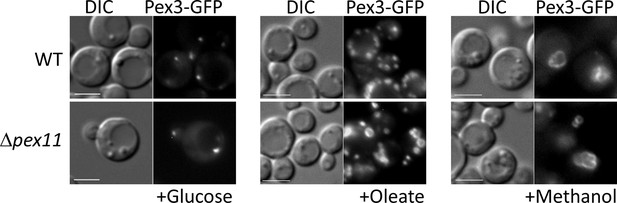
OXPHOS deficiencies affect peroxisome proliferation by downregulating genes controlled by the SNF1 signaling pathway
Figures
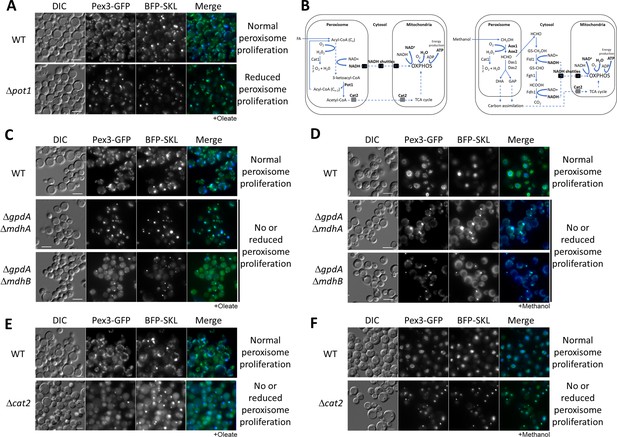
Peroxisome metabolites influence peroxisome size.
(A) Fluorescence microscopy of WT and Δpot1 mutant cells expressing Pex3-GFP driven by the PEX3 promoter and BFP-SKL driven by the GAPDH promoter, grown in oleate for 8 hr. (B) Brief description of FA β-oxidation, methanol metabolism, and NADH shuttling between peroxisomes and mitochondria. Cat1, catalase; Cat2, carnitine acetyl-CoA transferase; Pot1, 3-ketoacyl-CoA thiolase; Aox, alcohol oxidase; Das, dihydroxyacetone synthase; Fld1, formaldehyde dehydrogenase; Fgh1, S-formylglutathione hydrolase; Fdh1, formate dehydrogenase. (C–F) Fluorescence microscopy of WT, NADH shuttling and Δcat2 mutant cells expressing Pex3-GFP and BFP-SKL, grown in oleate and methanol for 8 hr, respectively. Bar: 5 μm.
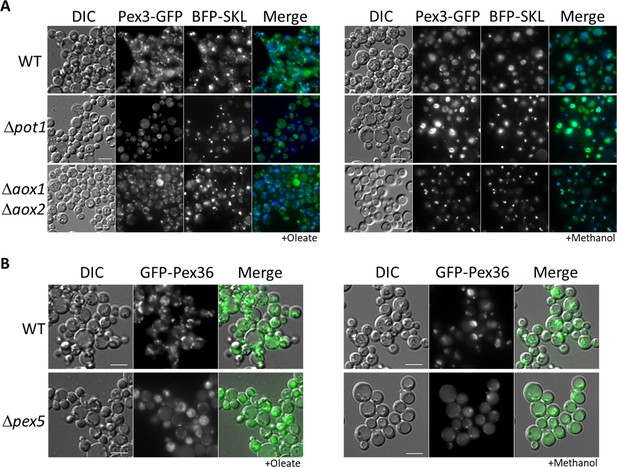
Peroxisome metabolites influence peroxisome size.
(A) Fluorescence microscopy of WT, Δpot1, and Δaox1 Δaox2 cells expressing Pex3-GFP and BFP-SKL driven by the PEX3 the GAPDH promoters, respectively. Cells were grown in oleate and methanol for 8 hr, respectively. (B) Fluorescence microscopy of WT and Δpex5 mutant cells expressing GFP-Pex36 from the PEX36 promoter. Bar: 5 μm.

NADH-shuttling mutants influence peroxisome size.
Fluorescence microscopy of WT and NADH-shuttling mutant cells expressing Pex3-GFP and BFP-SKL as in Figure 1—figure supplement 1. Cells were grown in oleate and methanol for 8 hr, respectively. Bar: 5 μm.
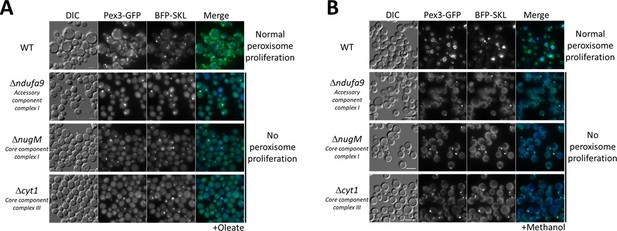
Dysfunctional mitochondria affect peroxisome proliferation.
Fluorescence microscopy of WT, Δndufa9, ΔnugM, and Δcyt1 mutant cells expressing Pex3-GFP and BFP-SKL, grown for 16 hr in (A) oleate and 8 hr in (B) methanol medium. Bar: 5 μm.

Altered mitochondrial respiration in mitochondrial CI mutants.
Equal amounts of WT, Δndufa9, and ΔnugM cells were grown as described in Materials and methods, and mitochondrial metabolic activity was measured in digitonin-permeabilized cells using the PM1 MicroPlates from Biolog Inc. Results are presented as the maximum metabolic rates (A.U.). Respiratory substrates are highlighted with boxes. As expected, mitochondrial substrates, producing NADH for CI such as L-lactic acid, citric acid, α-keto-glutaric acid, pyruvic acid, and others were properly metabolized in WT, but not in the mutant cells. In sharp contrast, CII substrates, α-glycerol-phosphate, succinic acid, and methyl succinate, exhibited unchanged rates of metabolism in the mutants. This result also suggests that CIII activity was not impaired in these CI mutants. Each maximum metabolic rate corresponds to mean ± standard deviation (SD) of triplicate values.
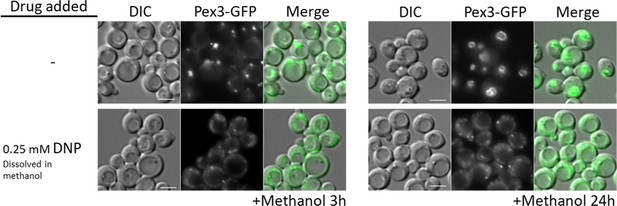
WT cells treated with the oxidative phosphorylation (OXPHOS) uncoupler, 2,4-dinitrophenol (DNP), share peroxisome proliferation defects with mitochondrial CI and CIII mutant cells.
Fluorescence microscopy of WT cells expressing Pex3-GFP, grown in methanol medium, with or without 0.25 mM DNP. Bar: 5 μm.

Cells treated with the oxidative phosphorylation (OXPHOS) uncoupler, 2,4-dinitrophenol (DNP), share same peroxisomal protein expression defects with OXPHOS mutants, without affecting Snf1 phosphorylation.
(A) Western blot for several peroxisomal proteins, as well as the total (T) the phosphorylated forms (P) of Snf1 in cells treated with and without DNP. Specific bands are indicated with an arrow. (B) Ponceau S staining was used as a loading control.
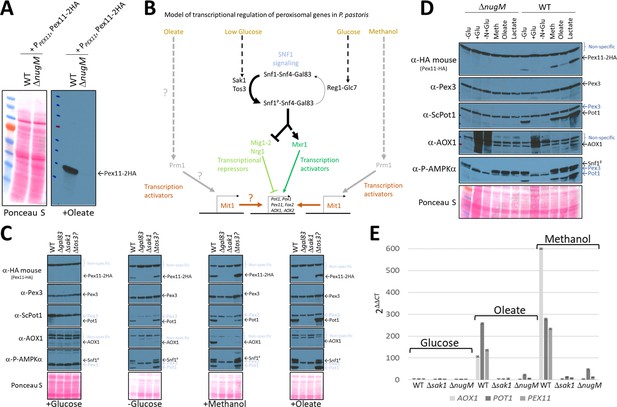
Oxidative phosphorylation (OXPHOS) mutants, like SNF1 mutants impair Pot1, Aox1, and Pex11 expression.
(A) Western blot of Pex11-2HA visualized with anti-HA antibodies in WT and ΔnugM mutant cells. (B) Model of transcriptional regulation of peroxisome genes regulated by SNF1 signaling. ? denotes unknown pathway for induction of Mit1 in oleate. (C, D) Western blots of several peroxisomal proteins and the phosphorylated form of Snf1 in WT, SNF1 complex mutants and/or ΔnugM mutant cells. Nonspecific bands and signal arising from previous western blots are indicated in blue font. Ponceau S staining was used as a loading control. (E) Relative expression of PEX11, POT1, and AOX1 after 4 hr incubation in the indicated carbon source using 18S ribosomal RNA (18S-rRNA) for qPCR normalization, and WT in glucose medium as reference and the 2 -ΔΔCT method for the analysis (Rao et al., 2013).
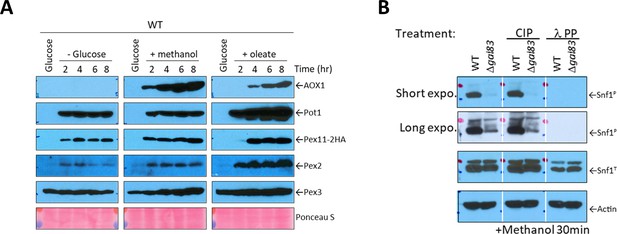
Peroxisomal protein expression under different environmental conditions and validation of the phospho-AMPKα (Thr172) antibody in P. pastoris.
(A) Western blots for several peroxisomal proteins at different time points from wild-type P. pastoris cells grown in glucose and shifted to no glucose (-glucose), oleate, or methanol medium. Ponceau S staining was used as a loading control. (B) Western blots from cell lysates of WT and Δgal83 cells after 30 min of methanol induction. The trichloroacetic acid precipitated protein pellets of both strains, equivalent to 0.5 OD600, were loaded in triplicate in a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel, transferred to a PVDF membrane and blocked with 5% BSA in TBS with 0.1% Triton X-100 (TBS-T) for 1 hr at room temperature. Each triplicate from the PVDF membrane was separated, placed in different boxes containing phosphatase buffer and incubated for 3 hr at 30°C in the absence of phosphatase, with 50 units of alkaline phosphatase (CIP; NEB # M0290) or with 1200 units of Lambda protein phosphatase (λ PP; NEB #P0753). After the incubation, the membranes were washed three times with TBS-T and incubated with the indicated antibodies. The phosphorylated Snf1 was detected using phospho-AMPKα (Thr172) antibody purchased from Cell Signaling Technology, Inc (#2535). Actin antibody was used as a loading control.
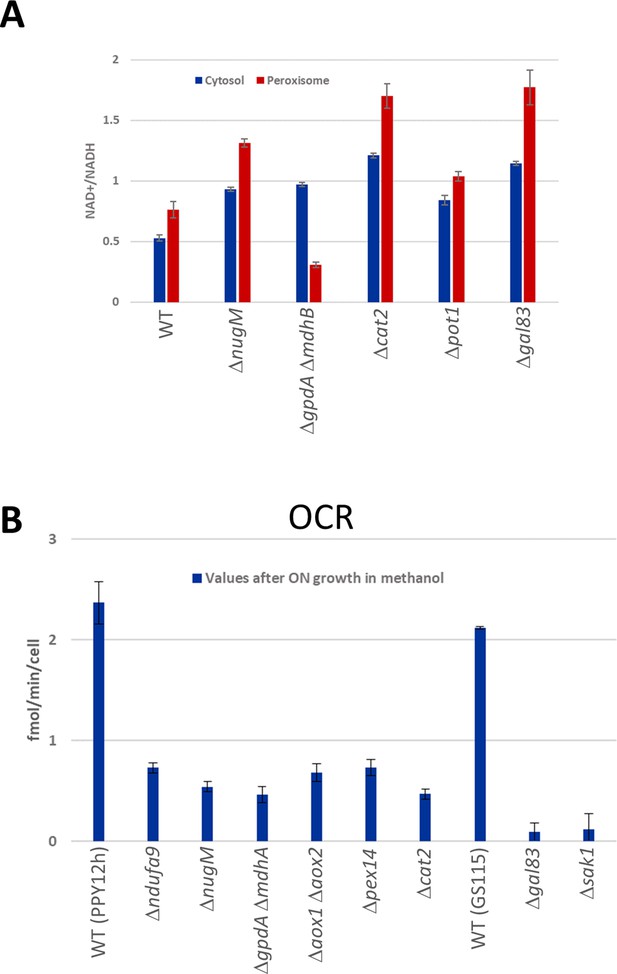
NAD+/NADH ratios and oxygen consumption rates (OCRs).
(A) NAD+/NADH ratios obtained using SoNar sensors from oleate-grown cells (Zhao et al., 2015), as described in Materials and methods in wild-type and indicated mutant strains. (B) Cell number corrected OCR data from methanol-grown cells for wild-type and mutants from two different parental strains (PPY12h and GS115). Mutants generated from PPY12h: Δndufa9, ΔnugM, ΔgpdAΔ mdhA, Δaox1 Δaox2, Δpex14, and Δcat2. Mutants generated from GS115: Δgal83 and Δsak1. Each point in A and B corresponds to mean ± standard deviation (SD) of triplicate values. The p values (calculated using JMP statistical discovery version 16) were < 0.01 in Student paired t-tests between WT and mutant strains. Further details of p values and n numbers can be found in Source data 3.
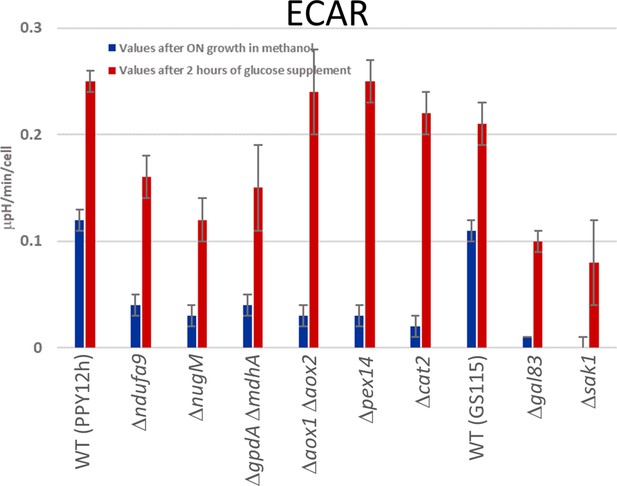
Extracellular acidification rates (ECARs).
ECAR data from methanol and after adding glucose (2% final concentration) for wild-type and mutant cells from two different parental strains (PPY12 or closely related to PPY12, and GS115). Mutants generated from PPY12: Δndufa9, ΔnugM, ΔgpdA ΔmdhA, Δaox1 Δaox2, Δpex14, and Δcat2. Mutants generated from GS115: Δgal83 and Δsak1. Values were normalized using cell numbers and each point corresponds to mean ± standard deviation (SD) of triplicate values. The p values (calculated using JMP statistical discovery version 16) were < 0.01 in Student paired t-tests between each strain grown in methanol versus those with glucose addition. Further details of p values and n numbers can be found in Source data 3.
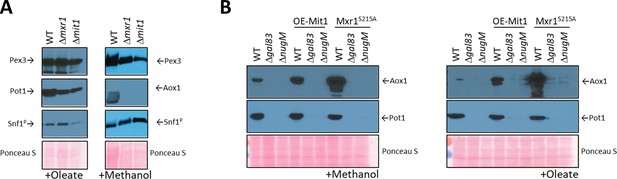
Analysis of transcriptional activators, Mxr1 and Mit1, of peroxisomal proteins.
(A) Western blots of Pex3, phospho-Snf1, and Pot1 or Aox1 in WT, Δmxr1, and Δmit1 mutant cells. (B) Western blots of Pot1 and Aox1 in WT, Δgal83, and ΔnugM mutant cells, either expressing or not expressing the active form of Mxr1 (Mxr1S215A). (C) Western blot of Pot1 in WT, Δgal83, and ΔnugM mutant cells, without or with overexpression of Mit1 (OE-Mit1). Ponceau S staining was used as a loading control.
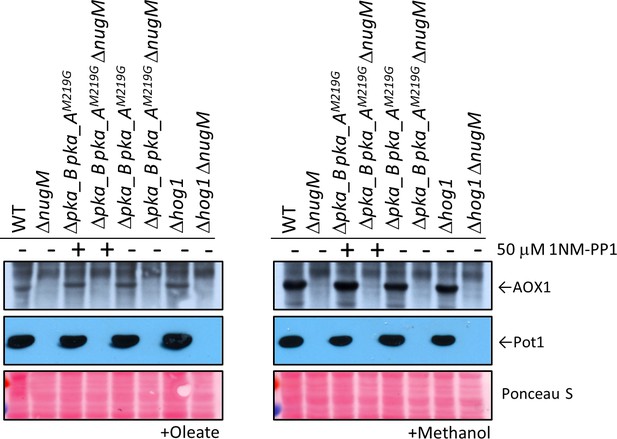
PKA inhibition or HOG1 deletion did not rescue peroxisome proliferation defect of ΔnugM mutant cells.
Western blot of Aox1 and Pot1 in WT and ΔnugM mutant cells, with and without PKA inhibition or HOG1 deletion. In P. pastoris, PKA consists of a regulatory subunit dimer (Bcy1) and two catalytic subunits (Pka_A, UniProt gene name: PAS_chr1-4_0357; Pka_B, UniProt gene name: PAS_chr3_0964). PKA inhibition was obtained by using a strain with a deletion of PKA_B gene and a point mutation in Pka_A (M219G) that renders the kinase sensitive to the drug, 1NM-PP1 (#529606, EMD Millipore, Germany), which was added when cultures were shifted to methanol or oleate medium. Ponceau S staining was used as a loading control.
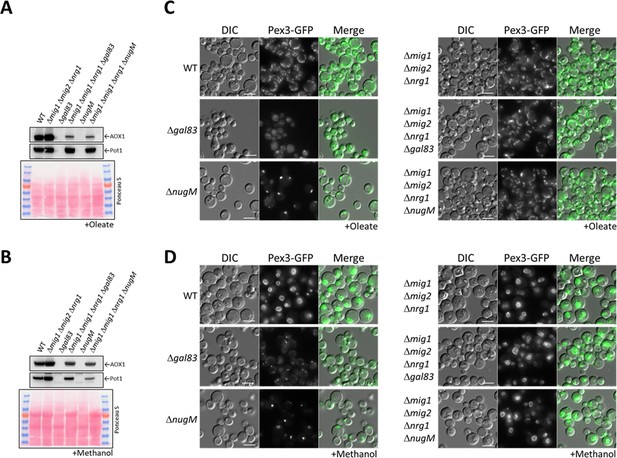
Deletion of transcriptional repressors regulated by SNF1 complex signaling rescues ΔnugM and Δgal83 mutant cells.
(A, B) Western blot of Aox1 and Pot1 in WT, ΔnugM and Δgal83 mutant cells, with and without deletions of genes (MIG1, MIG2, and NRG1) encoding the transcriptional repressors regulated by SNF1 signaling. Ponceau S staining was used as a loading control. (C, D) Fluorescence microscopy of WT, Δgal83, and ΔnugM mutant cells expressing Pex3-GFP, with and without deletions of genes (MIG1, MIG2, and NRG1) encoding the transcriptional repressors. Cells were grown in oleate and methanol for 16 hr, respectively. Bar: 5 μm.
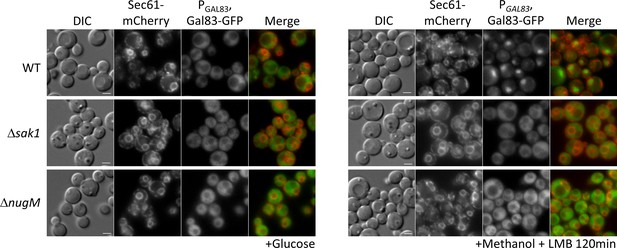
Gal83 nuclear localization during methanol adaptation is inhibited in ΔnugM and Δsak1 mutant cells.
Fluorescence microscopy of WT, Δsak1, and Δnugm mutant cells expressing Gal83-GFP driven by the GAL83 promoter in the presence of 200 ng/ml leptomycin B (LMB) and Sec71-mCherry as perinuclear ER marker. Bar: 5 μm.
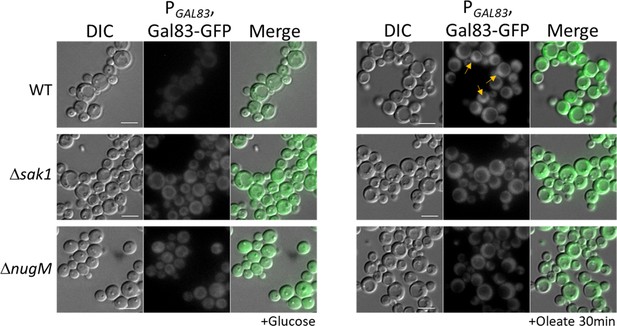
Gal83 nuclear localization during oleate adaptation is inhibited in ΔnugM mutant cells.
Fluorescence microscopy of WT, Δsak1, and ΔnugM mutant cells expressing Gal83-GFP driven by the GAL83 promoter. Bar: 5 μm.
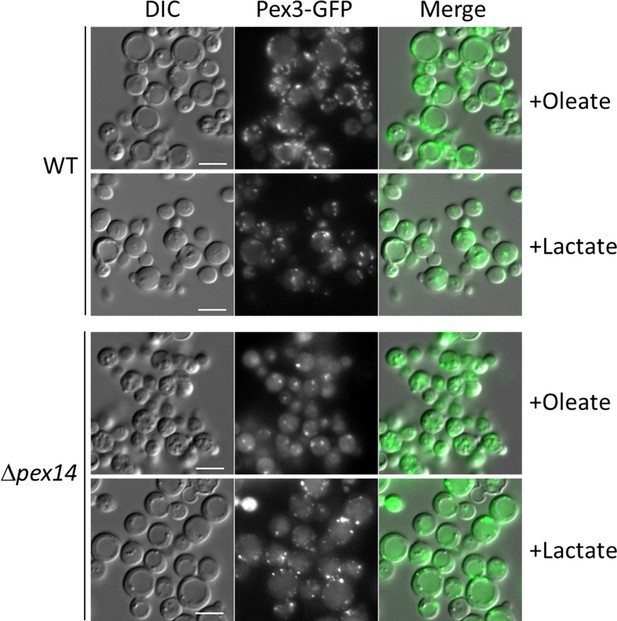
Cultivation of Δpex14 mutant cells in the respiratory medium, lactate, induces peroxisome proliferation.
Fluorescence microscopy of WT, Δpex14 mutant cells expressing Pex3-GFP in different media for 8 hr. Bar: 5 μm.
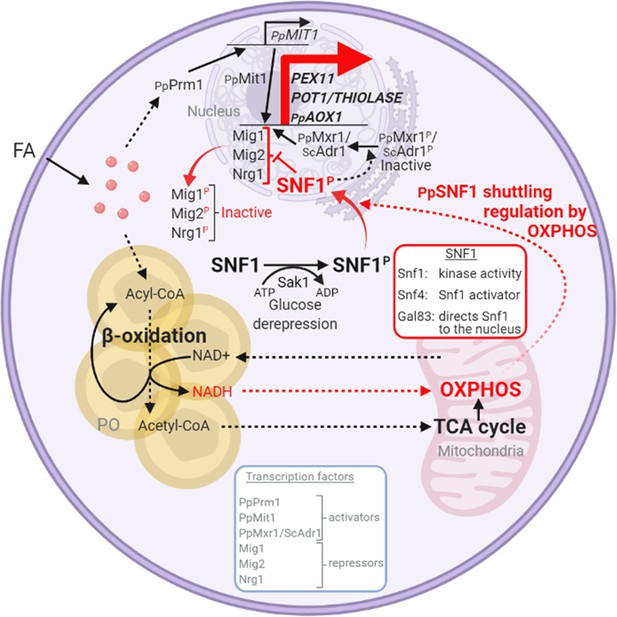
Interorganellar communication and signaling pathways in peroxisome proliferation, division, and matrix protein biogenesis.
Feedback loop between peroxisome and mitochondria is shown in red. Fatty acids (FA) uptake and its β-oxidation produce NADH equivalents and acetyl-CoA. However, the peroxisome membrane is impermeable to large hydrophilic solutes, including NAD+, NADH, NADP+, and NADPH, as well as ATP and acylated or unacylated coenzyme A (CoA) (Wanders et al., 2020). Consequently, NADH-shuttling proteins, working together in the peroxisomes, cytosol, and mitochondria, deliver NADH to mitochondria to feed oxidative phosphorylation (OXPHOS; Farré et al., 2021). Acetyl-CoA produced in peroxisomes is delivered to mitochondria via acetyl-carnitine produced in peroxisomes, and mitochondria use the TCA cycle and OXPHOS for full oxidation to CO2 and H2O (Wanders et al., 2020). The SNF1/AMP-activated protein kinase (AMPK) complex, which is sensitive to the cellular AMP:ATP ratio, maintains the balance between ATP production and consumption (Kayikci and Nielsen, 2015; Kim et al., 2013; Yan et al., 2018). Glucose deprivation, which reduces ATP production, activates Snf1 (by phosphorylation) via the action of the Sak1 kinase, and in P. pastoris, the nuclear translocation of the SNF1–Gal83 complex requires OXPHOS and ATP production. Peroxisome-associated proteins, such as the division protein, Pex11, and the matrix proteins, Pot1 and Aox1, are regulated negatively by transcriptional repressors, that compete with transcriptional activators, such as Mit1 and Mxr1 (equivalent to ScAdr1). Snf1 activation in the cytosol and SNF1–Gal83 entry into the nucleus removes, by phosphorylation of the appropriate proteins, the repression of expression of the peroxisome-associated proteins, while also activating the transcriptional activators. This simultaneous action of SNF1–Gal83 turns on the biogenesis of peroxisome-associated proteins, peroxisome proliferation, as well as division. In the ΔnugM, Δndufa9, Δgal83, Δpot1, Δaox1 Δaox2, and Δpex5 mutants of P. pastoris, or in the presence of 2,4-dinitrophenol (DNP), peroxisome proliferation, division, and the biogenesis of certain peroxisome-associated proteins in compromised. FA, fatty acids; PO, peroxisome; P, phosphorylation.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/75143/elife-75143-mdarchecklist1-v2.docx
-
Supplementary file 1
Strains and plasmids table.
- https://cdn.elifesciences.org/articles/75143/elife-75143-supp1-v2.docx
-
Supplementary file 2
RT-qPCR primers.
- https://cdn.elifesciences.org/articles/75143/elife-75143-supp2-v2.docx
-
Source data 1
Western blots and Ponceau S stained membranes (raw and annotated data).
- https://cdn.elifesciences.org/articles/75143/elife-75143-data1-v2.zip
-
Source data 2
Western blots and Ponceau S stained membranes (raw and annotated data).
- https://cdn.elifesciences.org/articles/75143/elife-75143-data2-v2.zip
-
Source data 3
Excel files containing the data for Figure 2—figure supplement 1, Figure 4E, Figure 5, and Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/75143/elife-75143-data3-v2.zip