Functional and structural segregation of overlapping helices in HIV-1
Figures
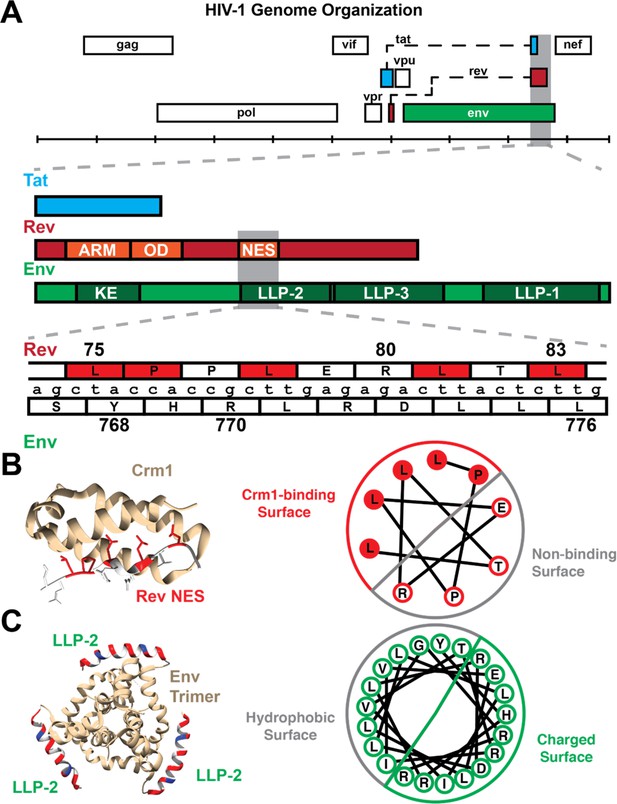
Organization of Rev NES/Env LLP-2 overlap.
(A) Genetic organization of the rev/env overlap. (Top) The full genome organization of HIV-1 (HXB2 numbering) demonstrates that the second exon of rev overlaps tat (second exon) and env. A detailed annotation of this overlap depicts the overlap between the quasi-helical NES of Rev with the helical LLP-2 of Env. The ARM (arginine-rich motif, nuclear localization sequence), OD (oligomerization domain), and NES (nuclear export sequence) are other functional domains of Rev that overlap with Env-gp41 (cytoplasmic tail), including the KE (Kennedy epitope) and two lentiviral lytic peptide motifs (LLP-3 in addition to LLP-2). (B) The structure of the Rev NES (red: binding residues, white: non-binding residues) bound to Crm1 (yellow) (PDB: 3nc0). A helical wheel diagram demonstrates how the arrangement of the 2D amino acid sequence creates a distinct binding surface in 3D. (C) Composite modeled structure (PDB: 6ujv) of the Env C-terminus trimer, including the first 21 residues of the LLP-2. The LLP-2 is labeled with the hydrophobic face in gray and the charged surface (positively charged residues in red and negative in blue). A corresponding helical wheel diagram derived from Murphy et al., 2017, for the Env LLP-2 is also shown with charged residues colored. Functionally important residues of Rev are labeled in red.
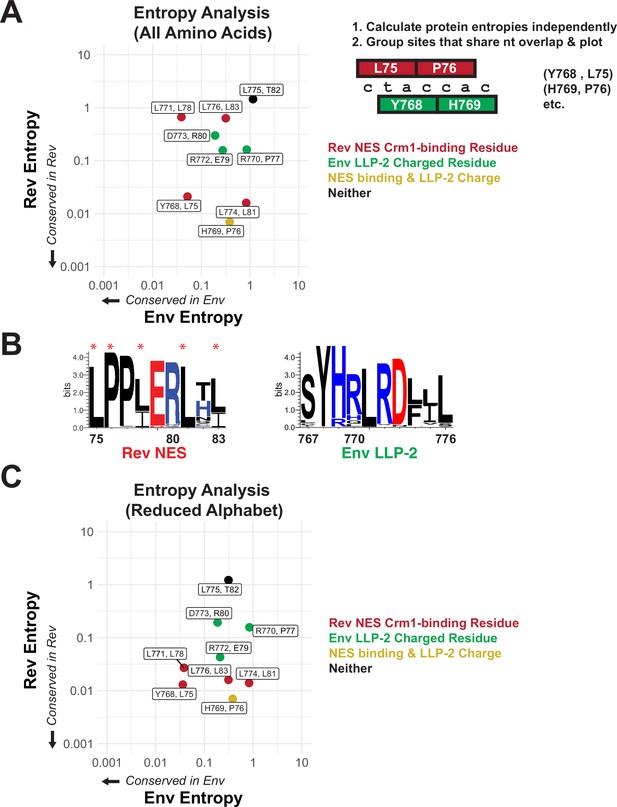
Rev nuclear export sequence (NES) and Env LLP-2 conservation.
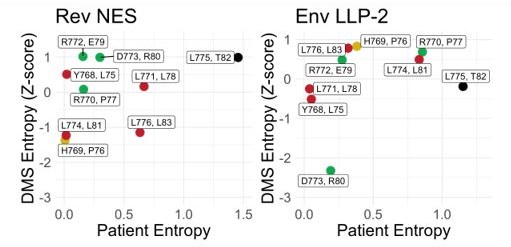
(A) Shannon entropies (low entropy = high conservation) from alignments of patient sequences (see panel B). Residues in Rev and Env are grouped by positions that share two nucleotides of overlap. Red points contain Crm1 contact residues, green points are charged LLP-2 residues, and yellow points contain both. (B) Sequence logos of Rev NES and Env LLP-2 showing sequence conservation at each position computed from a sequence alignment of patient data from the Los Alamos Database (https://www.hiv.lanl.gov/content/index; 2018). Positively charged residues (KRH) are in blue; negatively charged residues (DE) are in red. Functionally important residues of Rev are marked with a star. (C) Shannon entropies recomputed for a simplified amino acid alphabet (D=E, R=K, I=L and V, M, F=W & Y, N=Q, S=T). Coloring is as in 2A.
-
Figure 2—source data 1
Site-specific entropy values used to make graphs in Figure 2.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig2-data1-v3.xlsx
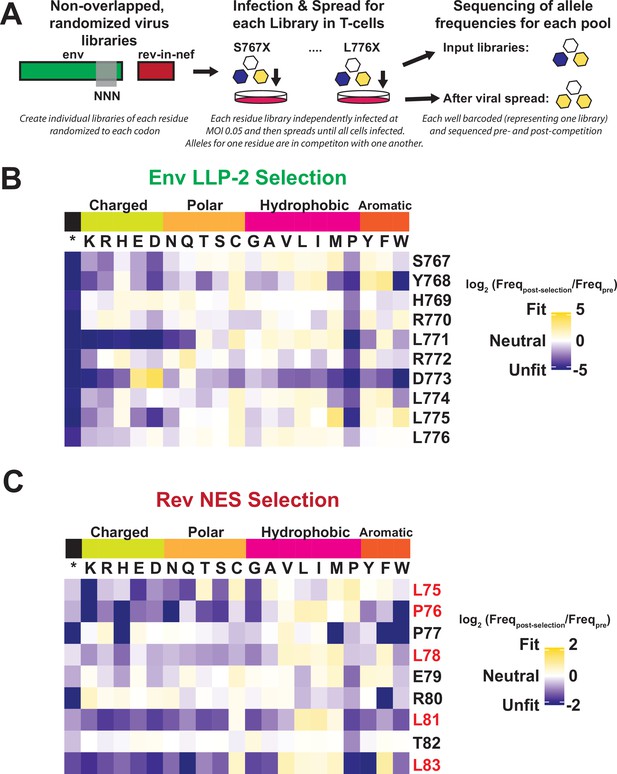
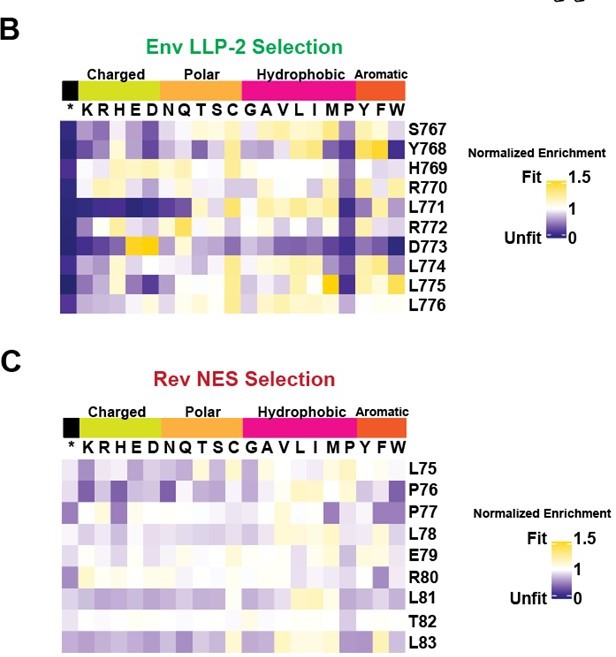
Deep mutational scanning of Rev nuclear export sequence (NES) and Env LLP-2.
(A) Schematic of the experiment. Non-overlapped viruses with rev inserted in the nef locus were created and the endogenous LLP-2 region randomized to NNN to create 10 independent libraries. These libraries were then arrayed and allowed to spread (~6 generations of viral spread) and next-generation sequencing was used to determine frequencies of each variant in a replication competition experiment and to calculate experimental fitness. (B) Fitness (blue = unfit, white = neutral, and gold = fit) for every allele in the region of the Env LLP-2 that overlaps the Rev NES. Row headers denote the reference sequence while column headers specify the allele. (C) Fitness for every allele in the region of the Rev NES that overlaps the Env LLP-2. Functionally important residues of Rev are labeled in red.
-
Figure 3—source data 1
Fold change values in allele frequency during deep mutational scanning experiment for Rev nuclear export sequence (NES) used to generate heatmaps.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Fold change values in allele frequency during deep mutational scanning experiment for Env LLP-2 used to generate heatmaps in Figure 3.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig3-data2-v3.xlsx
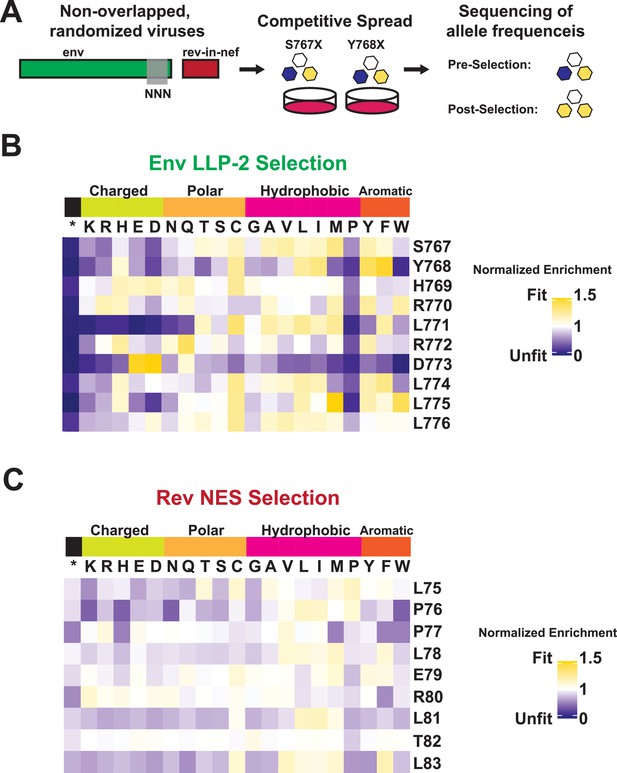
Normalized Enrichment Scores for deep mutational scanning of Rev nuclear export sequence (NES) and Env LLP-2.
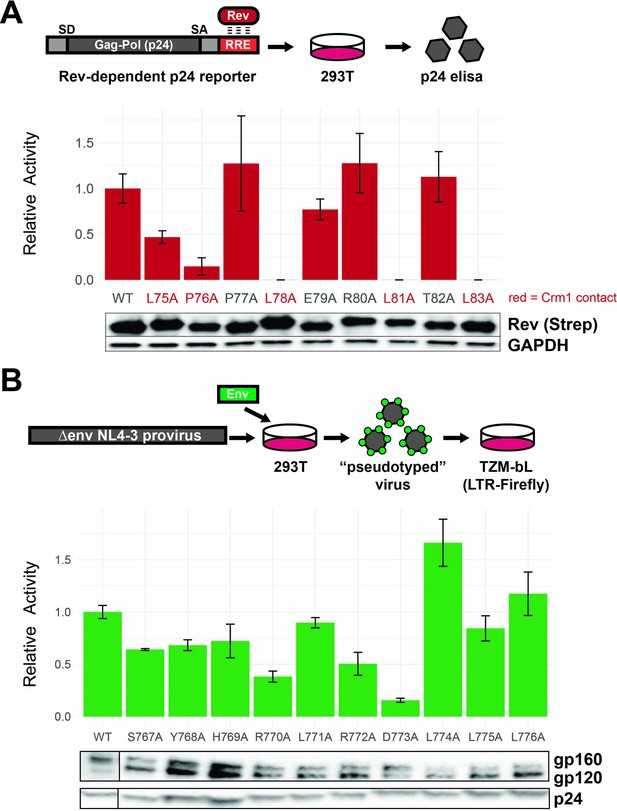
Functional dissection of Rev nuclear export sequence (NES) and Env LLPs.
(A) Schematic of the Rev reporter assay and activities of individual NES alanine mutants. To monitor Rev export activity, an RRE-containing gag-pol reporter system that produces viral capsid (p24) in a Rev-dependent manner was used. Data are mean ± standard deviation (s.d.) of three biological replicates. Western blot shows expression levels of Strep-tagged Rev variants and GAPDH as loading controls. Functionally important residues of Rev are labeled in red. (B) Schematic of the Env reporter assay in the TZM-bL system and results with individual Env LLP-2 alanine mutants. Data are mean ± s.d. of three biological replicates. Western blots show Env incorporation (gp120 antibody) into pseudoviruses and p24 as loading controls. For these experiments, we ablated the env reading frame by mutating the start codon and introducing multiple downstream stop codons. Plasmids were co-transfected into 293T cells with each alanine mutant to generate Env pseudotyped viruses. The resulting virions were used to infect TZM-bL reporter cells which express the HIV-1 surface receptors CD4, CCR5, CXCR4, and produce luciferase under the control of the tat-dependent HIV-1 LTR (Sarzotti-Kelsoe et al., 2014). A functional Env protein permits binding and entry and expression of Tat and thus reflects the number of infectious viral particles in a single round of infection, but the integrated virus lacks the env gene and cannot initiate additional rounds of replication. Incorporation of Env mutants into virions was also measured by Western blot.
-
Figure 4—source data 1
Reporter data used to generate graphs for Rev and Env alanine scans in Figure 4.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig4-data1-v3.xlsx
-
Figure 4—source data 2
Unedited Western blot for Figure 4.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig4-data2-v3.pdf
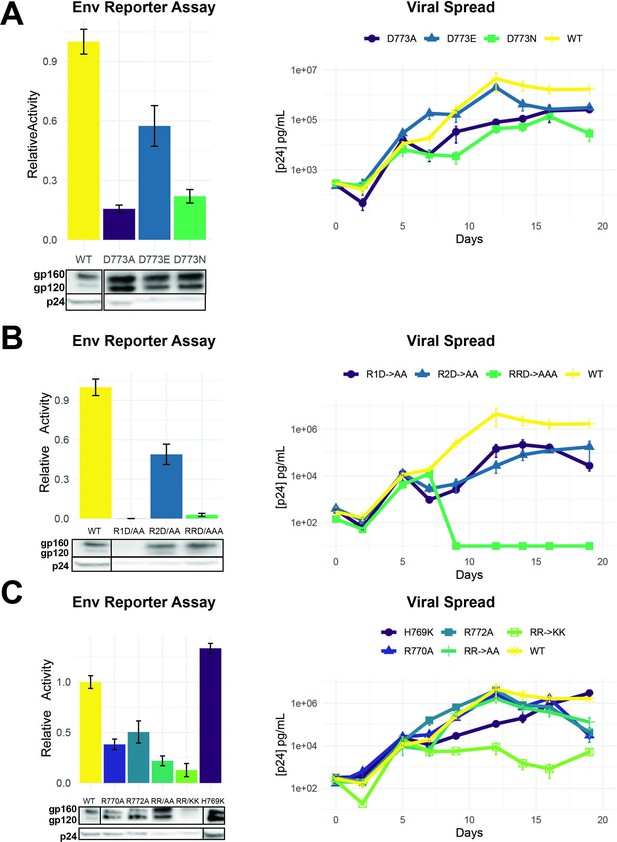
Importance of LLP-2 charged residues for replication.
(A) Left: Env functional assays as described in Figure 4 with different D773 mutations. Right: Viral replication kinetics in spreading assays using NL4-3 rev-in-nef proviral plasmids (Fernandes et al., 2016) engineered with Env D773 mutations. (B) Left: Env functional assays showing the role of three charged residues. Western blots show incorporation of Env mutants in virions (dark boxes represent a break in the lane loading presentation, with all samples loaded on a single gel). Right: Viral replication kinetics of viruses engineered with the mutations. R1D→ AA is a double mutant with R770/D773 changed to alanine, R2D→ AA is a double mutant with R772/D773 changed to alanine, and RRD→ AAA a triple mutant R770/R772/D773 changed to alanine. (C) Left: Env functional assays showing effects of positive charge mutations with corresponding Western blots of Env incorporation in virions. Right: Viral replication kinetics of viruses engineered with the mutations. Replication assays are represented on a log scale. All data are mean ± standard deviation (s.d.) of biological triplicates.
-
Figure 5—source data 1
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Viral spread data used to generate graphs for Env mutants in Figures 5 and 6.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig5-data2-v3.xlsx
-
Figure 5—source data 3
Unedited Western blot for Figure 5.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig5-data3-v3.pdf
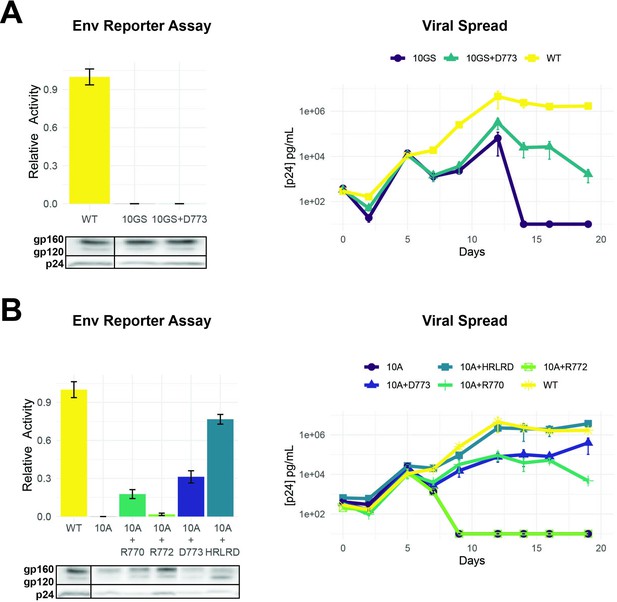
Importance of LLP-2 structure and charge for Env activity.
(A) Left: Env functional assays as described in Figure 4 when LLP-2 helical structure was disrupted by replacing 10 residues (767–776) with alternating glycine/serine residues (10GS) to create a flexible region. Western blots show expression of Env mutants (dark boxes represent a break in the lane loading presentation, with all samples loaded on a single gel). Right: Viral replication kinetics of wild-type (WT), 10GS, and 10GS + D262 viruses. (B) Left: Env functional assays with charge mutants within a 10-alanine (10A) context designed to maintain a helical structure with corresponding Western blots of Env incorporation in virions. Right: Viral replication kinetics with helical structure mutant viruses. Data points in both panels are mean ± s.d. of biological triplicates. Replication assays are represented on a log scale.
-
Figure 6—source data 1
Unedited Western blot for Figure 6.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig6-data1-v3.pdf
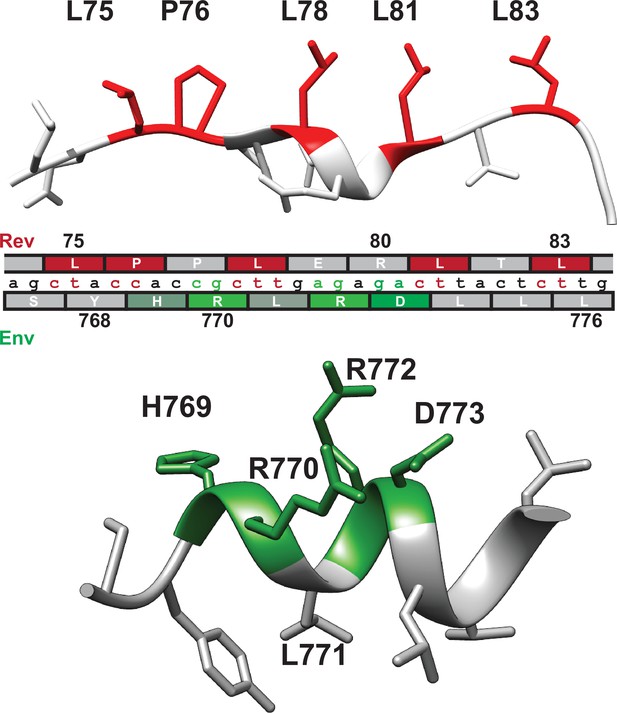
Nucleotide segregation and protein structure of overlapping Rev nuclear export sequence (NES) and Env LLP-2.
Functionally important residues of the Rev NES (red, CRM1-binding residues) and Env LLP-2 (green, charged residues) are highlighted in their respective structures (PDB: 3nc0 and PDB: 6ujv) and shown with the corresponding nucleotide sequence.
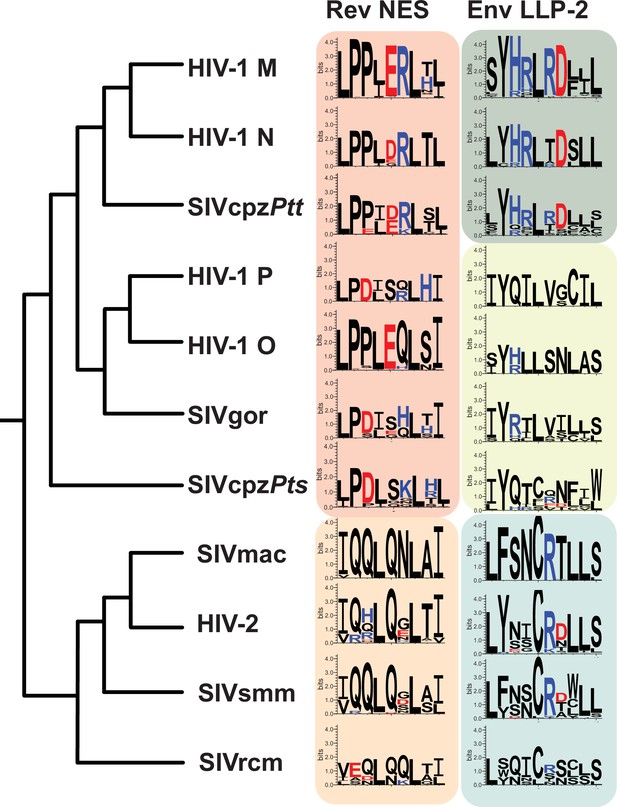
LLP-2/NES overlaps in primate lentiviruses reveals variation of NES and LLP-2.
Phylogenetic relationship of different primate lentiviruses, including four HIV-1 groups (M, N, O, P), SIVcpzPts (from eastern chimpazees), SIVgor (from gorillas), SIVcpzPtt (from central chimpanzees), HIV-2, SIVsmm (from sooty mangabeys), SIVmac (from macaques), and SIVrcm (from red-capped mangabey). Sequence logos showing conservation of the corresponding Rev NES and Env LLP-2 regions of each virus computed from their sequence alignments (extracted from 2018 curated amino acid alignments of HIV-1/SIVcpz, HIV-2/SIVsmm, and other SIV from the Los Alamos Database (hiv.lanl.org)). Positively charged residues (KRH) are indicated in blue and negatively charged residues (DE) in red. NES, nuclear export sequence.
-
Figure 8—source data 1
Number of sequences used to create each sequence logo in Figure 8 in addition to each sequence alignment.
- https://cdn.elifesciences.org/articles/72482/elife-72482-fig8-data1-v3.docx
Tables
| Rev NES | Env LLP-2 | |
|---|---|---|
| HIV-1 M | 4,054 | 7,004 |
| HIV-1 N | 9 | 11 |
| SIVcpzPtt | 12 | 15 |
| HIV-1 P | 5 | 4 |
| HIV-1 O | 48 | 48 |
| SIVgor | 6 | 8 |
| SIVcpzPts | 9 | 9 |
| SIVmac | 26 | 38 |
| HIV-2 | 49 | 75 |
| SIVsmm | 32 | 30 |
| SIVrcm | 6 | 7 |
Additional files
-
Supplementary file 1
Fasta alignment of Env LLP-2 sequences used for analysis in Figure 8.
- https://cdn.elifesciences.org/articles/72482/elife-72482-supp1-v3.fasta
-
Supplementary file 2
Fasta alignment of Rev nuclear export sequence (NES) sequences used for analysis in Figure 8.
- https://cdn.elifesciences.org/articles/72482/elife-72482-supp2-v3.fasta
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72482/elife-72482-transrepform1-v3.docx