Divergent acyl carrier protein decouples mitochondrial Fe-S cluster biogenesis from fatty acid synthesis in malaria parasites
Figures
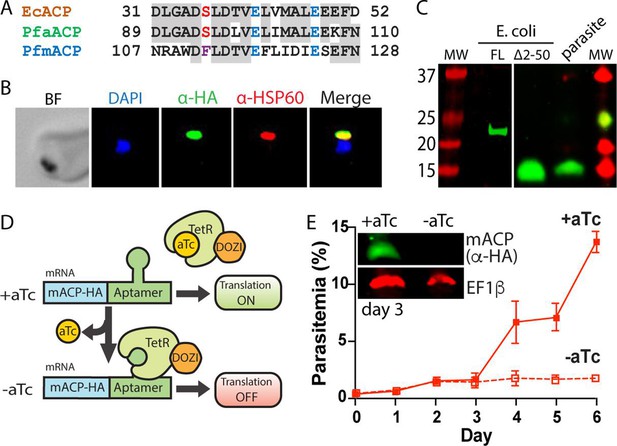
Mitochondrial ACP is essential for blood-stage P. falciparum.
(A) Partial sequence alignment of ACP homologs from E. coli and the P. falciparum apicoplast (aACP) and mitochondrion (mACP). The conserved Ser residue that is modified by a 4-Ppant group is in red, the divergent Phe residue of mACP is in purple, and conserved Glu residues are in blue. (B) Immunofluorescence microscopy of fixed Dd2 parasites episomally expressing mACP-HA2 and stained with DAPI (nucleus, blue), anti-HA (green), and anti-HSP60 (mitochondrion, red) antibodies (BF = bright field). (C) Anti-HA western blot (WB) analysis of cellular lysates of E. coli expressing full-length (FL) or truncated (∆2–50) mACP-HA2 and Dd2 P. falciparum parasites episomally expressing full-length mACP-HA2. The left and right images are from different WB experiments (10% gels) that were aligned by molecular weight (MW) markers. (D) Schematic depiction of protein translational regulation using the aptamer/TetR-DOZI system. Normal protein expression occurs in the presence but not absence of anhydrotetracycline (aTc). (E) Continuous growth assay of synchronous mACP-HA/FLAG-aptamer/TetR-DOZI parasites in the presence or absence of aTc. Data points and error bars are the average and standard deviation from two biological replicates. Inset is an anti-HA and anti-EF1β (37 kDa) WB of parasite lysates harvested on day 3 of the continuous growth assay. WBs were repeated 2–3 times.
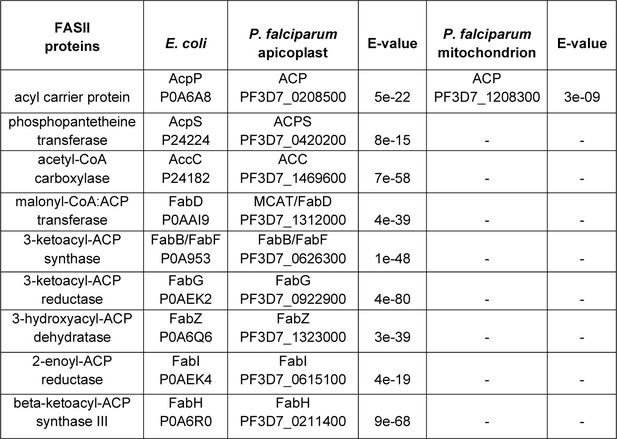
P. falciparum homologs of E. coli FASII proteins.
The e-value cut-off was 0.01.
-
Figure 1—figure supplement 1—source data 1
Source file for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig1-figsupp1-data1-v2.docx

Sequence and mass spectrometry (MS) analyses of ACP.
(A) Alignment of yeast ACP with P. falciparum apicoplast ACP (aACP) and mitochondrial ACP (mACP). (B) Sequence of P. falciparum mACP with peptide residues in red that were detected by tandem MS analysis of parasite samples in this study or available on PlasmoDB. The putative mACP processing site is shown, based on its proximity to the most N-terminal tryptic peptide detected (NSAFFSTEK) and alignment with the known mitochondrial ACP processing sites in yeast and humans (Vögtle et al., 2009; Vaca Jacome et al., 2015).
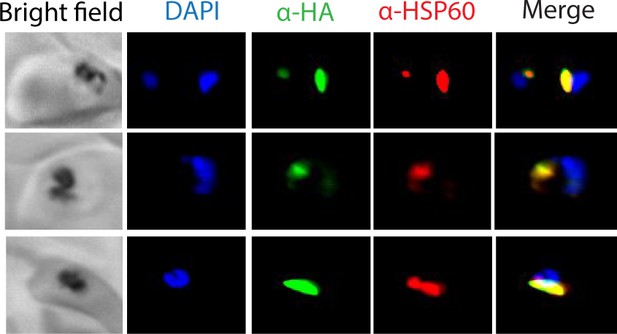
Additional immunofluorescence microscopy images mACP-HA2.
Epifluorescence microscopy images of fixed Dd2 parasites episomally expressing mACP-HA2 and stained with DAPI (nucleus, blue), anti-HA (green), and anti-HSP60 (red) antibodies.
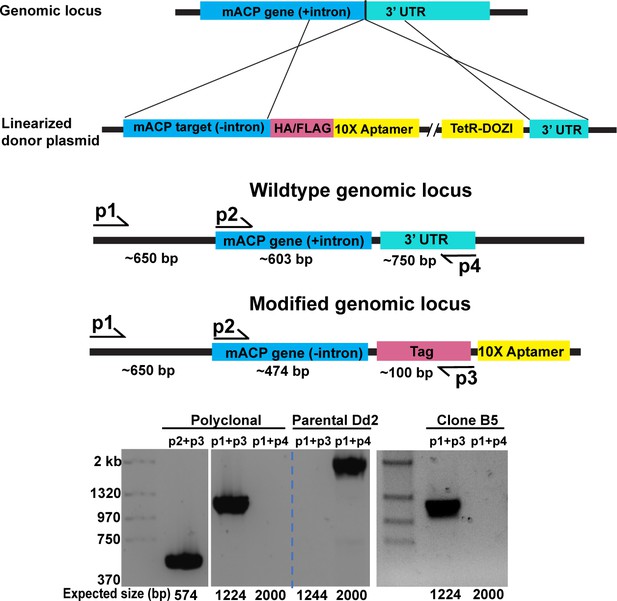
Schematic depiction of mACP gene editing and verifying genomic integration by PCR.

Giemsa-stained blood smears of mACP-aptamer/TetR-DOZI parasites cultured for 120 hr (5 days) ± anhydrotetracycline (aTc).
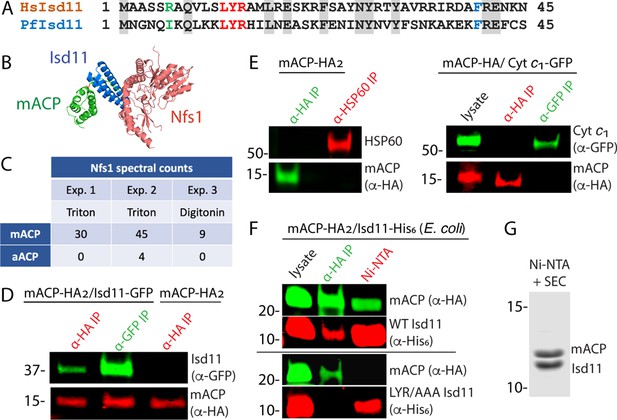
Mitochondrial ACP binds the Isd11-Nfs1 complex.
(A) Partial sequence alignment of human and P. falciparum Isd11. The conserved LYR sequence motif is in red, the conserved Phe residue is in blue, and residue 6 is in green. (B) X-ray structural model of the mACP-Isd11-Nfs1 complex (PDB entry 5USR). For simplicity, only a single copy of each protein is shown, rather than the functional dimer (Boniecki et al., 2017). (C) Table of spectral counts for P. falciparum Nfs1 detected by tandem mass spectrometry for anti-HA immunoprecipitation (IP) studies of lysates from Dd2 parasites episomally expressing mACP-HA2 or aACP-HA2. Parasites were lysed in either Triton X-100 or digitonin. Similar spectral counts for mACP and aACP bait proteins were detected in each experiment (Figure 2—figure supplement 5). (D) Anti-HA or anti-GFP co-immunoprecipitation (co-IP)/western blot (WB) studies of Dd2 parasites episomally expressing mACP-HA2 with or without Isd11-GFP. (E) Anti-HA, anti-HSP60, or anti-GFP co-IP/WB studies of Dd2 parasites episomally expressing mACP-HA2 or endogenously expressing mACP-HA (knockdown line) and episomally expressing cyt c1-GFP. (F) Anti-HA co-IP or nickel-nitrilotriacetic acid (Ni-NTA) pulldown and WB studies of E. coli bacteria recombinantly expressing full-length mACP-HA2 and Isd11-His6 (WT or LYR/AAA mutant) and probed with anti-HA and anti-His6 antibodies. All lanes were loaded with equivalent sample volumes, are from the same blot, and were processed identically but were cropped from non-contiguous lanes. (G) Coomassie-stained SDS-PAGE gel of recombinant His6-Isd11 and ∆2–50 mACP-HA2 purified from E. coli by Ni-NTA pulldown of His6-Isd11 and size-exclusion chromatography (SEC) after removal of affinity tags. Additional images for WB experiments are shown in Figure 2—figure supplement 3. WBs are representative of 2–3 replicate experiments with independent samples.

BLAST analysis of LYR-motif protein homologs found in P. falciparum.
The e-value cut-off was 0.01.
-
Figure 2—figure supplement 1—source data 1
Source file for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig2-figsupp1-data1-v2.docx

Specific enrichment of Nfs1 in anti-HA-tag IP of mACP-HA2 relative to anti-HA-tag IP aACP-HA2.
Specific enrichment in mACP sample was observed in three matched experimental samples, calculated as log2 of the spectral count ratio (IP:mACP/IP:aACP).
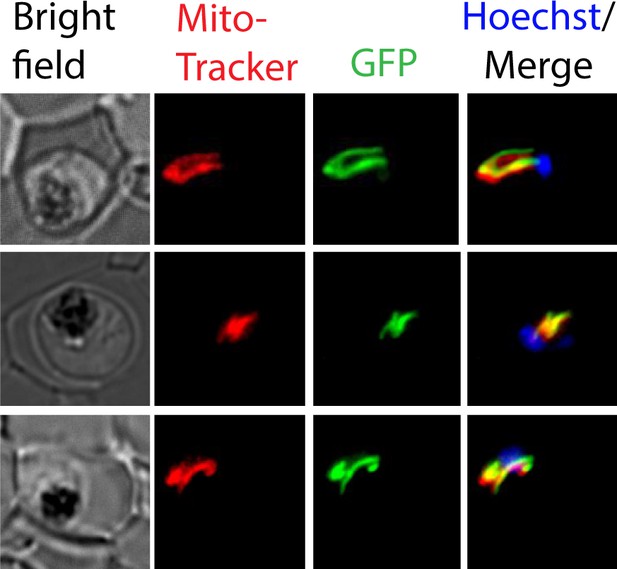
Co-localization of Isd11-GFP and MitoTracker in Dd2 parasites expressing mACP-HA2 and Isd11-GFP.
Live parasites were treated with 1 µg/mL Hoechst and 10 nM MitoTracker and imaged by epifluorescence microscopy.
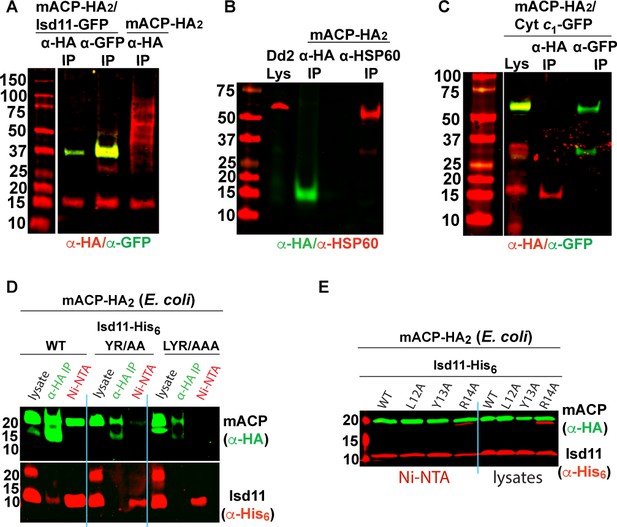
Additional images for western blot (WB) analyses.
Reciprocal immunoprecipitation (IP)/WB studies of Dd2 parasites expressing (A) mACP-HA2 with and without Isd11-GFP, (B) mACP-HA2 and HSP60, and (C) mACP-HA (endogenous) and cyt c1-GFP. Anti-HA IP or nickel-nitrilotriacetic acid (Ni-NTA) pulldown and WB studies of E. coli bacteria recombinantly expressing (D) full-length mACP-HA2 and Isd11-His6 (WT, YR/AA, LYR/AAA, L12A, Y13A, or R14A mutants) and (E) ∆2–50 mACP-HA2 (WT or F113A mutant) and Isd11-His6 and probed with anti-HA and anti-His6 antibodies. Blots were probed as labeled and as described in Materials and methods. Equivalent sample volumes were loaded in all lanes in a given gel.
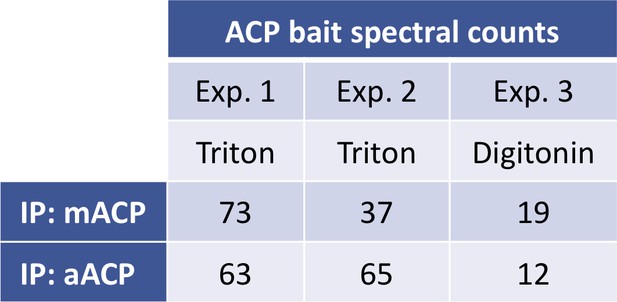
Spectral counts for P. falciparum mACP or aACP detected by tandem mass spectrometry.
Samples are for anti-HA IP studies of lysates from Dd2 parasites episomally expressing mACP-HA2 or aACP-HA2. Parasites were lysed in either Triton X-100 or digitonin.
-
Figure 2—figure supplement 5—source data 1
Source file for Figure 2—figure supplement 5.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig2-figsupp5-data1-v2.docx

Mass spectrometry (MS) analysis of recombinant parasite Isd11 and mACP expressed in bacteria.
Sequences are for P. falciparum Isd11 and mACP constructs that were heterologously coexpressed in E. coli, purified by nickel-nitrilotriacetic acid (Ni-NTA) and size-exclusion chromatography, and analyzed by tandem MS. Blue sequence reflects the overall coverage of residues for peptides that were detected by tandem MS analysis after tryptic digest. Lines represent individual peptides that were detected.

P. falciparum Isd11 does not stably bind to apicoplast ACP.
Anti-HA immunoprecipitation (IP) or nickel-nitrilotriacetic acid (Ni-NTA) pulldown and western blot of E. coli bacteria recombinantly expressing ∆2–40 apicoplast ACP-HA2 and Isd11-His6 and probed with anti-HA and anti-His6 antibodies.
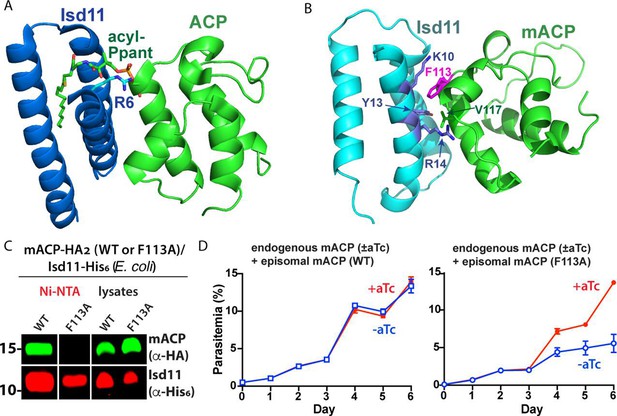
Structural modeling and functional tests of the divergent Phe113 residue of parasite mACP.
(A) X-ray crystallographic structure of human Isd11 bound to E. coli ACP highlighting the central role of the acyl-Ppant group of ACP and R6 residue of Isd11 in stabilizing this complex (PDB entry 5USR). (B) Rosetta-based, energy-minimized structural model of the P. falciparum mACP-Isd11 interface. Conserved electrostatic interactions that also contribute to the binding interface are shown in Figure 3—figure supplement 1. The Rosetta model is included as Figure 3—source data 1. (C) Nickel-nitrilotriacetic acid (Ni-NTA) pulldown and WB studies of E. coli bacteria recombinantly expressing ∆2–50 mACP-HA2 (WT or F113A mutant) and Isd11-His6 and probed with anti-HA and anti-His6 antibodies. All lanes were loaded with equivalent sample volumes, are from the same blot, and were processed identically but were cropped from non-contiguous lanes. The uncropped WB is shown in Figure 2—figure supplement 3. (D) Continuous growth assays of synchronous mACP-aptamer/TetR-DOZI parasites episomally expressing WT or F113A mACP-HA2 and grown in the presence or absence of anhydrotetracycline (aTc). Data points and error bars are the average and standard deviation from two biological replicates. The presence of an HA epitope tag on both the endogenous and episomal mACP prevented determination of endogenous mACP expression differences ± aTc by WB. WBs are representative of 2–3 replicate experiments with independent samples.
-
Figure 3—source data 1
PDB file of energy-minimized Rosetta model of the P. falciparum Isd11-mACP complex.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig3-data1-v2.pdb
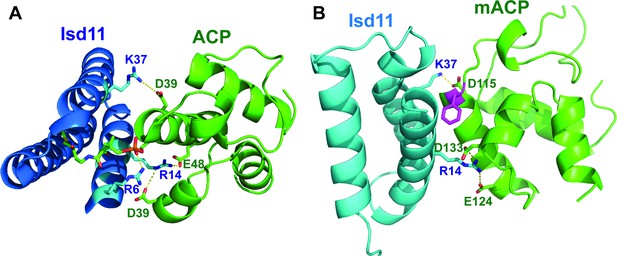
Conserved electrostatic interactions at the mACP-Isd11 interface.
(A) X-ray structure of human Isd11 bound to E. coli ACP (PDB 5USR). (B) Energy-minimized Rosetta model of P. falciparum Isd11 bound to mACP.

Uncropped image of western blot (WB) probing interaction of WT or F113A mACP-HA2 and Isd11-His6 in E. coli.
Anti-HA immunoprecipitation (IP) or nickel-nitrilotriacetic acid (Ni-NTA) pulldown and WB studies of E. coli bacteria recombinantly expressing ∆2–50 mACP-HA2 (WT or F113A mutant) and Isd11-His6 and probed with anti-HA and anti-His6 antibodies. Equivalent sample volumes were loaded in all lanes.

Loss of mACP specifically destabilizes Nfs1 and the Rieske Fe-S protein.
Western blot analysis of equivalent volumes of lysates from mACP-aptamer/TetR-DOZI Dd2 parasites without or with episomal expression of Rieske-HA2 or cyt c1-GFP that were grown ±aTc for 3 days and probed for cytosolic EF1α (~50 kDa) and (A) Nfs1, (B) HA-tag and HSP60, or or (C) GFP. For (B), the blot was first probed with rat anti-HA and rabbit anti-HSP60 1° antibodies, imaged after probing with anti-rat and anti-rabbit 2° antibodies, probed additionally with rabbit anti-EF1α antibody, and imaged again after probing with anti-rabbit 2° antibody. The diminished intensity of EF1α in the -aTc relative to +aTc samples likely reflects the slowing growth of these late second-cycle parasites cultured -aTc due to reduced fitness from loss of mACP. (D) Mean ± SEM of the integrated signal intensity for each indicated protein under ± aTc conditions relative to the loading control (EF1α or EF1β) and normalized to +aTc conditions. Black circles are individual data points from 2–3 replicate experiments with independent samples. Normalized signal intensity values for +aTc versus -aTc conditions were analyzed by two-tailed unpaired t-test to determine the following p values: Nfs1 (0.2), Rieske-HA2 (0.07), HSP60 (0.9), and cyt c1 (0.4). Uncropped blots are shown in Figure 4—source data 1.
-
Figure 4—source data 1
Raw integrated signal intensity values for western blot densitometry analysis.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig4-data1-v2.xlsx
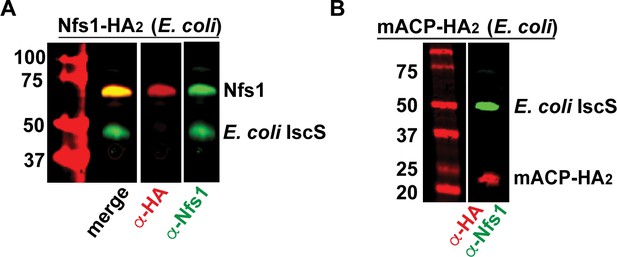
Validation of anti-Nfs1 antibody for specific recognition of the parasite Nfs1 homolog.
Western blot (WB) studies of E. coli bacteria recombinantly expressing P. falciparum Nfs1-HA2 (A) or full-length mACP-HA2 (B) and probed with anti-HA and anti-His6 antibodies. P. falciparum Nfs1-HA2 was recognized by both the anti-HA and anti-Nfs1 antibodies, whereas full-length mACP-HA2 was only recognized by the anti-HA antibody. In both lysates, the anti-Nfs1 antibody also recognized the endogenous E. coli IscS (~48 kDa) that is ~60% identical to human Nfs1.
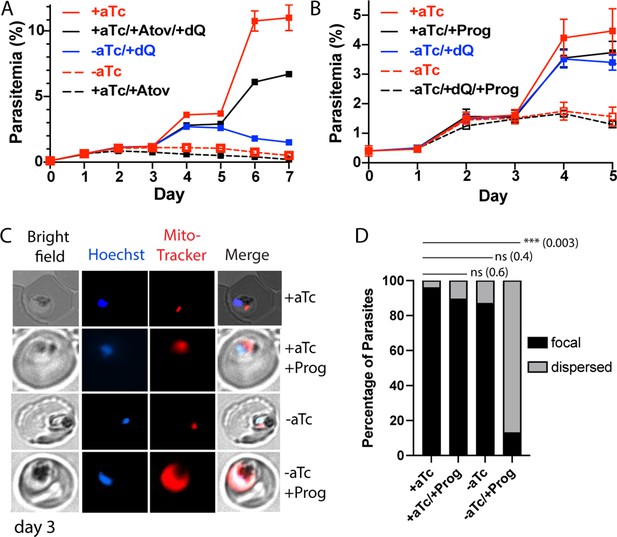
Loss of mACP causes electron transport chain (ETC) failure and sensitizes parasites to proguanil.
Continuous growth assays of synchronous mACP-aptamer/TetR-DOZI parasites grown (A) ± anhydrotetracycline (aTc), ± atovaquone (Atov, 100 nM), ± decyl-ubiquinone (dQ, 15 µM), and (B) ± proguanil (Prog, 1 µM). Data points and error bars are the average and standard deviation from 2 to 3 biological replicates. (C) Fluorescence microscopy images of live mACP-aptamer/TetR-DOZI Dd2 parasites cultured 3 days ±aTc, 2 days ± 5 µM proguanil, and stained with Hoechst or MitoTracker Red (10 nM). (D) Statistical analysis of MitoTracker signal for 40–50 total parasites from each condition in panel (C) from two independent experiments. MitoTracker signal was designated as focal if similar in size to nuclear signal (Hoechst stain) or dispersed if similar in size to the parasite cytoplasm (based on brightfield image). For clarity, error bars are not shown but standard errors of the mean were ≤10% in all cases. Cell percentage differences were analyzed by two-tailed unpaired t-test (p values in parentheses, ns = not significant). Parasite counts for individual experiments are given in Figure 5—source data 1.
-
Figure 5—source data 1
Raw parasite counts for individual microscopy experiments.
- https://cdn.elifesciences.org/articles/71636/elife-71636-fig5-data1-v2.xlsx
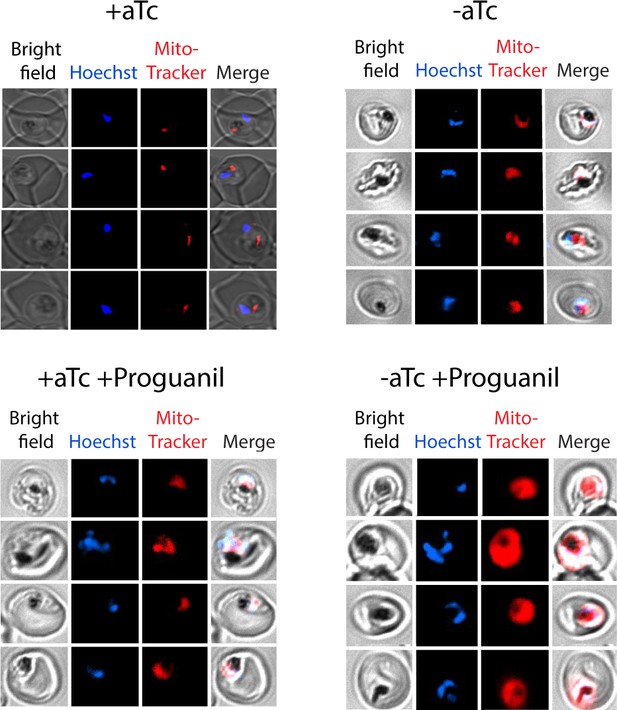
Additional live microscopy images of mACP-aptamer/TetR-DOZI parasites.
Parasites were cultured for 72 hr ± anhydrotetracycline (aTc) and for 24 hr ± proguanil (5 µM) and treated with 10 nM MitoTracker Red and Hoechst.
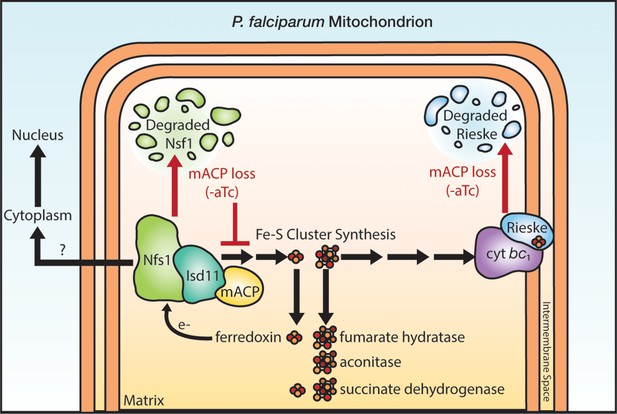
Schematic model for the impact of mACP knockdown (-aTc) on Fe-S cluster metabolism in P. falciparum parasites.
The question mark indicates uncertainty in the functional role of mitochondrial Nfs1 in supporting cytoplasmic Fe-S cluster biogenesis in parasites. For simplicity, the mACP/Isd11/Nfs1 complex is shown with only one monomer for each protein, rather than the functional dimer (Boniecki et al., 2017). Illustration by Megan Okada.

Sequence alignment of P. falciparum mACP (PF3D7_1208300) with homologs from T. gondii (TGME49_270310, e-value 2e-13), B. microti (BMR1_02g01455, e-value 7e-13), and V. brassicaformis (CEM12831, e-value 3e-16).
The divergent residue at the amino acid position of the canonical Ser (in mACP from yeast/humans) is colored red. T. gondii also encodes a second mACP homolog (TGME49_265538, e-value 2e-11), not shown in this alignment, with a Gly residue at the position of the canonical Ser.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Plasmodium falciparum) | Dd2 | PMID:1970614 | BEI ResourcesMRA-156 | |
| Cell line (Plasmodium falciparum) | Dd2 mACP-HA-FLAG 10X Aptamer/TetR-DOZI | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2Cyt c1-GFP (pTEOE) mACP-HA-FLAG 10X Aptamer/TetR-DOZI | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2Rieske-HA2 (pTEOE) mACP-HA-FLAG 10X Aptamer/TetR-DOZI | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2 mACP-HA2 (pTEOE) mACP-HA-FLAG 10X Aptamer/TetR-DOZI | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2F113A mACP-HA2 (pTEOE) mACP-HA-FLAG 10X Aptamer/TetR-DOZI | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2 mACP-HA2 (pTEOE) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2 aACP-HA2 (pTEOE) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line (Plasmodium falciparum) | Dd2 mACP-HA2 (pTEOE)Isd11-GFP (pRL2) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3Δ2–51 mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3Isd11-His6 (pET21d) mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3LYR12-14AAAIsd11-His6 (pET21d) mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3Isd11-His6 (pET21d)∆2–51 mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3Isd11-His6 (pET21d)F113A (∆2–51) mACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Cell line(Escherichia coli) | BL21/DE3Isd11-His6 (pET21d)∆2–40 aACP-HA2 (pET28a) | Made for this study | Described in Materials and methods. Can be obtained from Sigala lab. | |
| Chemical compound, drug | Anhydrotetracycline | Caymen Chemicals | Cat. no.10009542 | |
| Chemical compound, drug | Atovaquone | Caymen Chemicals | Cat. no.95233184 | |
| Chemical compound, drug | Decyl-ubiquinone | Caymen Chemicals | Cat. no.55486005 | |
| Chemical compound, drug | Proguanil | Sigma-Aldrich | Cat. no.637,321 | |
| Antibody | Anti-HA(rat monoclonal 3F10) | Sigma-Aldrich | Cat. no.11867423001,RRID:AB_390918 | (1:1000) |
| Antibody | Anti-HSP60(rabbit polyclonal) | Novus | Cat. no.NBP2-12734 | (1:1000) |
| Antibody | Anti-Nfs1(rabbit polyclonal) | Abcam | Cat. no. ab229829 | (1:1000) |
| Antibody | Anti-GFP(goat polyclonal) | Abcam | Cat. no. ab5450,RRID:AB_304897 | (1:1000) |
| Antibody | Anti-EF1α(rabbit polyclonal) | PMID:11251817 | (1:1000) | |
| Antibody | Anti-EF1β(rabbit polyclonal) | PMID:11251817 | (1:1000) | |
| Antibody | Anti-His6-DyLight680(mouse monoclonal) | Thermo Fisher | Cat. no.MA121315D680,RRID:AB_2536987 | (1:1000) |
Additional files
-
Supplementary file 1
PCR primers used for cloning and genotyping.
- https://cdn.elifesciences.org/articles/71636/elife-71636-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71636/elife-71636-transrepform1-v2.pdf





