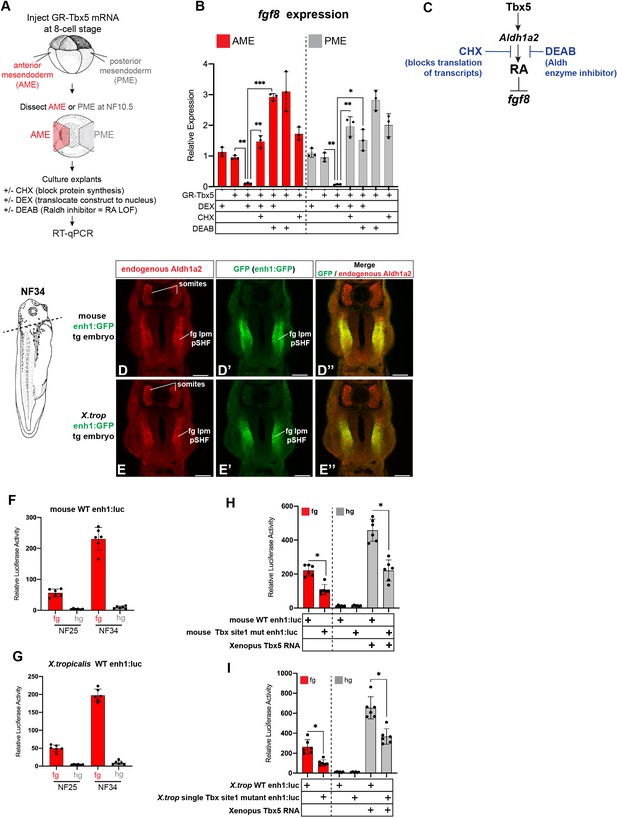
GR-Tbx5 indirectly suppresses fg8 in Xenopus via RA and additional analysis of the first intron Tbx5 enh1 enhancer.
(A–C) Experimental schematic of GR-Tbx5 direct/indirect target gene assay in Xenopus gastrula explants. (B) GR-Tbx5 suppresses fgf8 in both anterior and posterior tissue in a CHX sensitive manner demonstrating indirect suppression. The ability of GR-Tbx5 to suppress fgf8 is DEAB sensitive and thus is dependent on RA production. Graphs show mean relative expression ± standard deviation from N=3 biological replicates (four explants/replicate). Each black dot in the graphs represents a biological replicate (pool of n=4 explants). *p<0.05, **p<0.01, ***p<0.001, parametric two-tailed paired t-test relative to uninjected, untreated explants. (C) Model of the indirect regulation of fgf8 by Tbx5, which these experiments show is CHX and DEAB sensitive. (D, E) Immunostaining of stage NF34 transgenic X. laevis embryos shows that the enh1 enhancer drives GFP expression in the endogenous Aldh1a2 lpm domain but not in the somites. Scale bar=100 µM. (F, G) Both the mouse and X. tropicalis (X. trop) Aldh1a2 enh1 enhancers drive reporter activity in NF25 and NF34 foregut but not hindgut tissue, demonstrating spatial specificity. Graphs show mean relative luciferase activity ± standard deviation. N=6 biological replicates/time point, each containing n=5 embryos/replicate. *p<0.05, parametric two-tailed paired t-test. (H, I) Analysis of wild-type enh1 or enh1 with the single, perfectly conserved T-box/Tbx5 motif mutated. Mutation of the single T-box/Tbx5 motif in the mouse (H) or X. trop (I) Aldh1a2 enh1 enhancers significantly reduces enhancer ability to drive expression in the foregut as well as to be activated by exogenous Tbx5 in hindgut gain-of-function injections. Graphs show mean relative luciferase activity ± standard deviation. N=6 biological replicates/time point, each containing n=5 embryos/replicate. *p<0.05, parametric two-tailed paired t-test. ns, not significant; RA, retinoic acid.
-
Figure 4—figure supplement 1—source data 1
RT-qPCR source data of Xenopus anterior (AME) or posterior (PME) NF10.5 mesendoderm explants injected with GR-Tbx5 RNA and treated±DEX, CHX, or DEAB.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-figsupp1-data1-v2.xlsx
-
Figure 4—figure supplement 1—source data 2
Luciferase source data of mouse and Xenopus tropicalis enh1 reporter activity in foregut and hindgut at NF25, NF34.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig4-figsupp1-data2-v2.xlsx

Multiple species sequence alignment of the Aldh1a2 enh1 enhancers.
(A) Clustal DNA sequence alignment; putative T-box/Tbx5 motifs predicted by the CisBP tool (Weirauch et al., 2014) are shaded in aqua blue (CisBP score>10) and in yellow (CisBP score>8). Asterisks below the nucleotide alignment indicated conserved bases amongst all four species. Tbx motifs mutated and tested in this study are indicated in gray. Genomic co-ordinates of the Aldh1a2 enh1 enhancers used were: Xenopus tropicalis,> xenTrov9.1_dna range = chr3:89631924–89632943; Chicken,> galGal6_dna range = chr10:7578306–7579190;Mouse,> mm10_dna range = chr9:71241739–71242765 Human,> hg38_dna range = chr15:58038775–58039647.
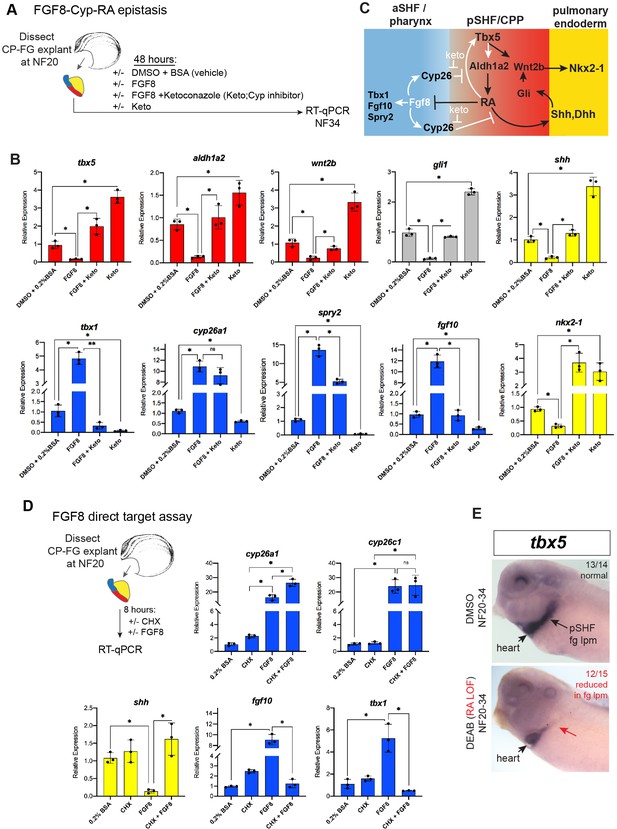
FGF8 gain-of-function (GOF) phenocopies Tbx5 loss-of-function in Xenopus.
(A) Schematic of FGF8 GOF assay in Xenopus cardiopulmonary foregut (CP-FG) explants dissected at NF20, and treated with vehicle controls (DMSO+0.2% BSA) or the indicated combinations of 100 ng/ml FGF8b and/or 0.5 µM ketoconazole (Cyp-inhibitor), harvested at NF34, and analyzed via RT-qPCR. (B) RT-qPCR showing mean relative expression of genes for pSHF (red), aSHF (blue), and pulmonary endoderm (yellow), ± standard deviation from N=3 biological replicates (four explants/replicate). *p<0.05, parametric two-tailed paired t-test. (C) Model depicting the observed FGF8 GOF results. White arrows indicate relationships tested in these experiments. (D) FGF8 direct target gene assay in Xenopus CP foregut explants, demonstrating that FGF8 directly activates cyp26a1, cyp26c1 and indirectly suppresses shh. Explants dissected at NF20 were pre-treated with 1 µM cycloheximide (CHX) for 2 hr prior to culture in 100 ng/ml FGF8b+CHX for 6 hr followed by RT-qPCR analysis. Graphs display mean relative expression ± standard deviation from N=3 biological replicates that contained four explants/replicate. *p<0.05, parametric two-tailed paired t-test. (E) RA signaling is required for the tbx5 expression in the fg lpm/pSHF domain, but not the heart. Embryos were cultured in 10 µM DEAB from NF20 to NF34 and assayed by in-situ hybridization. Number of embryos assayed and with the observed expression pattern is indicated. Also see Figure 5—figure supplement 1 and related source data files. aSHF, anterior second heart field; pSHF, posterior second heart field.
-
Figure 5—source data 1
RT-qPCR analysis of Xenopus foregut CP explants treated with FGF8±ketoconazole (Cyp26 inhibitor). Explants were dissected at NF20, cultured 48 hr±DMSO or recombinant Human FGF8a (200 ng/ml)±20 µM ketoconazole and assayed at NF34. RT-qPCR source data.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig5-data1-v2.xlsx
-
Figure 5—source data 2
RT-qPCR source data of Xenopus foregut cardiopulmonary explants treated with FGF8±CHX (cycloheximide).
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig5-data2-v2.xlsx
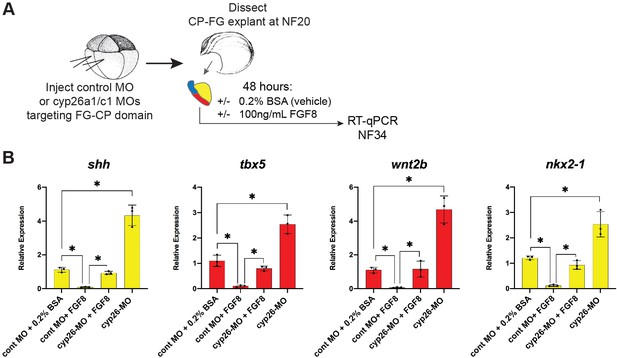
Cyp26a1/c1 morpholino (MO) knockdown phenocopies Cyp inhibitor treatment.
(A) Schematic of experiment testing FGF8-Cyp26 epistasis in Xenopus cardiopulmonary foregut (CP-FG) explants. Embryos were injected at the -cell stage with Cyp26a1+ Cyp26 c1 MOs, explants were then dissected at NF20, treated with vehicle controls (DMSO+0.2% BSA) or the indicated combinations of 100 ng/ml FGF8b, harvested at NF34, and analyzed via RT-qPCR. (B) FGF8 treatment suppresses the shh, tbx5, wnt2b, and nkx2-1 expression, and this inhibitory effect is blocked by Cyp26a1/c1 MO knockdown. RT-qPCR showing mean relative expression of the indicated genes, ± standard deviation. Each black dot in the graph represents a biological replicate (pool of n=4 foregut explants). *p<0.05, parametric two-tailed paired t-test.
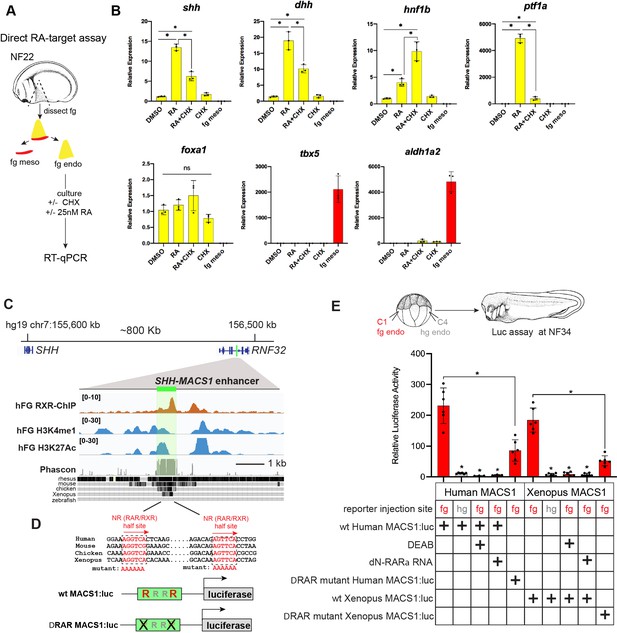
RA-RAR directly activates shh transcription in the Xenopus foregut endoderm via an evolutionarily conserved MACS1 enhancer.
(A) Schematic of direct RA target gene assay. Foregut endoderm (fg endo; yellow) was dissected from foregut mesoderm (fg meso; red) at NF25, pre-treated with 1 µM cycloheximide (CHX) for 2 hr prior to culture in 25 nM RA + CHX (or DMSO vehicle control) for 6 hr followed by RT-qPCR analysis. (B) RA directly activates shh and dhh expression in the presence of CHX. Graphs show mean relative expression ± standard deviation from N=3 biological replicates (four explants/replicate). Endoderm genes are shown in yellow, mesoderm makers in red confirm dissections. *p<0.05, parametric two-tailed paired t-test. (C) Genome browser of the human SHH locus showing the evolutionarily conserved MACS1 distal enhancer (green shading) embedded in an intron of the RNF32. Published ChIP-seq tracks of RXR, H3K4me1, and H3K27ac1 from hPSC-derived foregut endoderm (Vinckier et al., 2020, GSE104840; Wang et al., 2015, GSE54471). (D) MACS1 enhancer contains multiple RAR/RXR DNA-binding half sites, two of which are highly conserved. Schematics of the wild-type and mutant MACS1:luciferase reporter constructs. (E) Luciferase reporter assay in Xenopus show that the Human and X. tropicalis MACS1 enhancers are activated by RA via the RAR/RXR DNA-binding sites. 50 pg of MACS1:luciferase reporter +5 pg pRL-TK reporter were microinjected±250 pg of dominant-negative RARa RNA into either the C1 foregut (fg; red bars) or C4 hindgut (hg; gray bars) blastomeres and luciferase activity was assayed at NF34. 10 μM DEAB treatment was from NF20 to NF34. Mean relative luciferase activity ± standard deviation, from N=6 biological replicates/time point with five embryos/replicate. *p<0.05, parametric two-tailed paired t-test relative to WT MACS1:luc in the foregut (fg). Also see Figure 6—figure supplement 1, Figure 6—figure supplement 2 and related source data files. ns, not significant.
-
Figure 6—source data 1
RT-qPCR source data of Xenopus foregut endoderm explants treated for 2 hr in cycloheximide (CHX) prior to 6 hr in CHX +25 nM RA.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Luciferase source data of Xenopus tropicalis and human Shh MACS1-enhancer-reporter activity at NF34.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig6-data2-v2.xlsx

Multiple species alignment of the Shh MACS1 enhancers.
(A) Clustal DNA sequence alignment; putative RAR/RXR nuclear receptor motifs predicted by the CisBP tool (Weirauch et al., 2014) are shaded in aqua blue (CisBP score>10) and in yellow (CisBP score>8). Asterisks below the nucleotide alignment indicated conserved bases amongst all four species. RAR/RXR motifs mutated and tested in this study are indicated and boxed in red. Genomic co-ordinates of the Shh MACS1 (mammalian-amphibian-conserved sequence 1; Sagai et al., 2009) enhancers used in the multiple alignment were. Xenopus tropicalis: v9.1/xenTro9 genome build, chr6:9535614–9536245; Chick: galGal3 genome build, chr2:8370257–8370909; Human: hg19 genome build, chr7:156459384–156460049; Mouse: mm9 genome build, chr5:29538631–29539277.
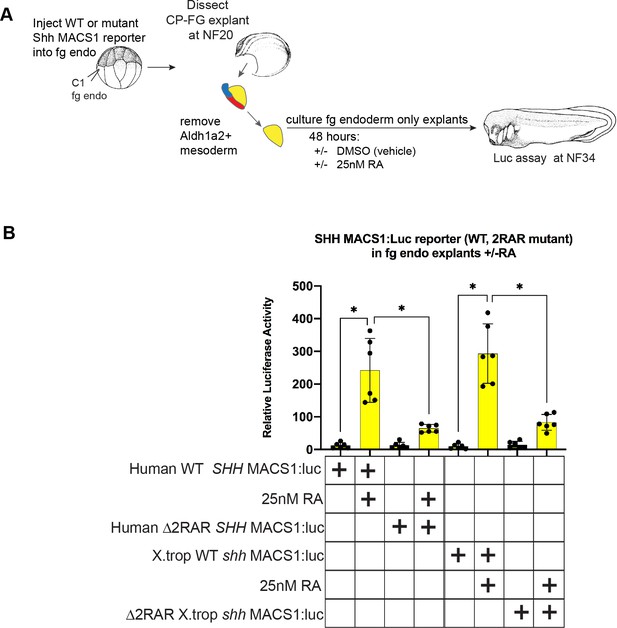
Conserved RAR/RXR sites in the Shh MACS1 enhancer are required for RA-mediated activation.
(A) Experimental schematic testing the ability of the wild-type (WT) or RAR/RXR mutant site reporters to respond to exogenous RA in Xenopus foregut endoderm explants. 50 pg of WT or mutant MACS1:luciferase reporter+5 pg pRL-TK reporter were microinjected into C1 foregut (fg) blastomeres; then at NF20 foregut explants were cut and the Aldh1a2+ fg lpm was manually removed, explants were treated with DMSO vehicle or 25 nM RA, and luciferase activity was assayed at NF34. Graphs show mean relative luciferase activity ± standard deviation; black dots in the graphs represent N=6 biological replicates, containing five embryos/replicate. Biological replicates contained pooled embryos from two separate fertilization/injection/dissections. *p<0.05, parametric two-tailed paired t-test. RA, retinoic acid.
-
Figure 6—figure supplement 2—source data 1
Luciferase source data of wild-type and mutant Shh-MACS1 reporters in Xenopus foregut endoderm explants ±25 nM RA.
- https://cdn.elifesciences.org/articles/69288/elife-69288-fig6-figsupp2-data1-v2.xlsx
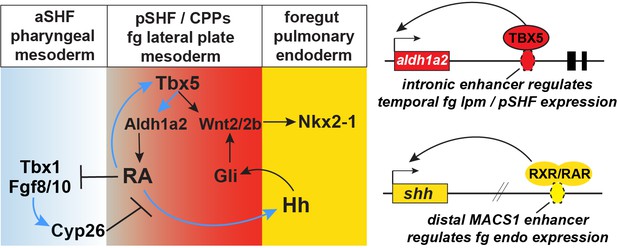
Model of the Tbx5 regulated GRN coordinating SHF pattering and pulmonary induction.
Our data indicate that between NF25and NF35 in Xenopus and around E9.5 in mice, Tbx5 directly maintains Aldh1a2 expression and a RA-Tbx5 positive feedback loop in the pSHF, which is necessary for Hh ligand expression, Wnt2/2b-dependent pulmonary fate induction, and SHF patterning. Blue arrows in the model indicate relationships demonstrated in this study. Tbx5/Aldh1a2-dependent RA signaling restricts FGF/Cyp activity in the aSHF, promotes pSHF identity, and drives expression of shh in pulmonary foregut endoderm. The aldh1a2 enh1 enhancer is directly regulated by Tbx5 and the shh MACS1 enhancer is regulated by RA/RXR/RAR. aSHF, anterior second heart field; GRN, gene regulatory network; pSHF, posterior second heart field.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background(Xenopus tropicalis, females) | Wild-typeadult females | Nasco | LM00823 | |
| Strain, strain background(X. tropicalis, males) | Wild-typeadult males | Nasco | LM00822 | |
| Strain, strain background(X. laevis, females) | Wild-typeadult females | Nasco | LM00531 | |
| Strain, strain background(X. laevis, males) | Wilt-typeadult males | Nasco | LM00715 | |
| Genetic reagent(X. tropicalis) | Xtr.Tg(WntREs: dEGFP)Vlemx | National Xenopus Resource (NXR) Center,Woods Hole, MA | RRID:NXR_1094 | X. tropicalis Wnt/Bcat reporter line |
| Genetic reagent(X. laevis) | Xla.Tg(WntREs: dEGFP)Vlemx, | NXR | RRID:NXR_0064 | X. laevis Wnt/Bcat reporter line |
| Genetic reagent(X. laevis) | Xla.Tg.(nkx2-5:GFP)Mohun | NXR | RRID:NXR_0030 | X. laevis Nkx2-5:GFP reporter line |
| Strain, strain background(Mus musculus) | CD-1 | Charles River Labs | Strain Code022RRID:IMSR_CRL:022 | WT mice |
| Genetic reagent(M. musculus) | Shhtm1(EGFP/cre)Cjt | Jax Labs | JAX: 005622RRID:IMSR_JAX:005622 | Shh:GFP mice |
| Cell line(M. musculus) | Tbx5OE-mESC line | Steimle et al., 2018 | Steimle et al., 2018 | |
| Chemical compound, drug | Doxycycline | Sigma-Aldrich | D9891 | |
| Antibody | (Rabbit polyclonal) anti-Aldh1a2 | Abcam | ab96060, RRID:AB_10679336 | IF (1:500) |
| Antibody | (MouseMonoclonal)Anti-Aldh1/2 | Santa Cruz Biotechnology | sc-166362, RRID:AB_2009458 | IF (1:500) |
| Antibody | (Mouse monoclonal) anti-Sox2 | Abcam | ab79351; RRID:AB_10710406 | IF (1:1000) |
| Antibody | (Rabbit polyclonal) anti-Nkx2-1 (H-190) | Santa CruzBiotechnology | sc-13040X; RRID:AB_793532 | IF (1:500) |
| Antibody | (Mouse monoclonal) anti-Fibronectin (4H2) | Developmental Studies Hybridoma Bank | DSHB #4H2; RRID:AB_2721949 | IF (1:2000) |
| Antibody | (Chicken polyclonal) anti-GFP | Aves Labs | GFP-1020; RRID:AB_10000240 | IF (1:1000) |
| Antibody | (Goat polyclonal)anti-Tbx5 | Santa Cruz Biotechnology | sc-17866, RRID:AB_2200827 | IF (1:300)ChIP: 5 µg |
| Recombinant DNA reagent | pI-SceI-d2EGFP plasmid | Addgene | Addgene_32674 | For meganuclease transgenics |
| Recombinant DNA reagent | pRL-TK(plasmid) | Promega | E2241 | |
| Recombinant DNA reagent | pGL4.23 luc2/miniP(plasmid) | Promega | E8411 | |
| Recombinant DNA reagent | pCS2+ GR-xTbx5 | Addgene | Addgene 117248 | |
| Recombinant DNA reagent | pCS2+ xTbx5 | Addgene | Addgene 117247 | |
| Recombinant DNA reagent | pCSf107mT-Gateway-3′myc | Addgene | Addgene 67617 | |
| Recombinant DNA reagent | pENTR223Human TBX5 | Horizon Discovery | OHS5894-202500411 | |
| Commercial assay or kit | Gateway LR Clonase II enzyme mix | Thermo Fisher Scientific | 11791020 | |
| Commercial assay or kit | mMessage mMachine SP6 RNA synthesis kit | Thermo Fisher Scientific | AM1340 | |
| Peptide, recombinant protein | FGF8b | R&D Systems | 423-F8-025 | |
| Peptide, recombinant protein | WNT2B | R&D Systems | 3900-WN-025 | |
| Commercial assay or kit | TRIzol | Thermo Fisher Scientific | 15596018 | |
| Commercial assay or kit | Direct-zolMiniprep plus kit | Thermo Fisher Scientific | R2070 | |
| Commercial assay or kit | SuperscriptVILO mastermix | Thermo Fisher Scientific | 11755050 | |
| Commercial assay or kit | PowerUP2× SYBR Green MasterMix | Thermo Fisher Scientific | A25742 | |
| Commercial assay or kit | Firefly Luciferase 2.0 kit | Biotium | 30085-1 | |
| Commercial assay or kit | Renilla Luciferase2.0 kit | Biotium | 30082-1 | |
| Chemical compound, drug | DEAB | Sigma-Aldrich | D86256 | |
| Chemical compound, drug | All-trans retinoic acid(RA) | Sigma-Aldrich | R2625 | |
| Chemical compound, drug | Ketoconazole | Tocris | Tocris#1103 | |
| Chemical compound, drug | Cycloheximide | Sigma-Aldrich | C4859 | |
| Chemical compound, drug | Dexamethasone | Sigma-Aldrich | Sigma D4902 | |
| Peptide, recombinant protein | CAS9 | PNA Bio | CP01-20 | |
| Sequence-based reagent | X. tropicalis tbx5 exon5 sgRNA | IDT DNA | GGGGTTCTGATATGAAGTGA | Steimle et al., 2018 |
| Sequence-based reagent | X. laevis Tbx5 MO1 | GeneTools | 5′-TTA GGA AAG TGT CTC TGG TGT TGC C -3′; | Brown et al., 2005 |
| Sequence-based reagent | X. laevis Tbx5 3 bp mismistach MO1 | GeneTools | 5′-TCA GTA AAG TAT CTC TGG TGT TGC C-3′ | This paper |
| Sequence-based reagent | X. laevis Tbx5 MO2 | GeneTools | 5′-CAT AAG CCT CCT CTG TGT CCG CCA T-3 | Brown et al., 2005 |
| Sequence-based reagent | X. laevis Tbx5 3 bp mismatch MO2 | GeneTools | 5′-TAT CAG ACT CCT CTG TGT CCG CCA T-3′ | This paper |
| Sequence-based reagent | X. laevis Aldh1a2-MO | GeneTools | 5′-GCA TCT CTA TTT TAC TGG AAG TCAT-3′ | Strate et al., 2009 |
| Sequence-based reagent | X. laevis Cyp26a1-MO | GeneTools | 5′-TAG TGA GCA GAG TAT ACA GAT CCA T-3′ | Janesick et al., 2013 |
| Sequence-based reagent | X. laevis Cyp26c1-MO | GeneTools | 5′-TAC AAG ATG TTC CTC CTT GAG ATC A-3′ | Yu et al., 2016 |
| Commercial assay, kit | Protein G-conjugated magnetic beads | Life Technologies | 1,003D | |
| Commercial assay, kit | NEBNext Ultra DNA Library Prep Kit | New England Biolabs | E7370S | |
| Commercial assay, kit | Sera-Mag magnetic beads | GE | 6515-2105-050-250 | |
| Software, algorithm | Morpheus | Broad Institute | https://software.broadinstitute.org/morpheusRRID:SCR_017386 | |
| Commercial assay, kit | SceI mega-nuclease enzyme | New England Biolabs | R0694S | Use within 1 month of purchase, store at –80°C |
| Commercial assay, kit | Dispase | Corning Life Sciences | 354235 | Use at 10 U/ml on Xenopus explants |
Additional files
-
Supplementary file 1
RT-qPCR primers used in this study for Xenopus and mouse.
- https://cdn.elifesciences.org/articles/69288/elife-69288-supp1-v2.xlsx
-
Supplementary file 2
Gene sets, utilized in Figure 1, from de Soysa et al., 2019 (GSE126128) single-cell RNA-seq studies of developing E7.75-E9.5 mouse embryos that transcriptionally define the anterior and posterior second heart field territories.
- https://cdn.elifesciences.org/articles/69288/elife-69288-supp2-v2.xlsx
-
Supplementary file 3
Gene sets, utilized in Figure 1, from Han et al., 2020 (GSE136689) single-cell RNA-seq studies of developing e8.75-e9.5 mouse embryos that transcriptionally define the cardiopulmonary progenitor (CPP)+ lung and ventral pharynx territories.
- https://cdn.elifesciences.org/articles/69288/elife-69288-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69288/elife-69288-transrepform1-v2.docx





