Regulation of protein complex partners as a compensatory mechanism in aneuploid tumors
Figures
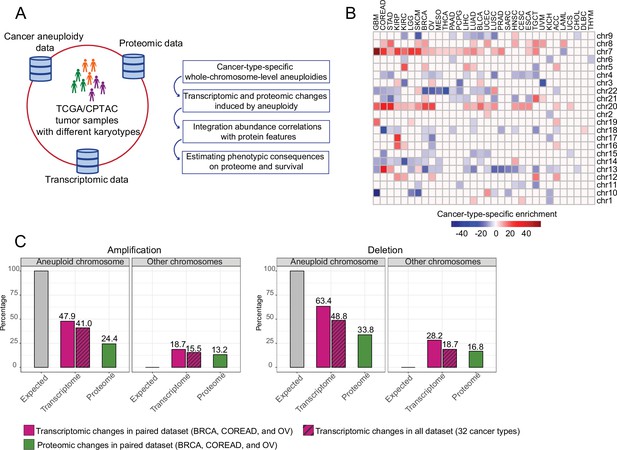
Transcriptomic and proteomic changes in aneuploid tumors.
(A) Data used in this study and schematic representation of the performed analyses. (B) Cancer-type-specific, whole-chromosome-level alterations across 32 cancer types. The color encodes the degree of their enrichment (standard residuals of the chi-square test multiplied by the alteration score [–1 in the case of deletion and 1 in the case of amplifications]). (C) Average percentage of differentially expressed genes or abundant proteins on aneuploid and other, non-aneuploid chromosomes (among the detected genes on the respective chromosomes).
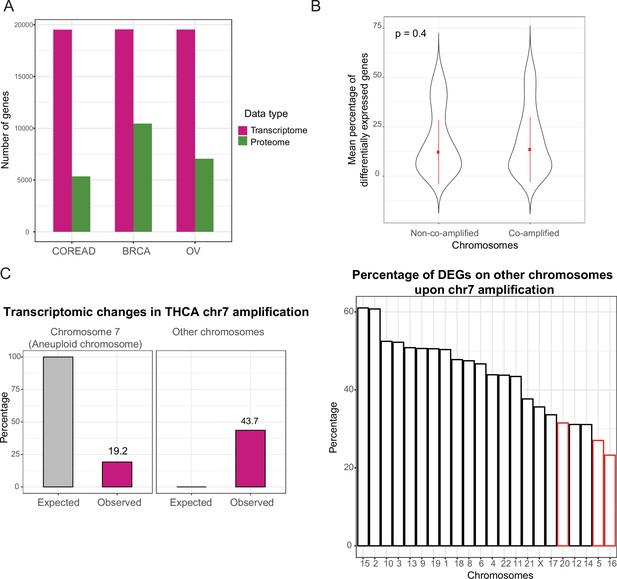
Proteome data coverage and differential expression changes on other chromosomes.
(A) The number of genes covered by transcriptomic and proteomic data for The Cancer Genome Atlas cancer patients comprising colorectal adenocarcinoma (COREAD), breast (BRCA), and ovarian (OV) cancer types. (B) Average percentage of differentially expressed genes on co-amplified and non-co-amplified chromosomes across 60 whole-chromosome-level amplifications. Paired Wilcoxon test was used to test differences between the groups. (C) Average percentage of differentially expressed genes on the aneuploid chromosome and other, non-aneuploid chromosomes (left), and percentages of differentially expressed genes on other chromosomes (right) in thyroid cancer (THCA) chromosome 7 amplification. Percentage was calculated as the ratio of differentially expressed genes from each chromosome to the total number of expressed genes on that chromosome. Red bars show the percentage for chromosome 5, 16, and 20 which are strongly co-amplified with chromosome 7 in THCA.
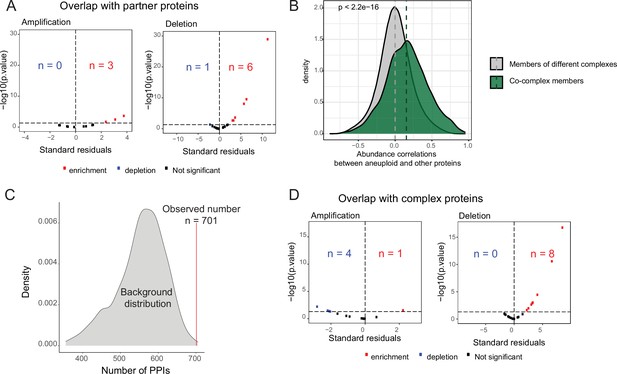
Enrichment of partners of aneuploid proteins in differentially abundant proteins on other chromosomes.
(A) Standard residuals and p-values for the overlap between co-complex members of differentially abundant proteins on aneuploid chromosomes and differentially abundant proteins on other chromosomes for 13 amplifications and 20 deletions. (B) Protein abundance correlations between differentially abundant proteins on aneuploid chromosomes and their co-complex and non-complex subunits. Correlations were calculated across cancer samples, separately for each cancer type, and then pooled. Wilcoxon test was used to determine whether two distributions are significantly different. (C) The number of protein-protein interactions (PPIs; n=701) between differentially abundant proteins of aneuploid chromosomes and those on other chromosomes against the background distribution for COREAD chromosome 7 amplification. (D) Standard residuals and p-values for the overlap between CORUM complex subunits and differentially abundant proteins on other chromosomes for 13 amplifications and 20 deletions.
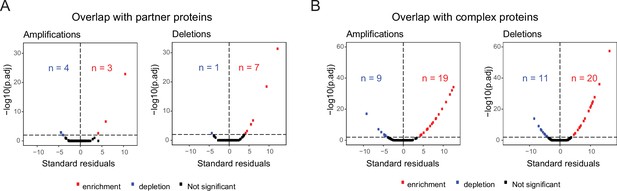
Transcriptome-level changes on other chromosomes.
Standard residuals and adjusted p-values for the overlap between (A) co-complex members of differentially expressed genes on aneuploid chromosomes and differentially expressed genes on other chromosomes, for 203 detected cancer-type-specific aneuploidies; 86 amplifications and 117 deletions, and (B) CORUM complex subunits and differentially expressed genes on other chromosomes.
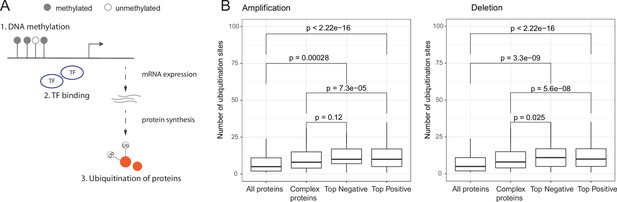
Post-translational regulation of co-complex members of aneuploid proteins.
(A) Overall representation of different levels of gene regulation. (B) Number of ubiquitination sites of all, human complex, and top positively and negatively correlated proteins. Wilcoxon test was used to test differences between groups. TF, transcription factors.
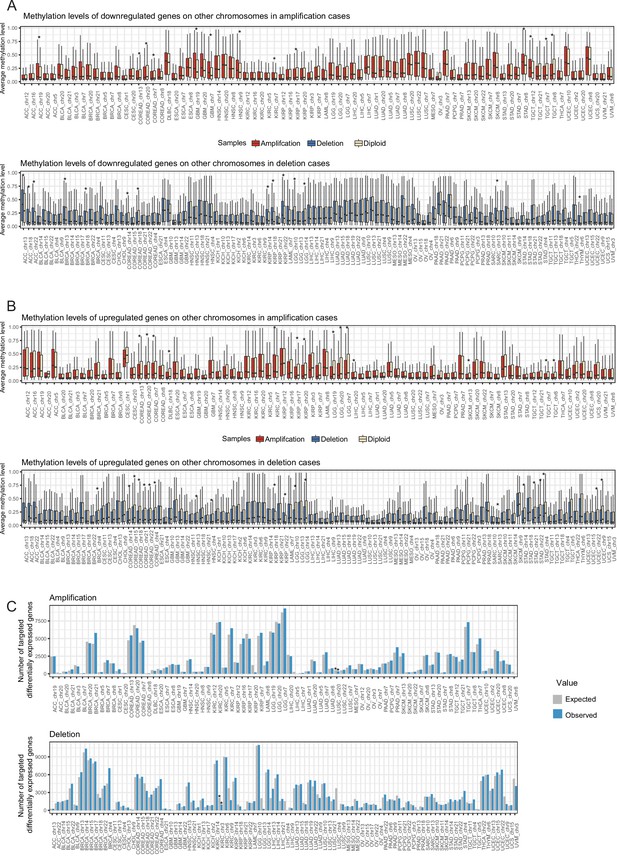
Expression changes on other chromosomes cannot be fully explained by transcriptional regulation.
Average methylation level of (A) downregulated and (B) upregulated genes in aneuploid vs diploid samples. Wilcoxon test was used to test differences in methylation level of genes between aneuploid and diploid samples (*: p<0.05). (C) The number of targets of differentially abundant transcription factors on aneuploid chromosomes among differentially expressed genes on chromosomes. Expected value and p-value were calculated using a randomization test (*: p<0.05).
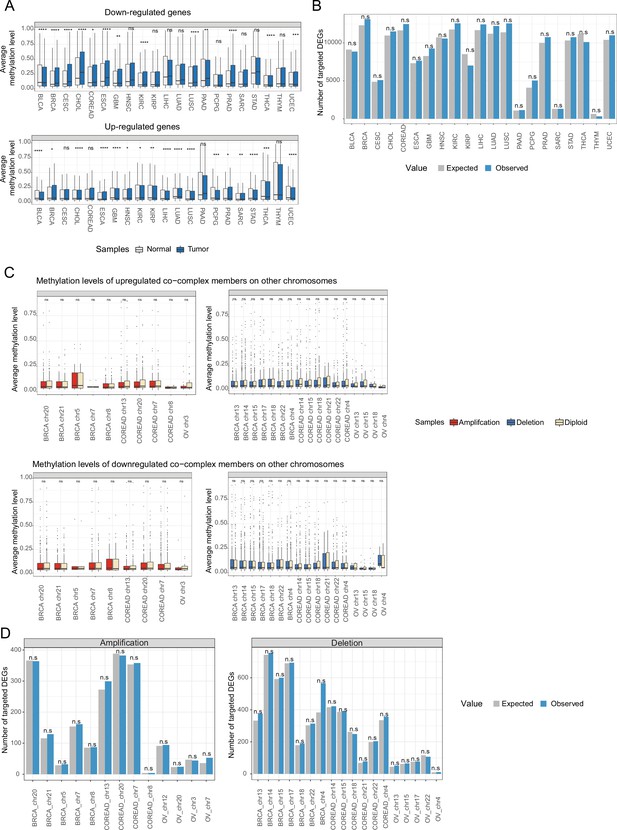
Transcriptional regulation on expression changes between tumor and normal, and of co-complex members of aneuploid proteins in aneuploid tumors.
(A) Average methylation level of down- and upregulated genes in tumor vs normal samples. (B) The number of targets of differentially expressed transcription factors (TFs) in tumors vs normal among differentially expressed genes. (C) Average methylation level of up- and downregulated co-complex members of aneuploid proteins on other chromosomes in aneuploid vs diploid samples. (D) The number of targets of differentially expressed TFs on aneuploid chromosomes among differentially expressed co-complex members of aneuploid proteins on other chromosomes. Wilcoxon test was used to test differences in methylation levels between sample groups, and expected value and p-value in B and D were calculated using a randomization test (*: p<0.05, n.s: not significant).
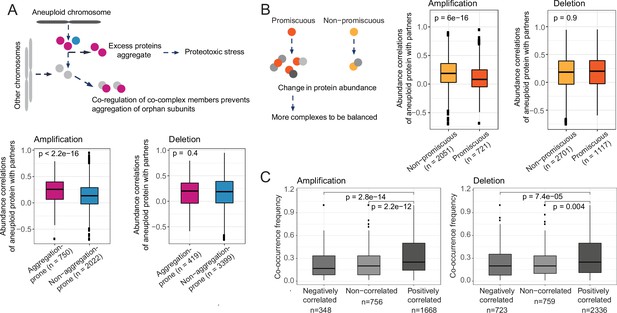
Compensatory mechanisms preventing aggregation and imbalances in complexes partly explain abundance changes on other chromosomes.
Protein abundance correlations between differentially abundant proteins on aneuploid chromosomes and their co-complex members on other chromosomes when aneuploid proteins are grouped as (A) aggregation-prone and non-aggregation-prone and (B) promiscuous and non-promiscuous. (C) Co-occurrence frequency of differentially abundant proteins on aneuploid chromosomes and their co-complex members on other chromosomes in different correlation groups, positively and negatively correlated and non-correlated co-complex members of aneuploid proteins. Wilcoxon test was used to test the differences between groups.
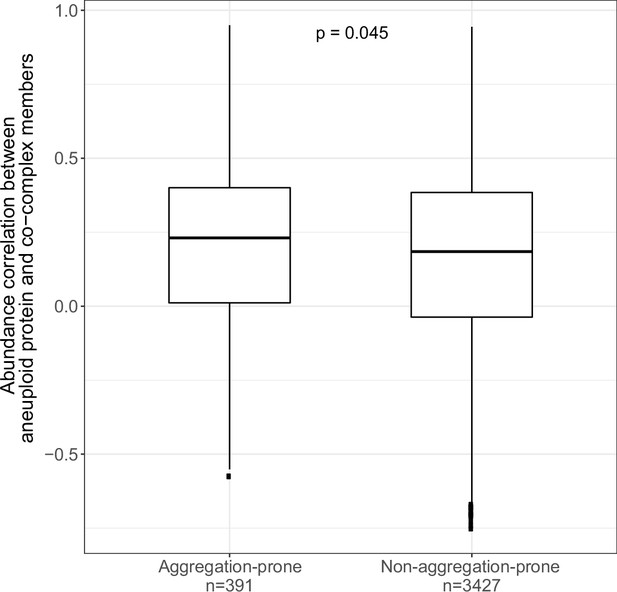
Aggregation propensity of co-complex members of aneuploid proteins in the case of deletions.
Protein abundance correlations between differentially abundant proteins on aneuploid chromosomes and their co-complex members on other chromosomes when co-complex proteins are grouped as aggregation-prone and non-aggregation-prone, in deletion cases. Wilcoxon test was used to test the difference between two groups.
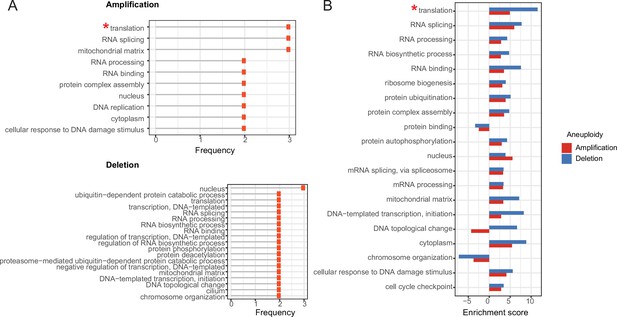
Enrichment of functional terms in complexes of top correlated proteins.
A) Most frequently enriched terms in the amplification and deletion cases. Frequency shows the number of aneuploidy cases in which the corresponding term is significantly enriched (* Enrichment is mostly driven by ribosomal genes). (B) Enrichment scores of enriched terms both in amplification and deletion cases. For the functional term that is enriched in more than one amplification/deletion cases, the enrichment score of the ones with the lowest p-value is displayed.
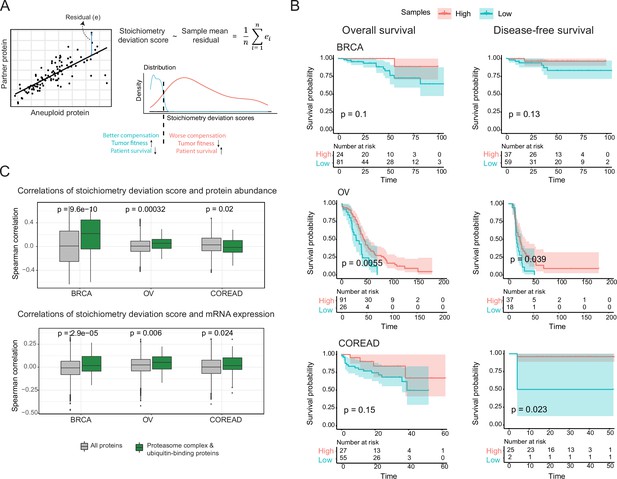
Consequences of stoichiometric compensation.
(A) Graphical representation for the calculation of stoichiometric deviation score for each sample (n=30 referring top 30 correlations). (B) Survival analysis results within each tissue. Survival analysis was done once with overall survival and once with disease-free survival. (C) Correlations between the stoichiometric deviation scores and protein abundance/mRNA expression of all proteins, and proteasome complex - ubiquitin binding proteins. Wilcoxon test was used to test differences between groups. BRCA, breast cancer; OV, ovarian cancer; COREAD, colorectal adenocarcinoma cancer.
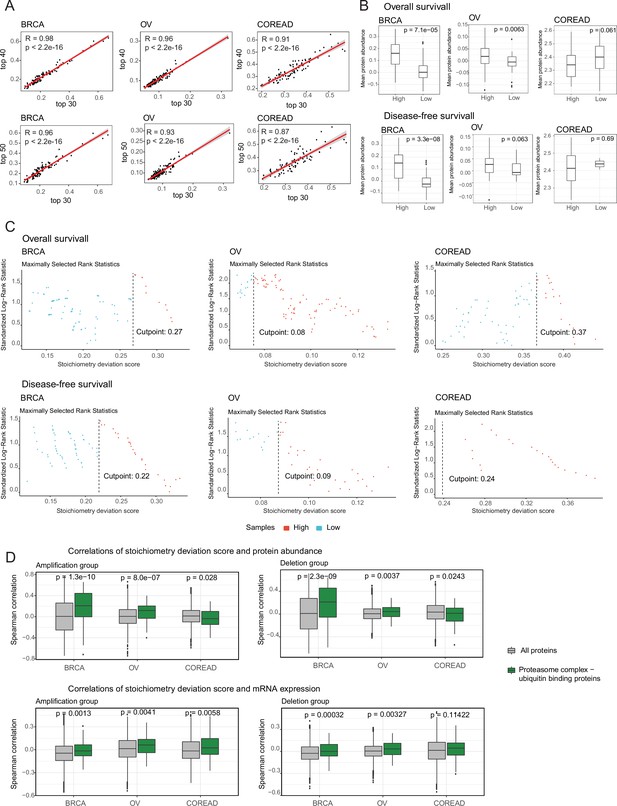
Association between the deviation from complex stoichiometry and survival probability.
(A) Correlations between the mean stoichiometric deviation scores across samples when different number of highly correlated protein pairs were considered. (B) Mean abundances of proteasome complex and ubiquitin-binding proteins in high and low samples. Samples were grouped once based on overall survival, and once based on disease-free survival. (C) Cutpoints to group samples as high and low based on stoichiometry deviation scores. (D) Correlations between the stoichiometric deviation scores and protein abundance/mRNA expression of all proteins, and proteasome complex - ubiquitin binding proteins when samples were separated as amplification and deletion groups. Wilcoxon test was used to test differences between groups. BRCA, breast cancer; OV, ovarian cancer; COREAD, colorectal adenocarcinoma cancer.
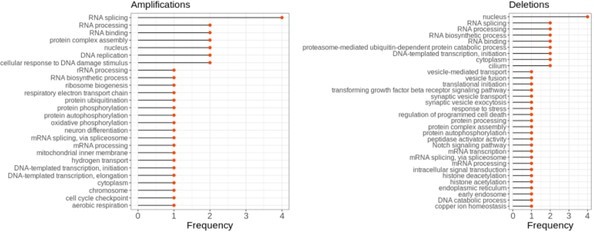
The enrichment of functional terms in complexes of top correlated partners of aneuploid proteins after removing ribosomal genes.
Frequency shows the number of aneuploidy cases in which the corresponding term is enriched.
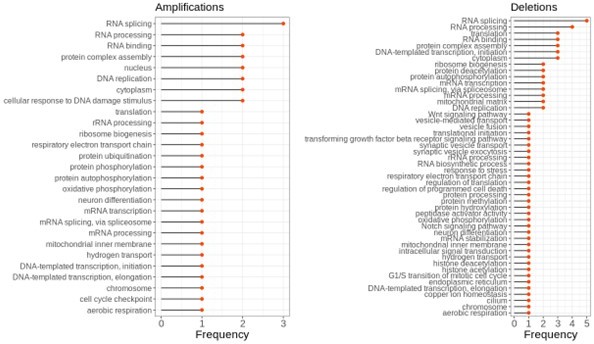
The enrichment of functional terms in complexes of top correlated partners of aneuploid proteins after removing NEDs.
Frequency shows the number of aneuploidy cases in which the corresponding term is enriched.
Additional files
-
Supplementary file 1
Whole-chromosome-level aneuploidy scores, cancer-type-specific whole-chromosome-level amplifications and deletions, and co-amplified chromosomes, related to Figure 1 and Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp1-v2.xlsx
-
Supplementary file 2
Frequently dysregulated genes on other chromosomes and their associated GO terms.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp2-v2.xlsx
-
Supplementary file 3
The number of differentially abundant proteins on other chromosomes in partners of differentially abundant aneuploid chromosomes, in all expressed proteins, and in CORUM subunits.
Standard residuals and p-values for the overlap between differentially abundant proteins on other chromosomes and partners of differentially abundant aneuploid proteins.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp3-v2.xlsx
-
Supplementary file 4
Network randomization results; the number of PPIs between differentially abundant proteins on aneuploid chromosomes and those on other chromosomes and their corresponding p-values, related to Figure 2C.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp4-v2.xlsx
-
Supplementary file 5
Protein-level Spearman correlations between differentially abundant proteins on amplified/deleted chromosomes and their co-complex members on other chromosomes, and groups of proteins and protein pairs, related to Figure 4.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp5-v2.xlsx
-
Supplementary file 6
Functional enrichment analysis of protein complexes; significantly enriched GO terms in different aneuploidy cases, their related p-values and enrichment scores, related to Figure 5.
- https://cdn.elifesciences.org/articles/75526/elife-75526-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75526/elife-75526-transrepform1-v2.docx






