A functional screen of RNA binding proteins identifies genes that promote or limit the accumulation of CD138+ plasma cells
Figures
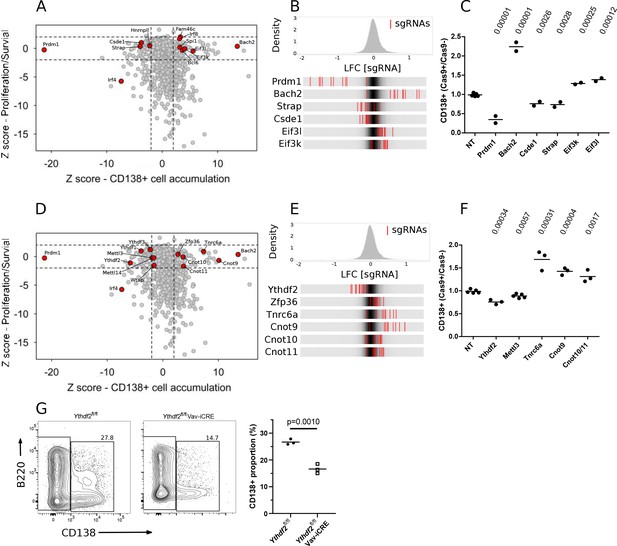
Genetic screen identifies modulators of B cell expansion and plasma cell accumulation.
(A) Dot plot representation of genetic screens of B cell proliferation/survival, and CD138 + cell accumulation: X-axis shows z-score of gene-level log2 fold change (LFC) for CD138 + cell accumulation (CD138 +v CD138- cells) and Y-axis shows z-score of gene-level LFC (day 8 v day 4 cells) for B cell expansion calculated by MAGECK. (B) Top: Distribution of enrichments of all sgRNAs (z-scores of sgRNA-level LFC: CD138 +v CD138- cells) for the CD138 + cell accumulation screen. Bottom: Enrichment for each of the ten sgRNAs, represented as red lines, targeting one of the indicated genes in comparison to the overall distribution of sgRNAs in the screen, depicted as the grey density in the middle of each bar. (C) The ratio of the proportion of cells expressing CD138 in Cas9 + cells and Cas9- cells transduced by viruses with non-targeting (NT), Prmd1, Bach2, Csde1, Strap, Eif3k, Eif3l targeting sgRNAs at day-8 of the in vitro B cell culture the data are representative of between two and four experiments performed on different days. Statistical significance was determined by two-tailed unpaired Student’s t-test, p values unadjusted for multiple testing are plotted. Each symbol is representative of a distinct sgRNA. (D) Same as in (A) with additional genes highlighted. (E) Same as in (B) with additional genes highlighted. (F) Same as in (C) with Ythdf2, Mettl3, Tnrc6a, Cnot9 targeting sgRNAs and paired sgRNAs against Cnot10/Cnot11. the data are representative of between two and four experiments performed on different days. (G) Left: representative flow cytometry for Ythdf2 CTL and Ythdf2 CKO and right: summary data of the proportion of cells expressing CD138 at day 8 of an in vitro culture of B cells from Ythdf2fl/fl mice (closed circles) or Ythdf2fl/fl-Vav1-iCre mice (open squares); the data are representative of three experiments performed on different days. Statistical significance was determined by two-tailed unpaired Student’s t-test. Each symbol is representative of cells from a single mouse.
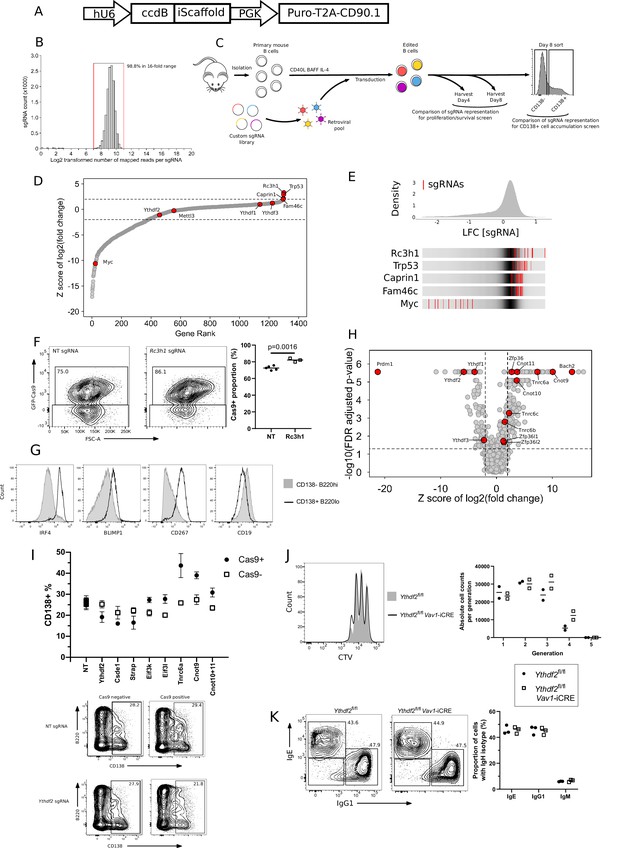
Custom sgRNA library targeting RBPs identifies regulators of B cell proliferation and survival.
(A) Schematic representation of the vector for the custom sgRNA library. (B) Distribution of the relative representation of sgRNAs within the sgRNA library. Red vertical lines indicate the upper and lower bounds of a 16-fold range in representation that includes 98.8% of all sgRNAs. (C) Schematic representation of the pooled CRISPR/Cas9 knockout screens in primary mouse B cells. Comparison of sgRNA representation between day 4 and day 8 identified genes involved in proliferation and/or survival. Comparison of representation between day 8 cells sorted as CD138 +ve or -ve identified genes promoting or inhibiting plasma cell accumulation. (D) Enrichment plot for the proliferation and/or survival screen. The Y-axis shows z-score of gene-level log2 fold change (LFC) (day 8 v day 4 cells). X-axis shows rank of genes by z-score of gene-level LFC. (E) Top: Distribution of enrichments of all sgRNAs (z-scores of sgRNA-level LFC: day 8 v day 4 cells) for the B cell proliferation and/or survival screen. Bottom: LFC (day 8 v day 4 cells) for all ten sgRNAs targeting the indicated genes. The grey gradient in the middle of each bar depicts the overall distribution. Red lines represent individual sgRNAs targeting a given gene. (F) Left: Representative flow cytometry and right: quantification of the proportion of cells in coculture that are expressing Cas9-GFP at day 8 of an in vitro culture. Each symbol represents an individual non-targeting (NT) sgRNA (closed circles) or Rc3h1 sgRNA (open squares). Statistical significance was determined by two-tailed unpaired Student’s t-test. (G) Representative flow cytometry of B220hi CD138- or B220lo CD138 +in vitro derived cells for expression of IRF4, BLIMP1, CD267 and CD19. (H) Volcano plot representation of CRISPR/Cas9 screen of plasma cell accumulation. X-axis shows z-score of gene-level LFC (CD138 +v CD138- cells) and the Y-axis shows FDR adjusted p-value calculated by MAGECK. (I) Top: The percentage of Cas9 +and Cas9- cells expressing CD138 transduced by viruses with non-targeting (NT), Ythdf2, Csde1, Strap, Eif3k, Eif3l, Tnrca, Cnot9 sgRNAs or paired sgRNAs against Cnot10/Cnot11 at day-8 of the in vitro B cell culture. The data are representative of between two and four experiments performed on different days. Each symbol is representative of a distinct sgRNA. Bottom: Representative flow cytometry showing B220 and CD138 staining of GFP+ (Cas9 expressing) and GFP- (Cas9 non-expressing) cells at day 8 of an in vitro culture following transduction with viruses expressing non-targeting sgRNA or sgRNA targeting Ythdf2. (J) Left: representative flow cytometry analysis of cell trace dilution of 72 hr anti-IgM stimulated B cells. Right: Enumeration of absolute cell number per generation. (K) Left: representative flow cytometry analysis for Ythdf2 CTL and Ythdf2 CKO of the proportions of cells expressing IgG1 and IgE at day 8 of an in vitro culture. Right: Quantification of the proportion of cells expressing each IgH isotype.
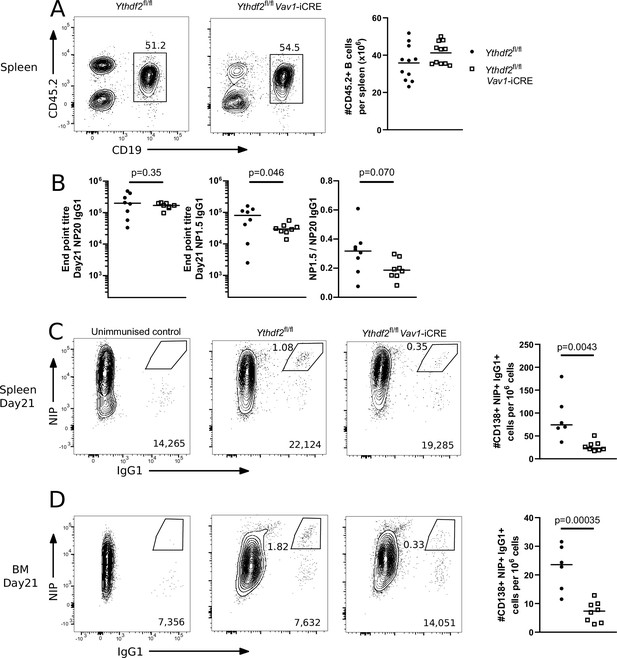
Analysis of µMT chimeras at day 21 after immunisation with NP-KLH in alum.
(A) Left: Representative flow cytometry analysis of cells expressing CD45.2 and CD19 in spleen of µMT chimeras 14 weeks after reconstitution. Numbers refer to the proportion of viable single cells within CD45.2 + CD19 + gate. Right: The number of cells expressing CD45.2 and CD19, n = 11. (B) Left: End point titre of serum for anti-NP20 IgG1 antibody. Center: End point titre of serum for anti-NP1.5 IgG1 antibody Right: Ratio of end point titres of serum for anti-NP1.5 and anti-NP20 IgG1 antibodies. Representative flow cytometry analysis of NIP and IgG1 intracellular staining of spleen (C) or bone marrow (D) cells expressing CD138 and CD267. The numbers in the bottom right corner of left-hand panels of C and D show the number of events plotted. The numbers adjacent to the gates indicate the proportion of NIP +IgG1 + cells. The right-hand panels of C and D show the number of cells expressing NIP and IgG1 per million viable cells as calculated from the indicated gates. Each symbol represents an individual Ythdf2fl/fl control (closed circles, n = 6) and Ythdf2fl/fl-Vav1-iCre knockout (open squares, n = 8) mouse. Statistical significance was determined by a two-tailed unpaired Student’s t-test. The data are pooled from two separate immunisation experiments.
Identification of CD138 +CD267 + splenic cells by flow cytometry.
(A) Representative flow cytometry plots of CD138, CD267 staining on splenic cells of μMT chimeras gated as CD45.2+. (B) Representative histograms and geometric mean fluorescent intensities (gMFIs) for CXCR4 signal in CD138 +CD267 + cells in the spleen of μMT chimeras 21 days after immunisation.
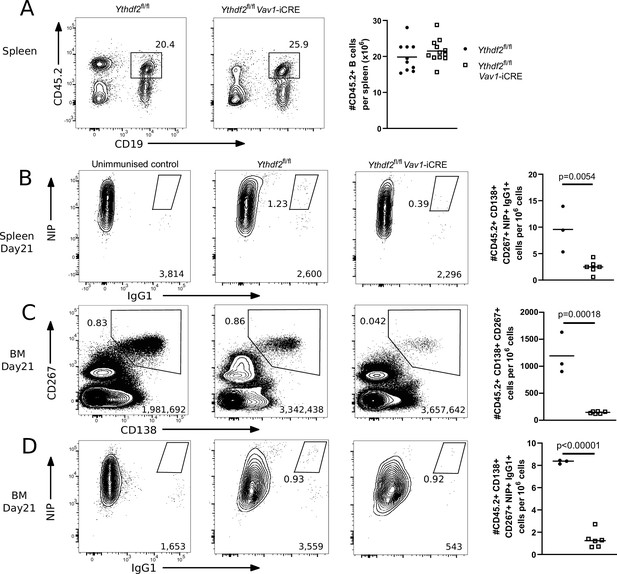
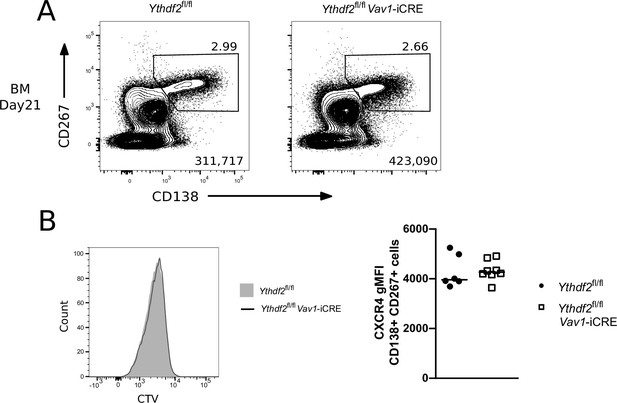
YTHDF2 deficient plasma cells fail to accumulate in the bone marrow.
(A) Left: Representative flow cytometry analysis of cells expressing CD45.2 and CD19 in spleen of competitive chimeras twelve weeks after reconstitution. Numbers refer to the proportion of viable single cells within CD45.2 + CD19 + gate. Right: The number of cells expressing CD45.2 and CD19, n ≥ 10. (B) Representative flow cytometry analysis of NIP and IgG1 intracellular staining on spleen cells gated as CD45.2 + CD138+, CD267 + cells at day 21. (C) Representative flow cytometry analysis of CD138 and CD267 staining on bone marrow cells gated as CD45.2 + at day 21. (D) Representative flow cytometry analysis of NIP and IgG1 intracellular staining on bone marrow cells gated as CD45.2 + CD138+, CD267 +at day 21. For each condition, the numbers in the bottom right corner of left-hand panels show the number of events plotted. The numbers adjacent to the gates indicate the proportion of cells within the gate. For each condition, the right-hand plots show the number of cells per million viable cells. Symbols represent data from an individual Ythdf2fl/fl control (closed circles, n = 3) and Ythdf2fl/fl-Vav1-iCre knockout (open squares, n = 6) mouse. Statistical significance was determined by two-tailed unpaired Student’s t-test. The data are from a single immunisation experiment.
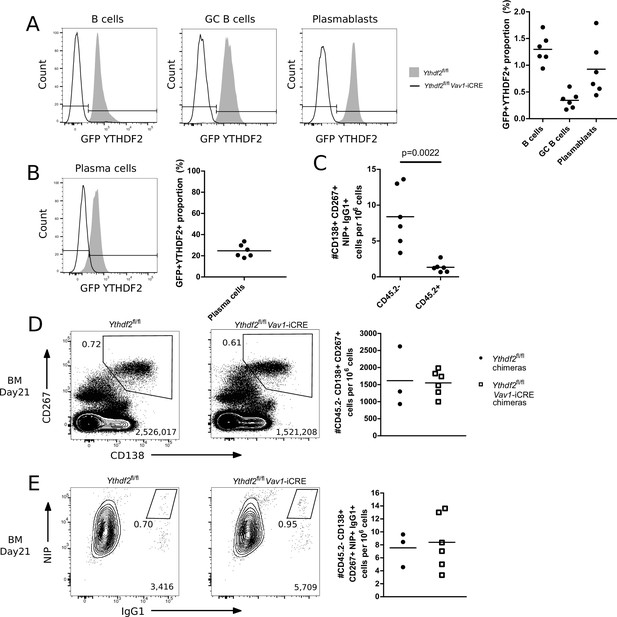
Absence of cells that escaped Cre mediated deletion.
(A) Left: Representative histograms of GFP-YTHDF2 fusion protein expression in surface stained CD45.2 positive total splenic B cells, splenic germinal centre B cells, splenic plasmablasts of competitive chimeras. Right: proportion of CD45.2 + cells within Ythdf2 CKO competitive chimeras expressing GFP-YTHDF2 +fusion protein, n = 6. (B) Left: Representative histograms of GFP-fluorescence in CD45.2 positive bone marrow plasma cells of competitive chimeras following fixation and intracellular staining for NIP and GFP. Right: proportion of CD45.2 + cells within Ythdf2 CKO competitive chimeras expressing GFP-YTHDF2 +fusion protein, n = 6. (C) Enumeration of CD138 +CD267 + NIP + IgG1 + cells that stained or lacked staining for CD45.2 in Ythdf2 CKO competitive chimeras at day 21, n = 6. (D) Representative flow cytometry analysis of CD138, CD267 staining on bone marrow cells of competitive chimeras gated as CD45.2- at day 21. (E) Representative flow cytometry analysis of NIP and IgG1 intracellular staining on bone marrow cells of competitive chimeras gated as CD45.2- CD138+, CD267 +at day 21. For each condition, the numbers in the bottom right corner of left-hand panels show the number of events plotted; the numbers adjacent to the gates indicate the proportion of cells within the gate. For each condition, the right-hand plots show the number of cells per million viable cells. Symbols represent data from an individual Ythdf2fl/fl control (closed circles, n = 3) and Ythdf2fl/fl-Vav1-iCre knockout (open squares, n = 6) mouse.
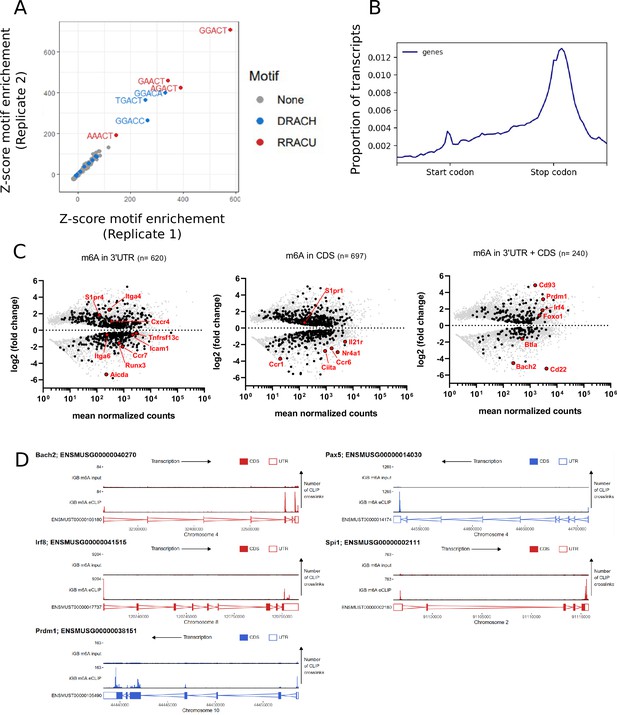
Negative regulators of B cell differentiation are methylated.
(A) Z-scores showing enrichment of five base motifs centred at in vitro derived B cell m6A-eCLIP crosslink sites relative to randomised control sites in each replicate, n = 2. (B) Metagene analysis of the location of m6A clusters throughout the B cell transcriptome. (C) Log2 fold change for differentially expressed (FDR-adjusted p-values < 0.05) methylated (black - identified using the CLIPper pipeline) and unmethylated (grey) transcripts between CD138 +B220 lo and CD138- B220hi in vitro cultured cells, n = 3. (Left) 3´UTR methylated transcripts, (centre) CDS methylated transcripts, (right) transcripts methylated in both their 3´UTR and CDS. (D) Methylation pattern of individual transcripts encoding regulators (Bach2, Pax5, Irf8, Spi1, Prdm1) of B cell differentiation.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72313/elife-72313-transrepform1-v2.pdf
-
Supplementary file 1
Supplementary tables.
- https://cdn.elifesciences.org/articles/72313/elife-72313-supp1-v2.xlsx





