Generation and diversification of recombinant monoclonal antibodies
Figures
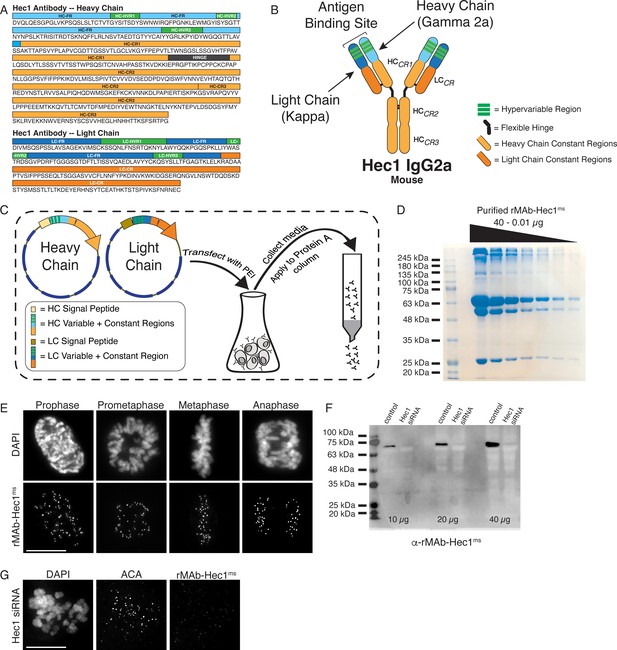
Generation of a recombinant Hec1 monoclonal antibody.
(A) Sequence data obtained for the Hec1 antibody is annotated for the heavy chain (HC) and light chain (LC) variable regions (HC = light blue; LC = dark blue), hypervariable regions (green), constant regions (HC = light orange; LC = dark orange), and the flexible hinge (dark gray). (B) rMAb-Hec1ms antibody structure and nomenclature. (C) Cloning, transfection, and purification scheme. Heavy and light chain coding regions were cloned into separate plasmids for transfection into human Expi293F cells. Cell media containing secreted antibodies was collected and applied to a Protein A Sepharose column. After column washing, antibodies were eluted using a low pH buffer. (D) Purified rMAb-Hec1ms antibody was serially diluted and run on a 12% SDS- polyacrylamide gel. The prominent band that runs above the 245 kDa molecular mass marker is likely a population of non-denatured antibody. The bands running at ~63 and ~50 kDa are glycosylated and non-glycosylated heavy chains, respectively. The antibody light chain runs at ~25 kDa. (E) HeLa cells immunostained with the purified rMAb-Hec1ms antibody. Cells were also stained with DAPI to detect chromosomes. (F) Immunoblot of control and Hec1 siRNA-depleted HeLa cell lysates. Increasing amounts of lysates are shown, and the blot is probed with the purified rMAb-Hec1ms antibody. (G) HeLa cell treated with Hec1 siRNA and stained with the rMAb-Hec1ms antibody and an ACA (anti-centromere) antibody to detect kinetochores. Cell is also stained with DAPI to detect chromosomes. Scale bars are 10 µm.
-
Figure 1—source data 1
Serially diluted, purified rMAb-Hec1-ms antibody analyzed by SDS-PAGE.
- https://cdn.elifesciences.org/articles/72093/elife-72093-fig1-data1-v3.tif
-
Figure 1—source data 2
Immunoblot of control and Hec1 siRNA- depleted HeLa cell lysates probed with purified rMAb-Hec1-ms antibody.
- https://cdn.elifesciences.org/articles/72093/elife-72093-fig1-data2-v3.tif
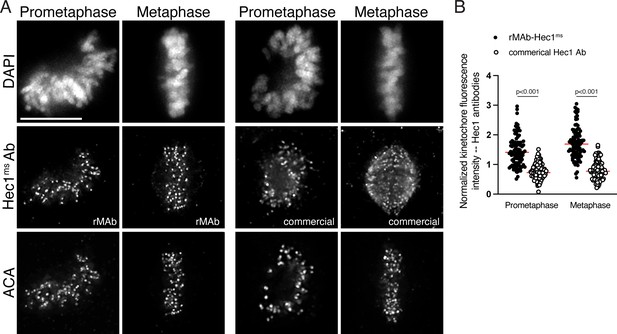
Sensitivity of commercial Hec1 monoclonal antibody compared to the recombinant Hec1 antibody.
(A) HeLa cells immunostained with the rMAb-Hec1ms antibody (left) and the commercial, traditionally generated Hec1 9G3 antibody (right). Cells were also stained with an ACA (anti-centromere) antibody to detect kinetochores and with DAPI to detect chromosomes. Cells in early prometaphase and metaphase are shown. (B) Quantification of kinetochore fluorescence intensities of cells immunostained with both antibodies. Values were normalized to the kinetochore fluorescence intensity values of the ACA antibody, which recognizes CENP-A, -B, and -C. For each condition, at least 20 kinetochores from at least five cells were measured. For each pair of measurements, a Student’s t-test was carried out to determine statistical significance. Scale bars in all immunofluorescence images are 10 µm.
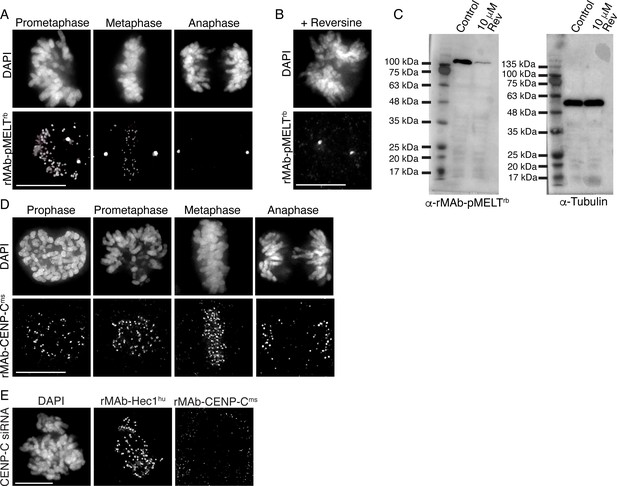
Characterization of recombinant antibodies to KNL1 pMELT and CENP-C.
(A) HeLa cells immunostained with rMAb-pMELTrb antibodies. (B) HeLa cell treated with 10 µM reversine and immunostained with rMAb-pMELTrb antibodies. (C) Immunoblots of lysates generated from untreated or reversine-treated HeLa cells expressing a 100 kDa fragment of KNL1 containing multiple pMELT domains, and probed with rMAb-pMELTrb antibodies (left) and tubulin antibodies as a loading control (right). (D) HeLa cells immunostained with rMAb-CENP-Cms antibodies. (E) HeLa cell treated with CENP-C siRNA and stained with rMAb-CENP-Cms antibodies and rMAb-Hec1hu antibodies to detect kinetochores. In all immunofluorescence images, cells were stained with DAPI to detect chromosomes. Scale bars are 10 µm.
-
Figure 2—source data 1
Immunoblots of lysates generated from untreated or reversine-treated HeLa cells expressing a 100 kDa fragment of KNL1 containing multiple MELT domains, and probed with either the rMAb-pMELT-rb antibody or a tubulin antibody.
- https://cdn.elifesciences.org/articles/72093/elife-72093-fig2-data1-v3.tif
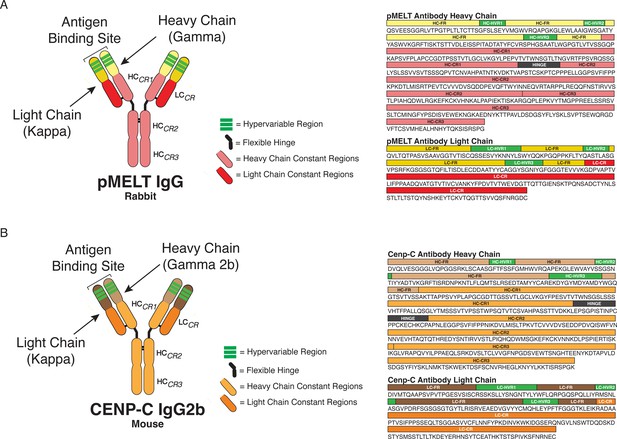
KNL1 pMELT and CENP-C antibody classification and domain architecture.
(A) Sequence data and domains for the KNL1 pMELT antibody are annotated for the heavy chain (HC) and light chain (LC) variable regions (HC = light yellow; LC = dark yellow), hypervariable regions (green), constant regions (HC = light pink; LC = red), and the flexible hinge (dark gray). (B) Sequence data and domains for the CENP-C antibody are annotated for the HC and LC variable regions (HC = light brown; LC = dark brown), hypervariable regions (green), constant regions (HC = light orange; LC = dark orange), and the flexible hinge (dark gray).
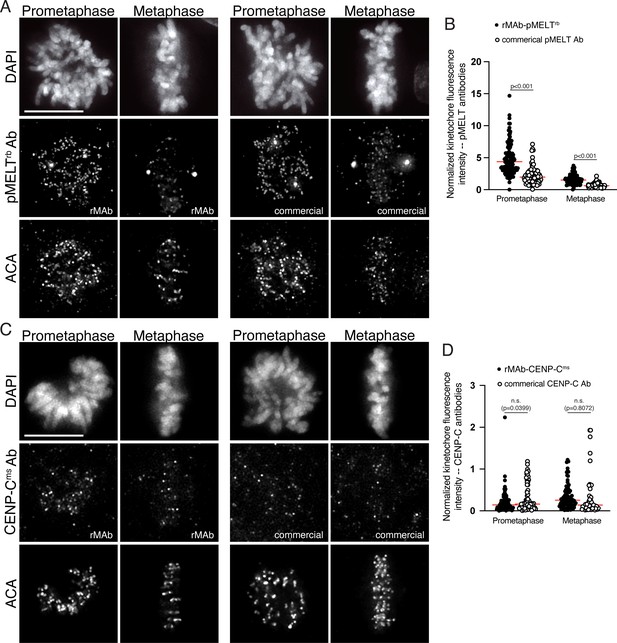
Sensitivity of commercial KNL1 pMELT and CENP-C monoclonal antibodies compared to the recombinant antibodies.
(A) HeLa cells immunostained with the rMAb-pMELTrb antibody (left) and the commercial, traditionally generated KNL1 pMELT antibody (right). Cells were also stained with an ACA antibody to detect kinetochores and with DAPI to detect chromosomes. Cells in early prometaphase and metaphase are shown. (B) Quantification of kinetochore fluorescence intensities of cells immunostained with both KNL1 pMELT antibodies. (C) HeLa cells immunostained with the rMAb-CENP-Cms antibody (left) and the commercial, traditionally generated CENP-C antibody (right). Cells were also stained with an ACA antibody to detect kinetochores and with DAPI to detect chromosomes. Cells in early prometaphase and metaphase are shown. (D) Quantification of kinetochore fluorescence intensities of cells immunostained with both CENP-C antibodies. For all quantifications, values were normalized to the kinetochore fluorescence intensity values of the ACA antibody which recognizes kinetochore proteins CENP-A, -B, and -C. For each condition, at least 20 kinetochores from at least five cells were measured. For each pair of measurements, a Student’s t-test was carried out to determine statistical significance. Scale bars in all immunofluorescence images are 10 µm.
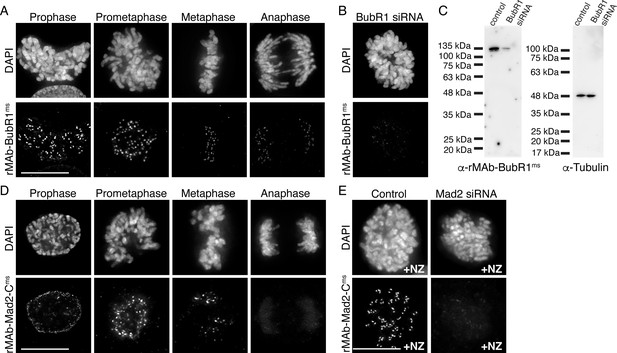
Characterization of recombinant antibodies to BubR1 and Mad2-C.
(A) HeLa cells immunostained with rMAb-BubR1ms antibodies. (B) HeLa cell treated with BubR1 siRNA and immunostained with rMAb-BubR1ms antibodies. (C) Immunoblots of lysates generated from control or BubR1 siRNA-treated HeLa cells and probed with rMAb-BubR1ms antibodies (left) and tubulin antibodies as a loading control (right). (D) HeLa cells immunostained with rMAb-Mad2-Cms antibodies. (E) HeLa cells (±Mad2 siRNA) pre-treated with 500 nM nocodazole for 12 hr and immunostained with rMAb-Mad2-Cms antibodies. In all immunofluorescence images, cells were stained with DAPI to detect chromosomes. Scale bars are 10 µm.
-
Figure 3—source data 1
Immunoblots of lysates generated from control or BubR1 siRNA-treated HeLa cells and probed with either the rMAb-BubR1-ms antibody or a tubulin antibody.
- https://cdn.elifesciences.org/articles/72093/elife-72093-fig3-data1-v3.tif
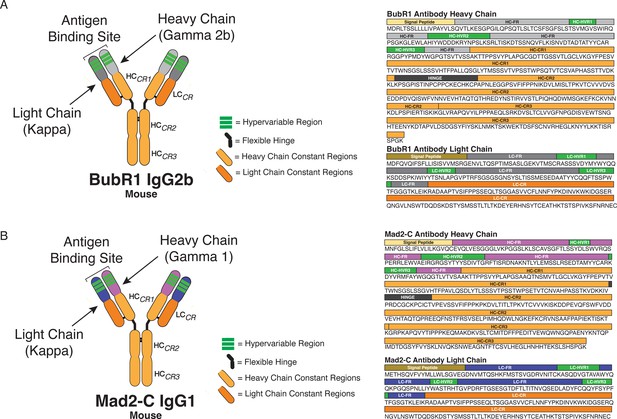
BubR1 and Mad2-C antibody classification and domain architecture.
(A) Sequence data and domains for the BubR1 antibody are annotated for the heavy chain (HC) and light chain (LC) variable regions (HC = light gray; LC = dark gray), hypervariable regions (green), constant regions (HC = light orange; LC = dark orange), and the flexible hinge (dark gray). (B) Sequence data and domains for the Mad2-C antibody are annotated for the HC and LC variable regions (HC = light purple; LC = dark purple), hypervariable regions (green), constant regions (HC = light orange; LC = dark orange), and the flexible hinge (dark gray). For each antibody sequence, the signal peptide domain is also highlighted.
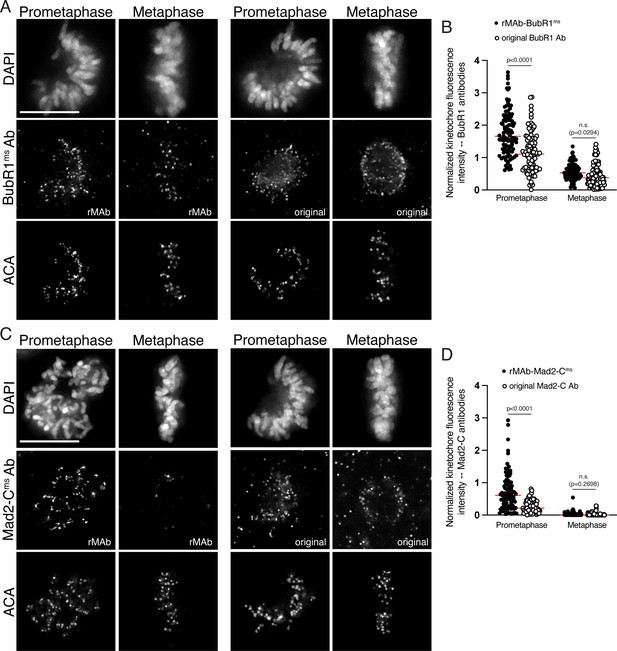
Sensitivity of original BubR1 and Mad2-C monoclonal antibodies compared to the recombinant antibodies.
(A) HeLa cells immunostained with the rMAb-BubR1ms antibody (left) and the original BubR1 antibody generated in mice and obtained from hybridoma cell lines (right). Cells were also stained with an ACA antibody to detect kinetochores and with DAPI to detect chromosomes. Cells in early prometaphase and metaphase are shown. (B) Quantification of kinetochore fluorescence intensities of cells immunostained with both BubR1 antibodies. (C) HeLa cells immunostained with the rMAb-Mad2-Cms antibody (left) and the original Mad2-C antibody generated in mice and obtained from hybridoma cell lines (right). Cells were also stained with an ACA antibody to detect kinetochores and with DAPI to detect chromosomes. Cells in early prometaphase and metaphase are shown. (D) Quantification of kinetochore fluorescence intensities of cells immunostained with both Mad2-C antibodies. For all quantifications, values were normalized to the kinetochore fluorescence intensity values of the ACA antibody, which recognizes kinetochore proteins CENP-A, -B, and -C. For each condition, at least 20 kinetochores from at least five cells were measured. For each pair of measurements, a Student’s t-test was carried out to determine statistical significance. Scale bars in all immunofluorescence images are 10 µm.
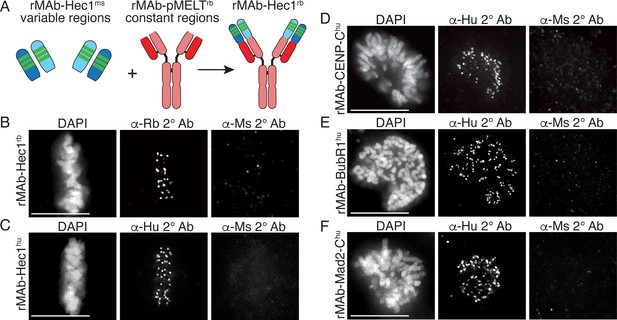
Generation of recombinant antibodies with modified species specificity.
(A) Schematic of ‘species swap’ approach. (B–F) HeLa cells stained with antibodies to rMAb-Hec1rb, panel B; rMAb-Hec1hu, panel C; rMAb-CENP-Chu, panel D; rMAb-BubR1hu, panel E; and rMAb-Mad2-Chu, panel F. For all panels B–F, cells were probed with the species-appropriate secondary antibody and also with secondary antibodies specific for the original species. Cells were also stained with DAPI to detect chromosomes. Scale bars are 10 µm.
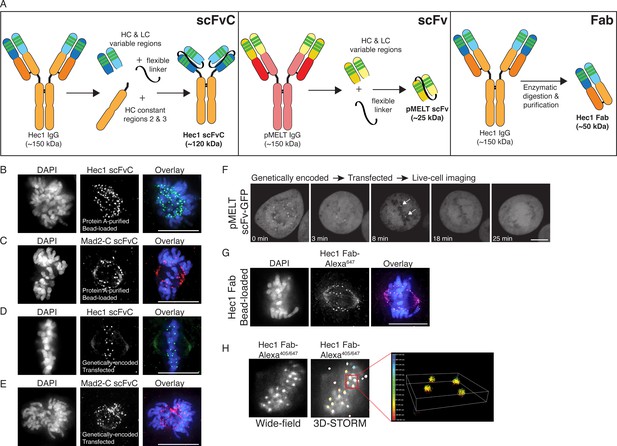
Generation of antibody fragments.
(A) Schematic illustrating the generation of three types of fragments: scFvC (left), scFv (center), and Fab (right). (B and C) Hec1 and Mad2-C scFvC fragments were purified on Protein A Sepharose columns and bead-loaded into HeLa cells. Cells were fixed and stained with anti-rabbit secondary antibodies and DAPI to detect chromosomes. (D and E) HeLa cells were transfected with the scFvC-Hec1 expression plasmid or the scFvC-Mad2-C expression plasmid. Cells were fixed and stained with anti-rabbit secondary antibodies and DAPI to detect chromosomes. (F) HeLa cells were transfected with the pMELT scFv-GFP expression plasmid and time-lapse imaged using confocal microscopy, and a representative cell is shown. At time = 0 min, many kinetochores are positive for scFv-pMELT-GFP, and by 18 min, when the cell has reached metaphase, no kinetochore-associated pMELT signals are detected. Arrows in the 8 min timepoint image point to the kinetochores which retain detectable MELT phosphorylation in late prometaphase. (G) Hec1 Fab were generated through proteolysis and directly labeled with an Alexa 647 fluorophore. HeLa cells were bead-loaded with Hec1 Fab647, stained with DAPI to detect chromosomes, and imaged. (H) Hec1 Fab were generated through proteolysis and directly labeled with both Alexa 405 and Alexa 647 fluorophores. HeLa cells were fixed, permeabilized, incubated with Hec1 Fab405/647, and subjected to both wide-field (left) and STORM imaging (right). Scale bars are 10 µm.
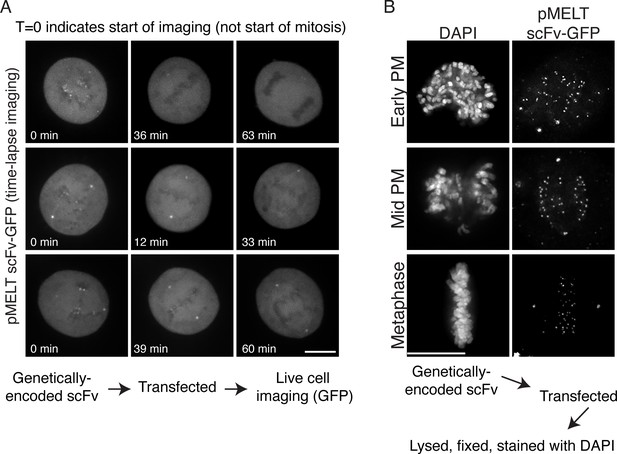
Time-lapse image stills and fixed cell images of cells expressing pMELT scFv-GFP.
(A) HeLa cells were transfected with the pMELT scFv-GFP expression plasmid and time-lapse imaged using confocal microscopy, and several examples are shown. Times are shown in minutes, and t = 0 indicates the start of filming, not the start of mitosis. (B) HeLa cells were transfected with the scFv-pMELT-GFP expression plasmid, lysed, fixed, and processed for fixed cell imaging. Representative cells in early prometaphase, mid prometaphase, and metaphase are shown. Cells are stained with DAPI to detect chromosomes. Scale bars are 10 µm.
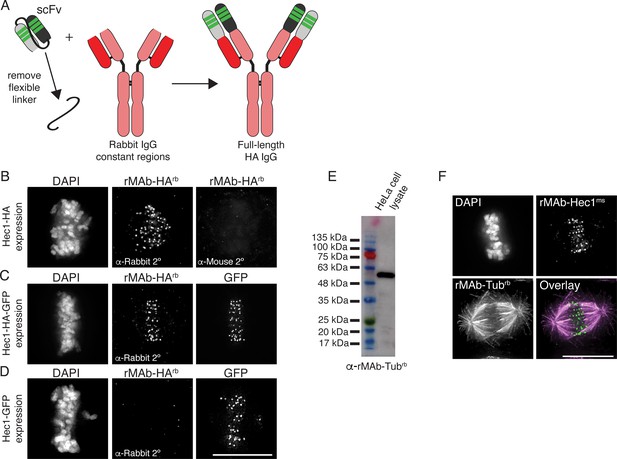
Generation of full-length, bivalent antibodies from fragment sequences.
(A) Schematic illustrating the generation of a reverse engineered, full-length HA-tag antibody from an scFv. (B–D) HeLa cells were transfected with Hec1-HA (B), Hec1-HA-GFP (C), or Hec1-GFP (C), and immunostained with rMAb-HArb antibodies. In panels C and D, GFP was also imaged. (E) Immunoblot of HeLa lysate probed with reverse engineered, full-length tubulin antibody, rMAb-Tubrb. (F) HeLa cells immunostained with rMAb-Hec1ms and rMAb-Tubrb. In all immunofluorescence images, cells were stained with DAPI to detect chromosomes. Scale bars are 10 µm.
-
Figure 6—source data 1
Immunoblot of HeLa cell lysate probed with the rMAb-Tub-rb antibody.
- https://cdn.elifesciences.org/articles/72093/elife-72093-fig6-data1-v3.tif
Tables
Yields from representative preparations of recombinant antibodies and antibody fragments.
Each antibody listed in the table was purified from a 30 ml culture of Expi293F cells.
| Recombinant Ab | Yield (mg) |
|---|---|
| rMAb-Hec1ms | 1.1 |
| rMAb-Hec1rb | 0.2 |
| rMAb-Hec1hu | 1.0 |
| rMAb-pMELTrb | 1.9 |
| rMAb-CENP-Cms | 0.2 |
| rMAb-CENP-Chu | 0.1 |
| rMAb-BubR1ms | 0.7 |
| rMAb-BubR1hu | 0.3 |
| rMAb-Mad2-Cms | 0.4 |
| rMAb-Mad2-Chu | 0.3 |
| rMAb-Tubrb | 2.0 |
| rMAb-HArb | 0.2 |
| scFvC-Hec1rb | 0.2 |
| scFvC-Mad2-Crb | 0.1 |
Descriptions of antibody-related plasmids generated in this study.
Plasmids will be made available by contacting the corresponding author. Use of the plasmids and sequence information is for non-commercial purposes only.
| Plasmid name | Description |
|---|---|
| pDL001_Hec1-ms_IgG_HC | Hec1 heavy chain variable and constant regions (mouse) + exogenous N-term signal peptide |
| pDL002_Hec1-ms_IgG_LC | Hec1 light chain variable and constant region (mouse) + exogenous N-term signal peptide |
| pDL003_Hec1-rb_IgG_HC | Hec1 heavy chain variable region + rabbit heavy chain constant regions + exogenous N-term signal peptide |
| pDL004_Hec1-rb_IgG_LC | Hec1 light chain variable region + rabbit light chain constant region + exogenous N-term signal peptide |
| pDL005_Hec1-hu_IgG_HC | Hec1 heavy chain variable region + human heavy chain constant regions (UniProt P01857) + exogenous N-term signal peptide |
| pDL006_Hec1-hu_IgG_LC | Hec1 light chain variable region + human light chain constant region (UniProt P01834) + exogenous N-term signal peptide |
| pDL007_pMELT-rb_IgG_HC | KNL1 pMELT heavy chain variable and constant regions (rabbit) + exogenous N-term signal peptide |
| pDL008_pMELT-rb_IgG_LC | KNL1 pMELT light chain variable and constant region (rabbit) + exogenous N-term signal peptide |
| pDL009_CENPC-ms_IgG_HC | CENP-C heavy chain variable region + mouse heavy chain constant regions (from Hec1 sequence) + exogenous N-term signal peptide |
| pDL010_CENPC-ms_IgG_LC | CENP-C light chain variable region + mouse light chain constant region (from Hec1 sequence) + exogenous N-term signal peptide |
| pDL011_CENPC-hu_IgG_HC | CENP-C heavy chain variable region + human heavy chain constant regions (UniProt P01857) + exogenous N-term signal peptide |
| pDL012_CENPC-hu_IgG_LC | CENP-C light chain variable region + human light chain constant region (UniProt P01834) + exogenous N-term signal peptide |
| pDL013_BubR1-ms_IgG_HC | BubR1 heavy chain variable and constant regions (mouse); (contains endogenous N-term signal peptide) |
| pDL014_BubR1-ms_IgG_LC | BubR1 light chain variable and constant region (mouse); (contains endogenous N-term signal peptide) |
| pDL015_BubR1-hu_IgG_HC | BubR1 heavy chain variable region + human heavy chain constant regions (UniProt P01857) (contains endogenous N-term signal peptide) |
| pDL016_BubR1-hu_IgG_LC | BubR1 light chain variable region + human light chain constant region (UniProt P01834) (contains endogenous N-term signal peptide) |
| pDL017_Mad2C-ms_IgG_HC | Mad2-C heavy chain variable and constant regions (mouse) (contains endogenous N-term signal peptide) |
| pDL018_Mad2C-ms_IgG_LC | Mad2-C light chain variable and constant region (mouse) (contains endogenous N-term signal peptide) |
| pDL019_Mad2C-hu_IgG_HC | Mad2-C heavy chain variable region + human heavy chain constant regions (UniProt P01857) (contains endogenous N-term signal peptide) |
| pDL020_Mad2C-hu_IgG_LC | Mad2-C light chain variable region + human light chain constant region (UniProt P01834) (contains endogenous N-term signal peptide) |
| pDL023_Tub-rb_IgG_HC | α-Tubulin heavy chain variable region + rabbit heavy chain constant regions + exogenous N-term signal peptide |
| pDL024_Tub-rb_IgG_LC | α-Tubulin light chain variable region + rabbit light chain constant region + exogenous N-term signal peptide |
| pDL025_HA-rb_IgG_HC_15F11 | HA-tag heavy chain hypervariable regions grafted into framework 15F11 scaffold (Zhao et al., 2019) + rabbit heavy chain constant regions + exogenous N-term signal peptide |
| pDL026_HA-rb_IgG_LC_15F11 | HA-tag light chain hypervariable regions grafted into framework 15F11 scaffold (Zhao et al., 2019) + rabbit light chain constant regions + exogenous N-term signal peptide |
| pDL027_scFvC_Hec1-rb | Hec1 heavy and light chain variable regions connected by linker + rabbit heavy chain constant regions 2 and 3 + exogenous N-term signal peptide |
| pDL028_scFvC_Mad2C-rb | Mad2-C heavy and light chain variable regions connected by linker + rabbit heavy chain constant regions 2 and 3 + exogenous N-term signal peptide |
| pDL029_scFv_pMELT_GFP_NoSP | KNL1 pMELT heavy and light chain variable regions connected by linker + GFP (no signal peptide included) |
| pDL030_scFvC_Hec1-rb_NoSP | Hec1 heavy and light chain variable regions connected by linker + rabbit heavy chain constant regions 2 and 3 (no signal peptide included) |
| pDL031_scFvC_Mad2C-rb_NoSP | Mad2-C heavy and light chain variable regions connected by linker + rabbit heavy chain constant regions 2 and 3 (no signal peptide included) |





