Lactoferricins impair the cytosolic membrane of Escherichia coli within a few seconds and accumulate inside the cell
Figures
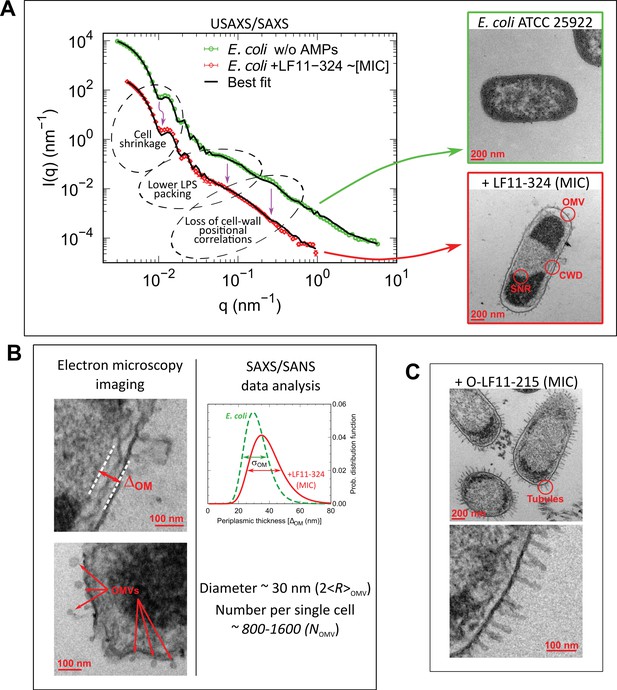
Overview of combined X-ray scattering and electron-microscopy measurements.
(A) Mapping the main structural changes in E. coli ATCC 25922 (green symbols) upon 1 hr incubation with LF11-324 (red symbols) as observed by (ultra) small-angle X-ray scattering (USAXS/SAXS) and transmission electron microscopy (TEM). Scattering data of E. coli ATCC 25922 are from Semeraro et al., 2021b and have been obtained at 10-fold higher sample concentration, leading to the observed offset of scattered intensities. Black lines are the best fits using Equation 6. OMV: outer membrane vesicle formation; CWD: cell-wall damaging; SNR: phase separation of the nucleoid region. Error bars are given by the experimental error of the measurements. (B) TEM examples of membrane detachment and OMV formation due to LF11-324, and respective ensemble results from scattering data analysis for the distance distribution between inner and outer membranes. (C) Bacteria upon 1 hr incubation with O-LF11-215, showing the formation of tube-like protrusions.
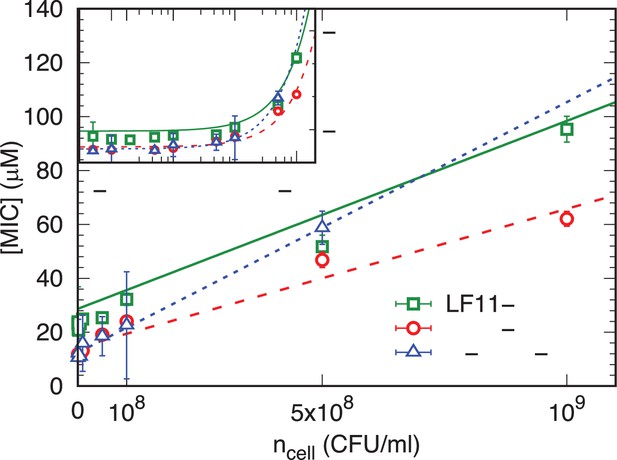
Cell number-dependent minimum inhibitory concentration (MIC) plots for different peptides.
MIC values as a function of for LF11-215 (green squares), LF11-324 (red dots), and O-LF11-215 (blue triangles) and best fits using Equation 1 and Equation 2. Error bars represent the absolute deviation from at least 3 independent experiments.
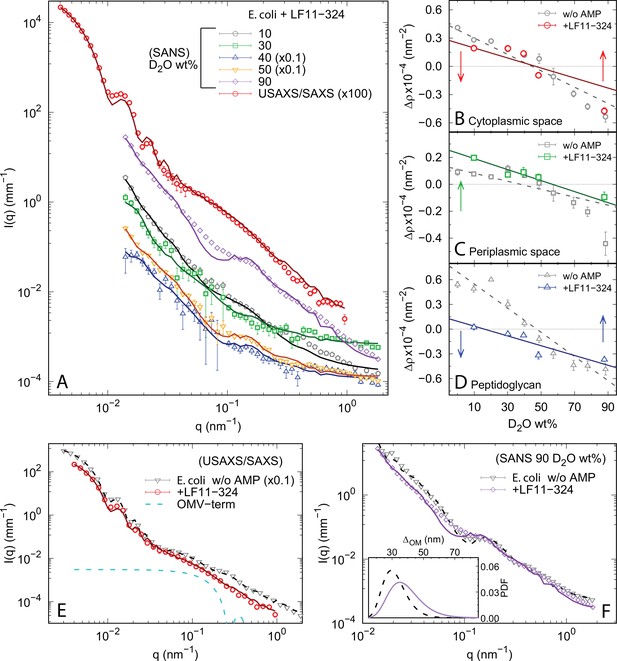
Comparison between (ultra) small-angle X-ray scattering (USAXS/SAXS) and contrast-variation small-angle neutron scattering (SANS) data and details of the scattering data analysis.
(A) X-ray and neutron scattering data of bacterial systems after 1 hr incubation with LF11-324 (end states) at the minimum inhibitory concentration (MIC) (SANS) and 1.2× MIC (SAXS). Lines are the best fits using Equation 6. (B–D) Scattering length contrasts as a function of D2O concentration (wt%) in the medium for the cytoplasm, , periplasm, , and peptidoglycan layer, . Gray symbols are the values reported in the absence of peptides (data adapted from Semeraro et al., 2021b). Solid and dashed lines correspond to linear regressions. (E) Comparison between SAXS curves from bacterial end states and a reference sample without peptides (data adapted from Semeraro et al., 2021b to match the bacterial concentration). The blue dashed line represents the additional term, whereas the fits refer to Equation 6. (F) Comparison between SANS curves from bacterial end states and a reference sample without peptides (data adapted from Semeraro et al., 2021b) in the case of 90 D2O wt%. Lines are the best fits with Equation 6. The inset shows the log-normal probability density function (PDF) of the inter-membrane distance with (dashed purple line) and without LF11-324 (solid black line). (E, F) The intensities are on absolute scale and normalized by the cell concentration. Data w/o antimicrobial peptides (AMPs) in panels (B–D) have been adapted from Figure 5 of Semeraro et al., 2021b. Data w/o AMPs in panels (E, F) have been adapted from Figure 3 of Semeraro et al., 2021b. Error bars in panels (A, E, F) are given by the experimental error of the measurements; error bars in panels (B-D) represent the associated standard deviations of the adjustable parameters obtained from the analysis of scattering curves.
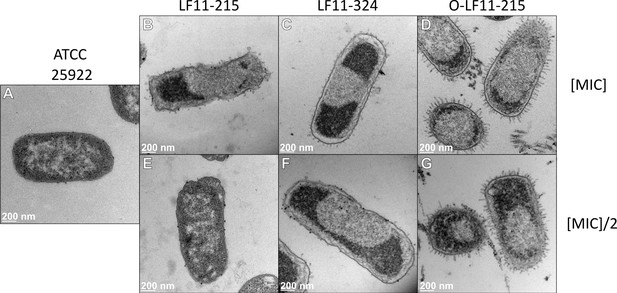
Transmission electron microscopy (TEM) observations for LF11-215, LF11-324, and O-LF11-215 at the minimum inhibitory concentrations (MICs) and sub-MICs.
TEM images of E. coli ATCC (A) and end states in the presence of LF11-215 (B, E), LF11-324 (C, F), and O-LF11-215 (D, G). All systems were probed at the MICs (B–D) and half the MICs (E–G).
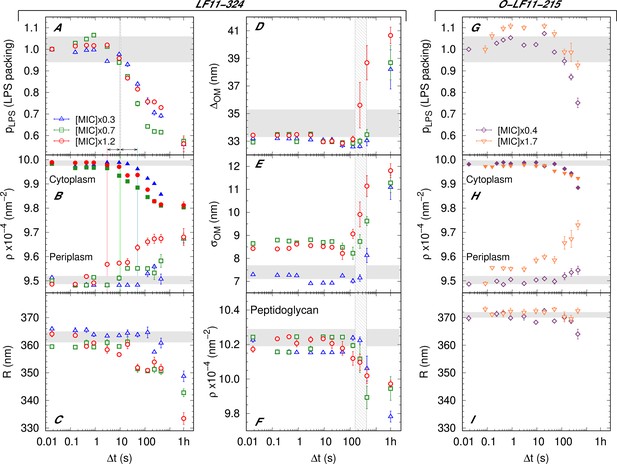
Kinetics of the bacterial structural response upon addition of peptide.
(A–F) Kinetics of the bacterial structural response to attack by LF11-324; results for three different peptide concentrations are shown. Lipopolysaccharide (LPS) packing (A); cytoplasm and periplasm scattering length density (SLD) (B); minor radius of the cell (C); intermembrane distance (∼ periplasm thickness) (D) and its deviation (E); and peptidoglycan SLD (F). (G–I) Bacterial response to O-LF11-215 at two concentrations. LPS packing (G); cytoplasm and periplasm SLDs (H); and minor ellipsoidal radius of the cell (I). Thick gray bands mark the degree of confidence from bacterial systems w/o peptides (see Table 1 and Semeraro et al., 2021b), except for (C) and (I), where they refer to the average of the current cell radii at . Fluctuations of initial values can be due to biological diversity. The vertical gray grid (A, D–F) indicates the time range of local (A) and macroscopic (D–F) cell-wall damage. Note that this range does not depend on peptide concentration. Colored lines in (B) mark the concentration-dependent lower boundary for the onsets of leakage. Results at hr refer to end states, when available. Error bars are given by the associated standard deviations of the adjustable parameters obtained from the analysis of scattering curves.
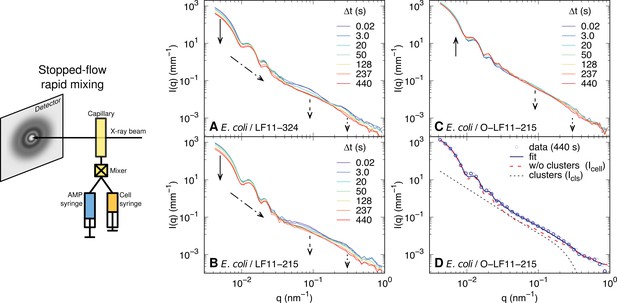
Schematic of the stopped-flow rapid mixing (ultra) small-angle X-ray scattering (USAXS/SAXS) experiments, including selected scattering patterns.
Schematic of the stopped-flow rapid mixing setup used for USAXS/SAXS experiments and kinetic changes of USAXS/SAXS curves of bacterial samples upon mixing with LF11-324 at minimum inhibitory concentration (MIC) ×1.2 (A), LF11-215 at MIC ×1.6 (B) and O-LF11-215 at MIC ×1.7 (C). The arrows highlight the most significant variations of intensity, such as the decrease of forward scattering (A, B); the evolution of the low-q oscillation profile (A–C); the disappearance of the feature at (A–C); the decrease of (A, B) and of (C) at . (D) Example of the combination of and in the case of applying O-LF11-215 (see Equation 6).
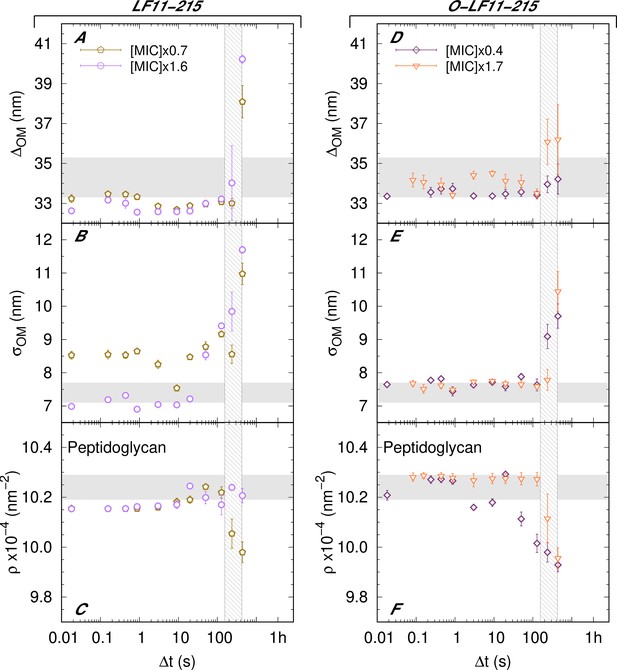
Kinetics of the adjustable parameters for LF11-215 and O-LF11-215 systems.
(A–C) Kinetics of the adjustable parameters upon mixing with two concentrations of LF11-215. These are the intermembrane distance (~ periplasm thickness) (A), its deviation from the average value (B), and the peptidoglycan scattering length density (SLD) (C). (D–F) Kinetics of the adjustable parameters upon mixing with two concentrations of O-LF11-215. The parameters are the intermembrane distance (~ periplasm thickness) (D), its deviation (E), and peptidoglycan SLD (F). Thick gray bands mark the degree of confidence from bacterial systems w/o peptides (see Table 1 and Semeraro et al., 2021b). The vertical gray grid in (A–F) is an approximated, concentration-independent time range during which the cell-wall damage occurs. Error bars are given by the associated standard deviations of the adjustable parameters obtained from the analysis of scattering curves.
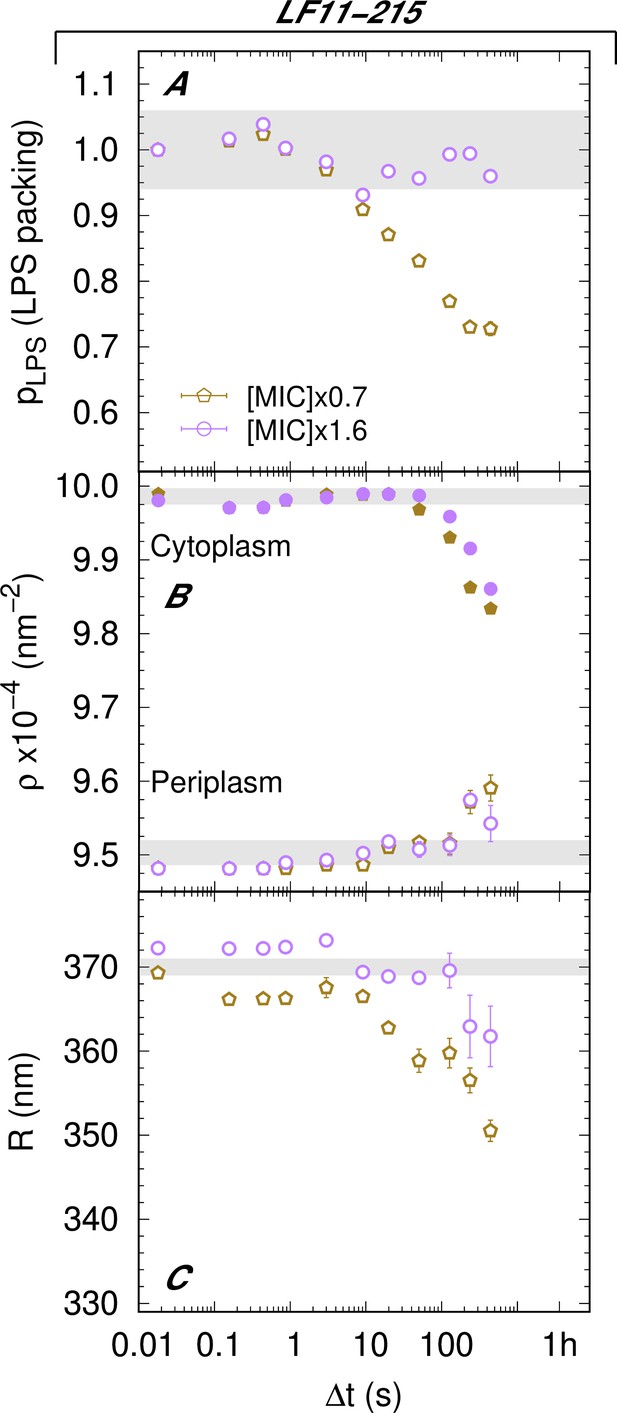
Kinetics of the adjustable parameters for LF11-215 systems.
(A–C) Kinetics of the adjustable parameters upon mixing with two concentrations of LF11-215. These parameters are the lipopolysaccharide (LPS) packing (A); the cytoplasm and periplasm scattering length density (SLD) (B); and the minor radius of the cell (C). Thick gray bands mark the degree of confidence from bacterial systems w/o peptides (see Table 1 and Semeraro et al., 2021b), except for (C), where they refer to the average of the current cell radii at . Error bars are derived from the associated standard deviations of the adjustable parameters obtained from the analysis of scattering curves.
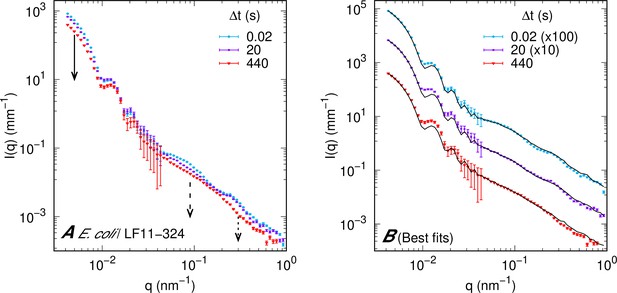
Representative scattering patterns including errors and fitted curves.
(A) Representative (ultra) small-angle X-ray scattering (USAXS/SAXS) curves of bacterial samples upon mixing with LF11-324 at minimum inhibitory concentration (MIC) ×1.2, highlighting intensity changes and comparison with errors. (B) Best fits of the curves shown in panel (A). Error bars are given by the experimental error of the measurements and proportional to the square root of the scattering intensity.
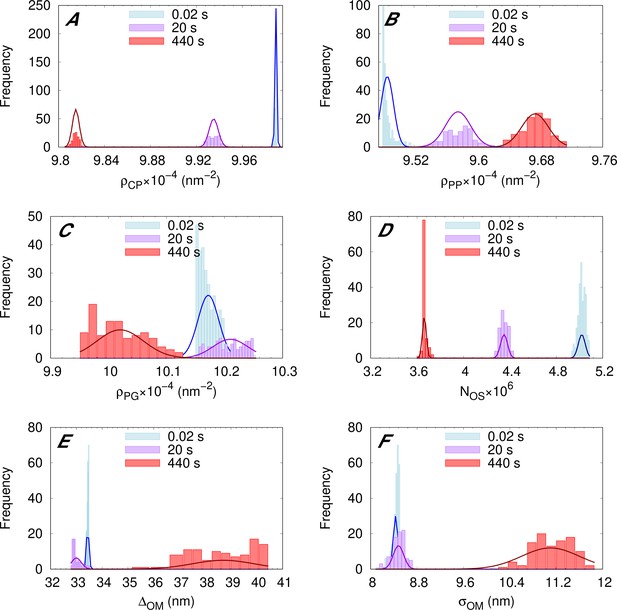
Representative distributions of the adjustable parameters at different time points.
Exemplary distributions obtained from our data analysis for adjustable parameters for LF11-324 system at minimum inhibitory concentration (MIC) ×1.2. Results refer to 0.02 s after mixing (light blue), 20 s (purple), and 440 s (red). Solid Gaussian curves are guides for the eye and show the center of mass and standard deviation of each distribution, which we report as parameter value and corresponding uncertainty.
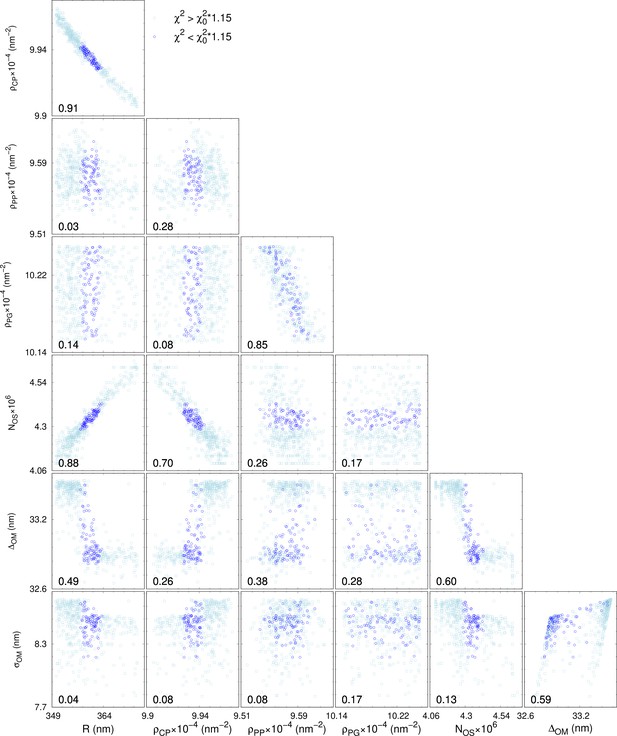
Representative series of correlation plots and coefficients of the adjustable parameters.
Selected example of the list of correlation plots and associated correlation coefficients (defined in the range 0–1). The adjustable parameters refer to LF11-324 system at minimum inhibitory concentration (MIC) ×1.2, 20 s after mixing. Coefficients and blue dots display the results for which is the chosen threshold that allows to discards local -minima. Light-blue squares show the remaining -landscape.
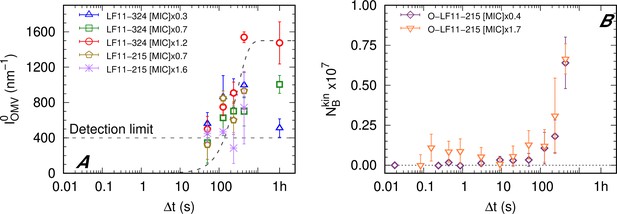
Kinetics of outer membrane vesicle formation and O-LF11-215 absorption.
(A) Kinetics of the forward intensity of outer membrane vesicle (OMV) scattering for different concentrations of LF11-324 and LF11-215. The dashed horizontal line represents the detection/’visibility’ limit, below which in the entire q-range. The dashed exponential curve is a guide for the eyes. (B) Evolution of the number of partitioned peptides per cell for two O-LF11-215 concentrations, as calculated from the analysis of . Error bars are derived from the associated standard deviations of the adjustable parameters obtained from the analysis of scattering curves.
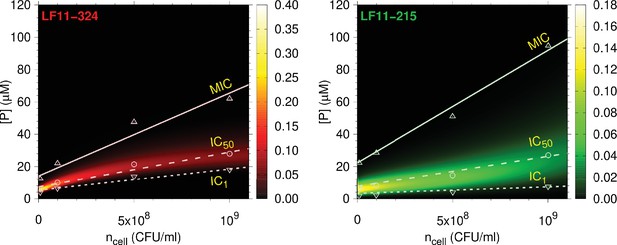
Full partitioning maps for LF11-324 and LF11-215.
Amount of LF11-324 or LF11-215 required to attain growth-inhibited fractions of either 0.999 (minimum inhibitory concentration [MIC], up triangles), 0.5 (circles), or 0.01 (down triangles) in E. coli ATCC 25922 as a function of . Lines are fits with Equation 1. These data are overlaid with a surface plot of the associated killing probability density function. The color scales indicate the corresponding magnitudes.
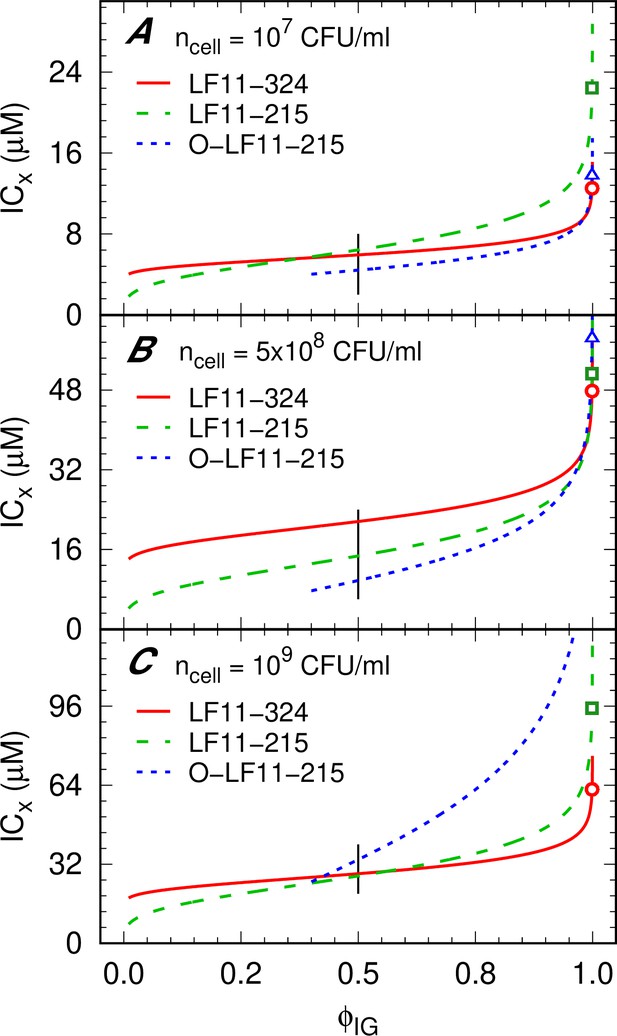
Inhibitory concentration (ICx) as a function of (inverse cumulative distribution function [CDF]).
(A–C) ICx as a function of inhibited fractions [inverse CDF, ] for different peptide and cell concentrations. Low values for O-LF11-215 were not accessible due to the high noise-to-signal ratio. Symbols mark the minimum inhibitory concentrations (MICs) for LF11-324 (circles), LF11-215 (squares), and O-LF11-215 (triangles), and black lines mark the range of IC50. The level of confidence is not displayed for the sake of clarity. ICx values have an about 10% relative error.
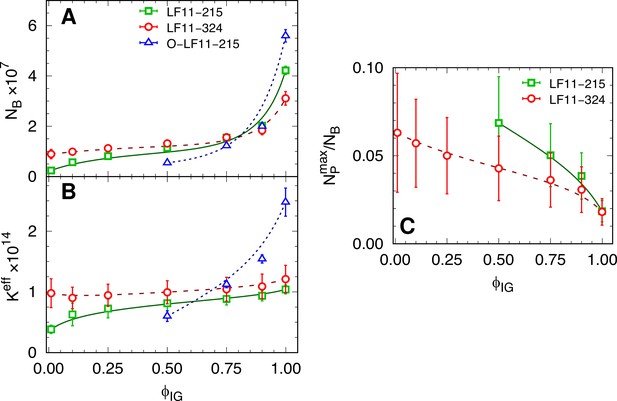
Partitioning parameters as a function of cell-growth inhibition.
(A, B) and values as a function of inhibited fraction. In the case of O-LF11-215, . (C) Ratio between the maximum number of peptides on the outer leaflet and total number of partitioned peptides, , as a function of inhibited fraction. Lines are guides for the eye. Error bars in panel (A and B) are given by the associated standard deviations of the adjustable parameters obtained from the analysis of the equi-activity assay. In panel (C) the errors are given by combining errors shown in panel (A) with the experimental error propagated from -potential measurements (see Figure 5—figure supplement 1).
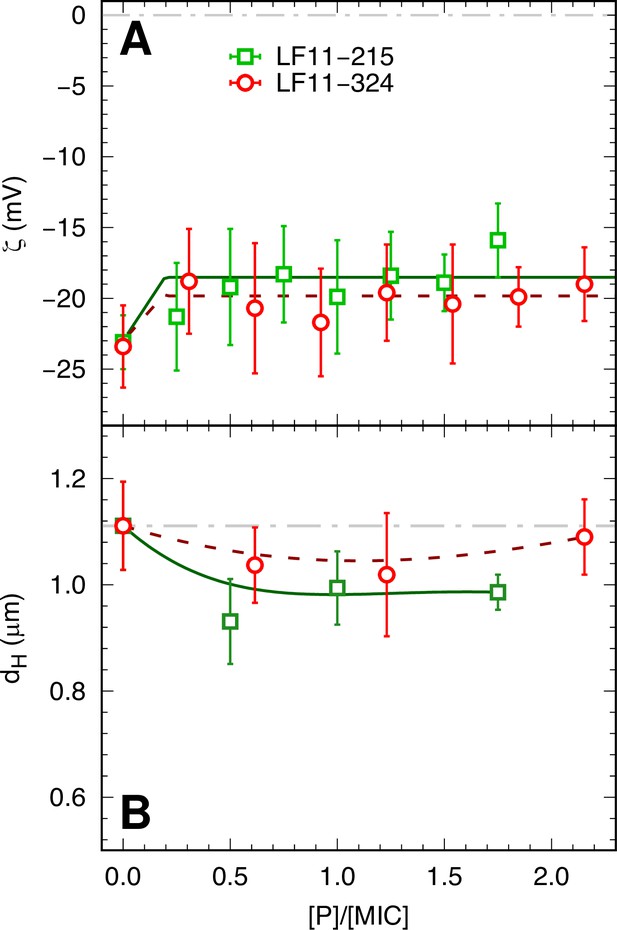
ζ-potential and size measurements of LF11-215 and LF11-324.
ζ-potential (A) and size (B) measurements as a function of peptide concentration (normalized by the respective minimum inhibitory concentrations (MICs) of LF11-215 and LF11-324 peptides). Lines are guides for the eye. Error bars are the given by the median-absolute-deviation around the median of at least 18 different measurements.
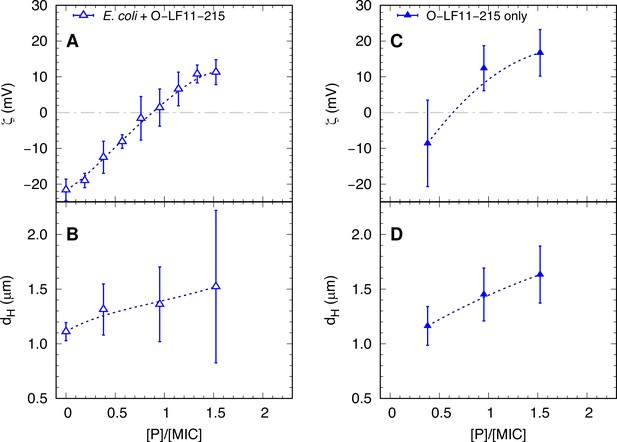
ζ-potential and size measurements of O-LF11-215.
Comparison between ζ-potential (A, C) and size (B, D) measurements of O-LF11-215 antimicrobial peptide (AMP) alone and mixed with E. coli as a function of peptide concentration (normalized by the minimum inhibitory concentration [MIC] of O-LF11-215). This partitioning behavior was also mirrored in the dependence of . was nearly constant for LF11-324, increased only slightly for LF11-215, and showed the largest variation for O-LF11-215, reaching about 2.5 times higher levels than the other two peptides (Figure 5B). The approximate equal values of LF11-324 and LF11-215 for demonstrate that both peptides partition about equally well into E. coli, not only at the MIC, but in a wide range of values. Error bars are the given by the median-absolute-deviation around the median of at least 18 different measurements.
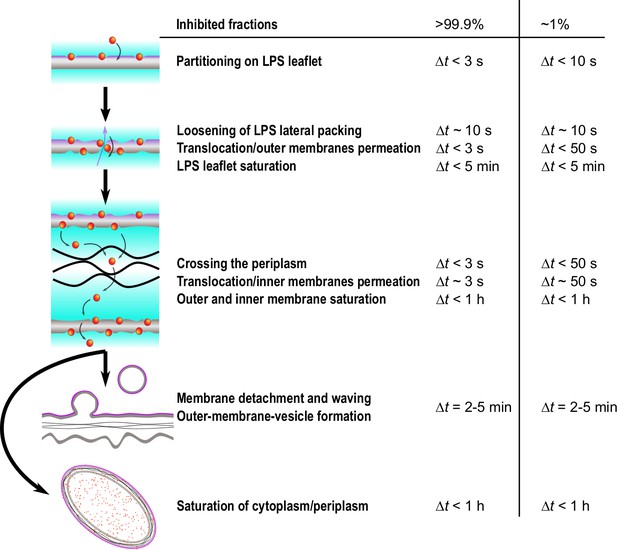
Simplified time sequence of LF11-215 and LF11-324 mode of action.
The measured time onsets and boundaries refers to LF11-324 at minimum inhibitory concentration (MIC) ×0.3 and MIC ×1.2, which correspond to measured inhibited fractions of ~1 and > 99.9%, respectively. The outer leaflet is affected by peptides within the first seconds after their attack. Then, depending on antimicrobial peptide (AMP) type and concentration, a number of rare translocation events, coupled with leakage, take places over a broad time range. When both membranes are saturated with peptides (exact time not determined), the cell wall breaks down, leading to outer membrane vesicle (OMV) formation (), detachment of outer and inner membranes and waving (). Simultaneously, AMPs accumulate in internal compartments and reach saturation levels within less than 1 hr.
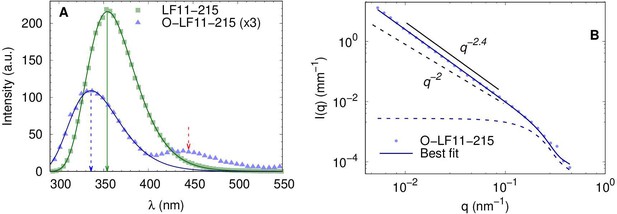
Trp-fluorescence and SAXS analyis of O-LF11-215 clustering.
(A) Trp fluorescence data of LF11-215 (green squares) and O-LF11-215 (blue triangles) at 100 µg/ml (LF11-324 are not shown to avoid redundancy). Data were fitted with Equation 4. Arrows mark the maxima positions of the fluorescence and phosphorescence bands. (B) Small-angle X-ray scattering (SAXS) data of O-LF11-215 at 400 µg/ml. The fit was performed with Equation 5. Additional lines highlight the obtained intensity decay of slope –2.4 (solid line), typical of mass fractals, as opposed to –2.0 (dashed line), which would correspond to either Gaussian chains or planar structures. Note that the slope is conserved in a q-range larger than one order of magnitude.
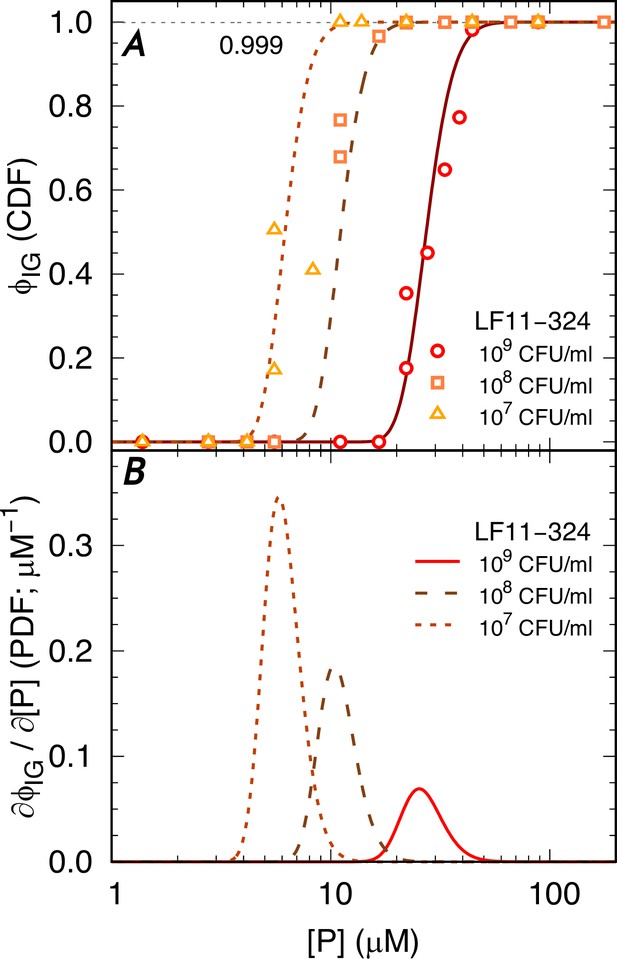
Statistical analysis of the AMP-induced bactericidal events.
(A) Selected data for LF11-324 and corresponding fits with the Gompertz function. (B) Corresponding probability density functions (PDFs).
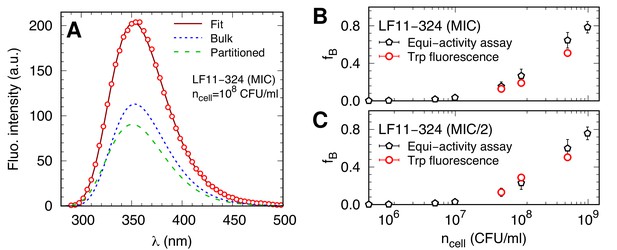
AMP-partitioning study in live bacteria based on Trp-fluorescence.
(A) Example of Trp fluorescence analysis in LF11-324 systems. The solid line is the best fit, and the dotted and dashed lines represent the Trp emissions from peptide in bulk and cell-associated, respectively. Data were fitted with Equation 4. (B, C) Comparison between values obtained from the Trp fluorescence analysis (red dots) and the equi-activity analysis from the susceptibility assay (black pentagons). Error bars in panel (B and C) are the associated standard deviations of the adjustable parameters obtained from the analysis of the equi-activity assay (black pentagons) and Trp-fluorescence (red dots).
Tables
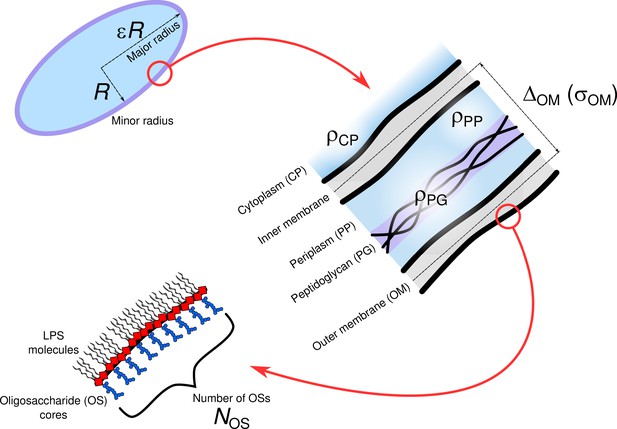
Change of E. coli structure due to LF11-324 () as observed from USAXS/SAXS/SANS data analysis.
Values are the difference between end- and initial state. See a graphical scheme of the adjustable parameters in Figure 1—figure supplement 4, and Semeraro et al., 2021b for a more detailed schematic.
| Parameters | Values | Description |
|---|---|---|
| –0.17 ± 0.02*; –0.14 ± 0.05† | ρCP → SLD of cytoplasmic space | |
| 0.18 ± 0.06*; 0.12 ± 0.04† | ρPP → SLD of periplasmic space | |
| 6 ± 3 | ΔOM → inter-membranes distance | |
| 3.4 ± 1.7 | σOM→ SD around ΔOM | |
| –0.27 ± 0.07*; –0.59 ± 0.14† | ρPG→ SLD of peptidoglycan layer | |
| –0.44 ± 0.08*; –0.26 ± 0.11† | pLPS → LPS packing parameter | |
| –27 ± 7 | R → minor radius of the cytoplasmic space |
-
Differences in X-ray and neutron SLDs are due to different physical interactions with matter. In case of , this originates from a biological variation of different bacterial cultures.
-
USAXS/SAXS: (ultra) small-angle X-ray scattering; SANS: small-angle neutron scattering; SLD: scattering length density; LPS: lipopolysaccharide.
-
*
From SAXS.
-
†
From SANS (SLDs were obtained by extrapolating to 0 wt% D2O); see Figure 1—figure supplement 2B–D.
List of fixed parameters for the combined analysis of USAXS/SAXS and contrast variation SANS data of E. coli.
| Description | Fixed parameters | Values |
|---|---|---|
| Center-to-center distance between the head-group layers in the CM | 3.73 | |
| Center-to-center distance between the head-group layers in the OM | 3.33 | |
| Width of the head-group layers for both CM and OM | 0.75 | |
| Center-to-center distance between the PG layer and the OM | 16.7 | |
| Width of the PG layer | 6.0 | |
| Average SLD of the tail group layer in the CM | 8.31*/0.022† | |
| Average SLD of the tail group layer in the OM | 8.86*/0.012† | |
| Ratio between major and minor radii | 2.0 | |
| Effective radius of gyration of each OS core | 0.45 |
-
USAXS/SAXS: (ultra) small-angle X-ray scattering; SANS: small-angle neutron scattering; SLD: scattering length density.
-
CM: Cytoplasmic membrane; OM: Outer membrane; PG: peptidoglycan; OS: Oligosaccharides.
-
*
X-ray SLDs.
-
†
Neutron SLDs.
List of fixed and D2O-dependent parameters for the combined analysis of USAXS/SAXS and contrast variation SANS data of E. coli.
The average SLD of both CM and OM head-group layers, , the SLD of the buffer solution, , and the product of the each OS core volume and its contrast relative to the buffer, .
| Fixed parameters | Values | |||||
|---|---|---|---|---|---|---|
| Neutrons (wt% D2O) | ||||||
| X-rays | 10 | 30 | 40 | 50 | 90 | |
| 12.9 | 1.56 | 2.20 | 2.52 | 2.84 | 4.11 | |
| 9.476 | 0.135 | 1.54 | 2.20 | 2.81 | 5.54 | |
| 10.7 | 3.83 | 2.32 | 1.68 | 0.69 | –2.44 | |
-
USAXS/SAXS: (ultra) small-angle X-ray scattering; SANS: small-angle neutron scattering; SLD: scattering length density.