Generation of vascularized brain organoids to study neurovascular interactions
Figures
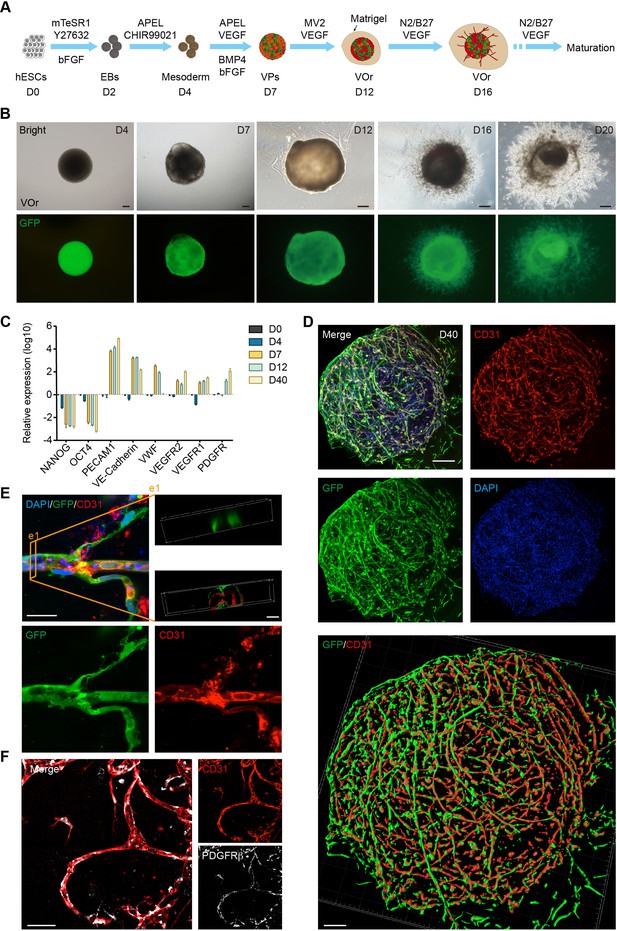
Generation of an in vitro model of vessel organoids (VOr).
(A) Schematic view of the methods for generating VOrs from GFP-hESC. EBs: embryonic bodies; VPs: vascular progenitors; VOr: vessel organoid; hESC: human embryonic stem cell. (B) Different developmental stages of VOrs from day (D) 4 to D20. Top, right field; bottom, GFP. Scale bar, 200 μm. (C) qPCR analysis for expression of stem markers (NANOG, OCT4) and vessel markers (PECAM1, VE-Cadherin, VWF, VEGFR1, VEGFR2, PDGFR) in developing VOrs, using GAPDH as internal control. Data are presented as mean ± SEM (n = 3 independent experiments), error bars indicate SEM. (D) Immunostaining of GFP and CD31 in D40 VOrs. Scale bar, 200 μm. Bottom: Imaris reconstruction of VOrs showing integrated vasculature structures. (E) Immunostaining of GFP and CD31 for the vascular structures in VOrs. Scale bar, 20 μm. Top right: section view in VOr showing the lumen structure. (F) Immunostaining of CD31 and PDGFRβ for endothelial cells and pericytes, respectively. Scale bar, 50 μm.
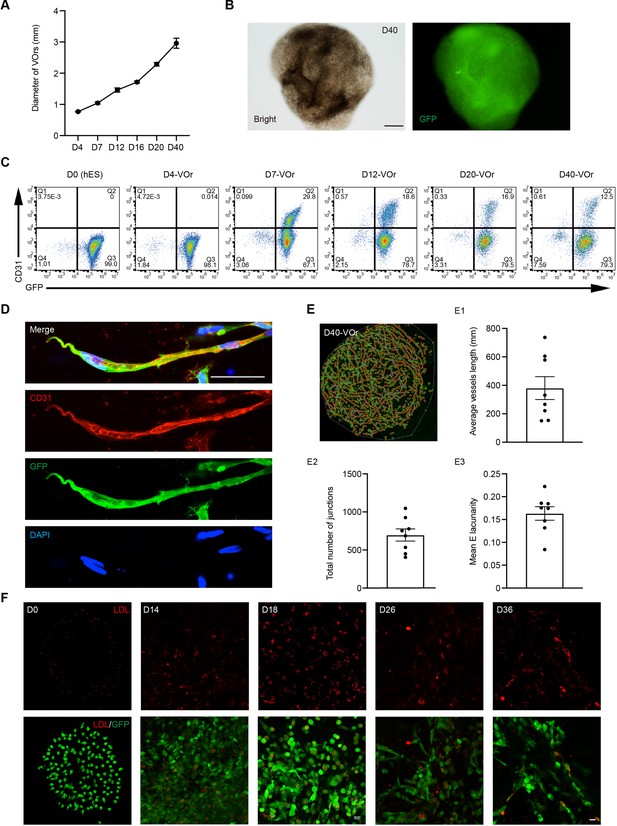
Vessel organoids (VOrs) recapitulate human vessel development.
(A) Quantification of the VOr diameter from day (D) 4 to D40. Data are mean ± SEM of 10–21 organoids at each time points. (B) Morphological appearance of VOrs at D40. Scale bar, 500 μm. (C) Flow cytometry plots of temporal development of CD31+ GFP+ cells in differentiating VOrs. (D) Immunostaining of GFP and CD31 for the vascular angiogenesis structures. Scale bar, 20 μm. (E) Quantification of the average vessel length (E1), total number of junctions (E2), and mean E lacunarity (E3) for D40 VOrs. Error bars indicate SEM. (F) Uptake of acetylated low-density lipoprotein (DiI-Ac-LDL) of VOrs at indicated time points. Scale bar, 20 μm.
Phosphate-buffered saline (PBS) fluid was microinjected into the vessel-like lumen in day (D) 40 vessel organoid (VOr) with continuous pressure, showing liquid flow and vessel wall expansion without leakage.
Electrode inner diameter: 500 nm. Scale bar, 2 μm.
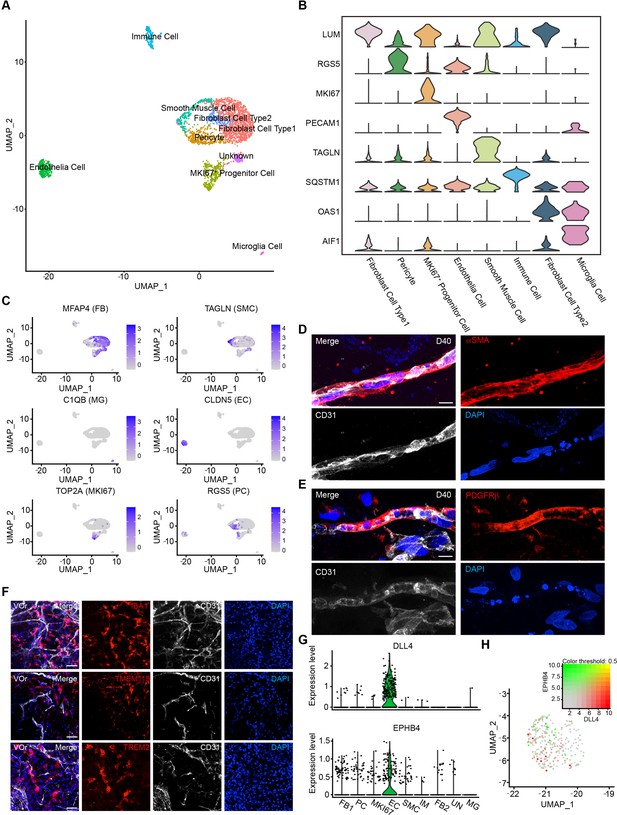
Single-cell transcriptomic analysis of vessel organoids (VOrs).
(A) UMAP plot showing the nine major cell types isolated from day (D) 40 VOrs. (B) Violin plots showing the expression value of the typical markers in each cluster. (C) Expression pattern of cell-type-specific markers in VOrs. Relative expression level is plotted from gray (low) to blue (high) colors. (D) Immunostaining of αSMA for representing the smooth muscle cells in VOrs. Scale bar, 10 μm. (E) Immunostaining of PDGFRβ for representing the pericytes in VOrs. Scale bar, 10 μm. (F) Immunostaining of microglia markers (IBA1, TREM2, TMEM119) and endothelial marker CD31 in VOrs at D40. Scale bar, 20 μm. (G) Violin plots showing the expression value of the venous marker EPHB4 and arterial marker DLL4 in endothelial cell (EC) clusters. (H) Expression pattern of arterial and venous markers in EC clusters. Relative expression level is plotted from gray to green (EPHB4) or red (DLL4) colors.
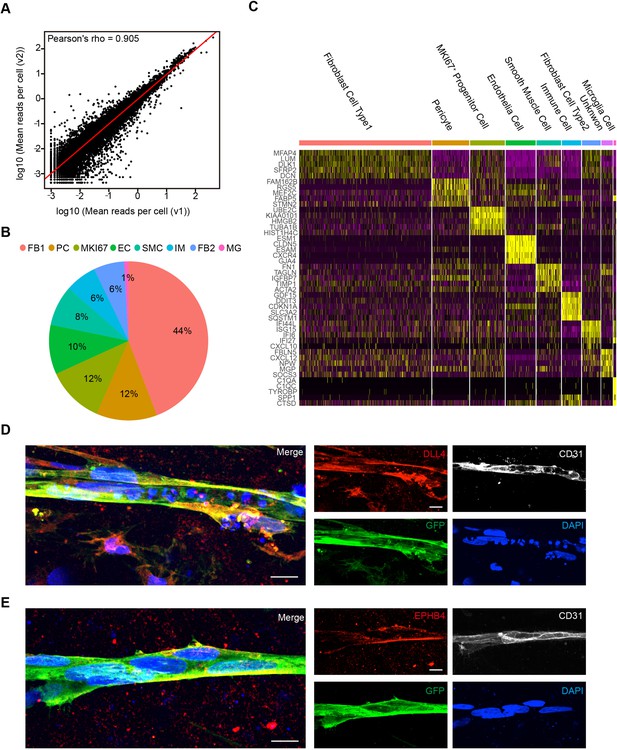
Cell-type analysis for vessel organoids (VOrs) by scRNA-seq and immunostaining.
(A) Correlation analysis of scRNA-seq data from two batches of VOr samples. (B) Proportions of cell types among all the cells from VOrs. (C) Heatmap showing the top five most enriched genes for each cell type. (D) Immunostaining of DLL4 for labeling the arterial endothelial cells in VOrs. Scale bar, 10 μm. (E) Immunostaining of EPHB4 for labeling the venous endothelial cells in VOrs. Scale bar, 10 μm.
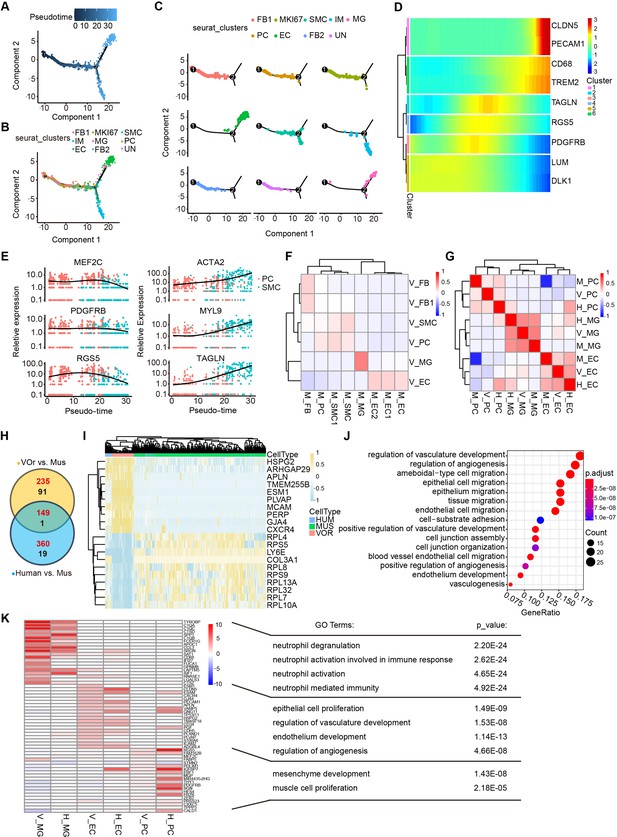
Cell fate trajectory analysis in vessel organoids (VOrs) and the comparison with cell types in vivo.
(A) Single-cell trajectories by monocle analysis showing developmental stage of the VOrs. (B) Clusters in UMAP showing trajectory track. (C) Developmental trajectory of indicated cell clusters in VOrs. (D) Heatmap showing the expression level of the main cell type-specific markers with pseudo-time. (E) Expression of markers in smooth muscle cell (SMC) and pericyte (PC) with pseudo-time. (F) Correlation analysis of cell clusters (endothelial cell [EC], microglia [MG], PC, SMC, fibroblast [FB]) between VOrs and mouse brain. V, data from VOrs; M, data from mouse. (G) Correlation analysis of cell clusters (EC, MG, PC) among VOrs, mouse, and human brain single-cell data. V, data from VOrs; M, data from mouse; H, data from human. (H) Venn diagram showing the differentially expressed genes (DEGs) in EC clusters for VOr and human samples compared to mouse samples. Red for upregulated genes, black for downregulated genes. (I) Heatmap showing the top enriched DEGs in the EC cluster for VOrs samples compared to mouse sample (fold change > 1.25 and p<0.05). (J) Gene Ontology (GO) analysis of the 149 upregulated DEGs in (H) (p-value<0.1 and false discovery rate [FDR]<0.05). (K) Top 20 marker genes for VOrs in the main clusters (EC, PC, MG) (fold change > 1.25 and p<0.05) compared to human sample, with significant pathways by GO analysis (p-value<0.1 and FDR < 0.05). V, data from VOrs; H, data from human.
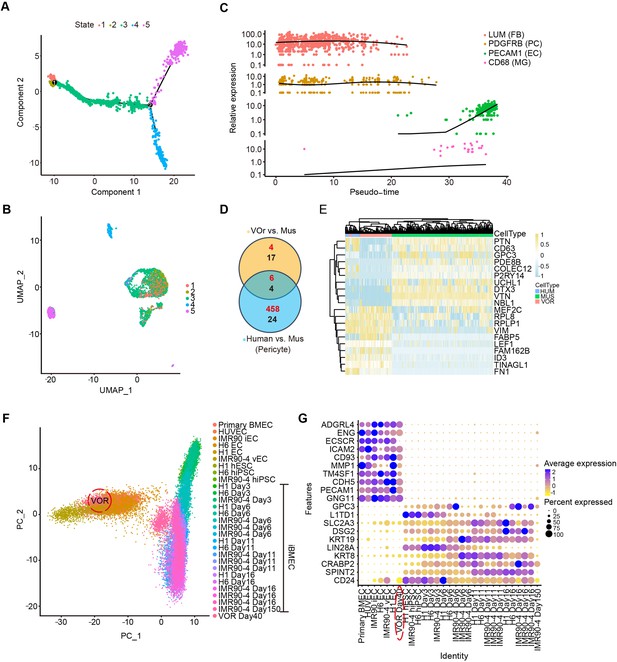
Cell types in vessel organoids (VOrs) are similar to that of human samples in vivo.
(A) Trajectory analysis showing five main developmental stages. (B) UMAP plot showing five developmental stages of VOrs. (C) Expression of markers in four main clusters with pseudo-time. (D) Venn diagram showing the differentially expressed genes (DEGs) in pericyte (PC) clusters for VOr and human samples compared with mice samples. Red for upregulated genes, black for downregulated genes. (E) Heatmap showing the top enriched common DEGs in the PC cluster for VOrs samples compared to mouse sample (fold change > 1.25 and p<0.05). (F) Principal component analysis for relative relationship of 29 distinct cell samples from published datasets, including primary endothelial cells (ECs), induced ECs (iECs), and neuroectodermal epithelial lineage-induced brain microvascular endothelial cells (Epi-iBMECs) at various days of differentiation and human pluripotent stem cells (hPSCs). ECs in VOrs are highlighted to emphasize the similarity to ECs rather than iBMECs. (G) Heatmap illustrating differences in expression levels of both endothelial- and epithelial-specific genes in all cell samples from (F). The VOr sample (circled) shows more EC-specific gene expression rather than epithelial-specific expression.
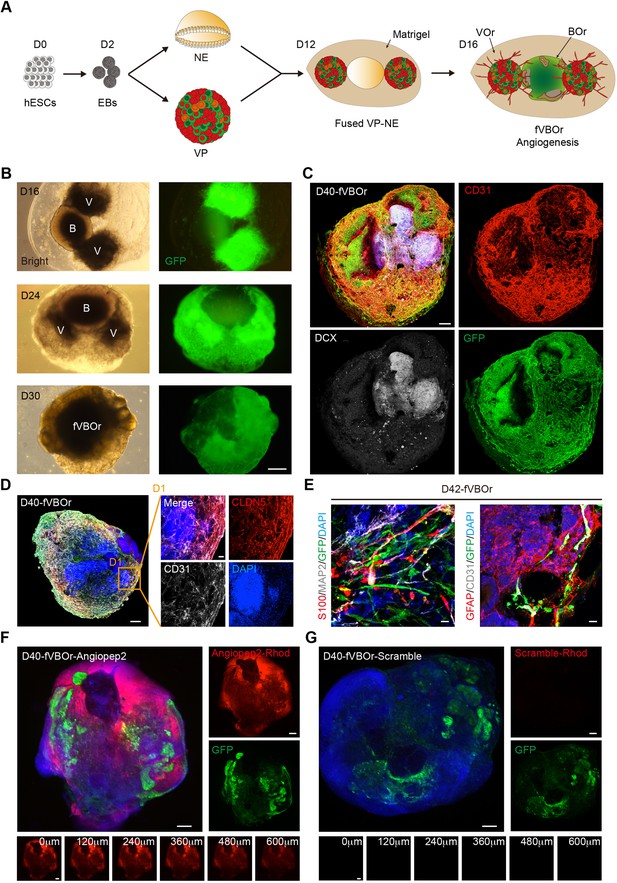
Generation of fused vasculature and brain organoids (fVBOrs) with blood–brain barrier (BBB) structure.
(A) Schematic view of the method for generating fVBOrs. EBs, embryonic bodies; NE, neuroepithelium; VP, vascular progenitor; VO, vessel organoid; BOr, brain organoid. (B) fVBOrs at different developmental stages. Scale bar, 500 μm. V, VOr; B, BOr. (C) Immunostaining of CD31 and DCX for labeling vessels and neurons, respectively, in day (D) 40 fVBOrs. Scale bar, 200 μm. (D) Immunostaining of CLDN5 for labeling tight junctions in fVBOrs. Scale bar, 200 μm. D1, enlarged area. (E) Immunostaining for markers of astrocytes (S100/GFAP), neurons (MAP2), endothelial cells (CD31), and vessel structures (GFP) in fVBOrs. Orange arrows indicate astrocytes end feet. Scale bar, 20 μm. (F, G) Confocal fluorescence images showing the transport of rhodamine-labeled angiopep-2 (Angiopep-2–Rhod), rhodamine–scramble peptide (Scramble–Rhod) in fVBOrs. Scale bar, 200 μm. Bottom, z-stack images of rhodamine signals.
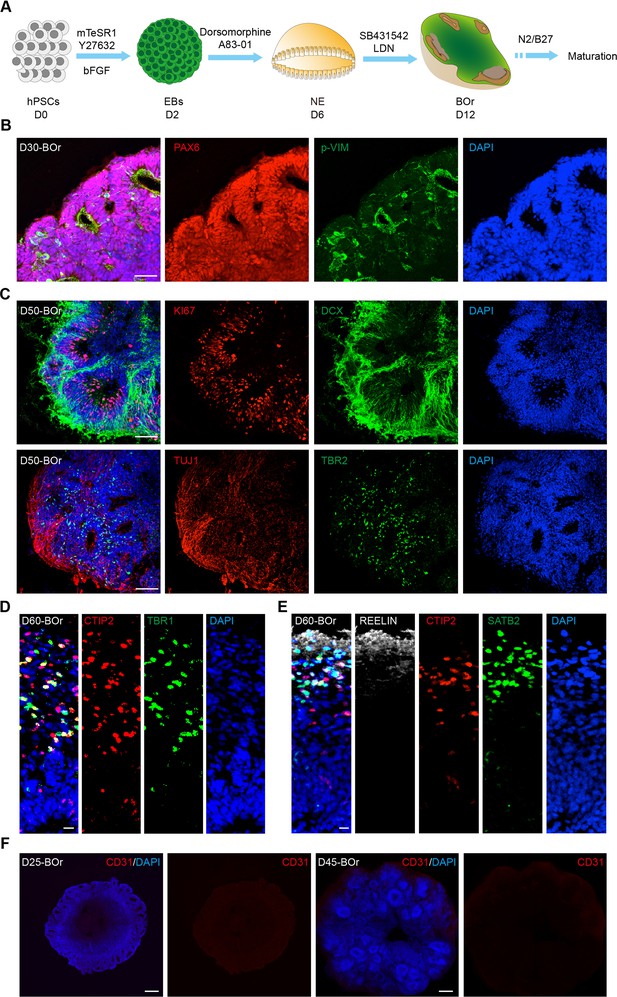
Generation of human brain organoids (BOrs).
(A) Schematic workflow for generating BOrs from human embryonic stem cell (hESC). EBs, embryonic bodies; NE, neuroepithelium. (B) Immunostaining for cortical progenitor markers PAX6 and phospho-vimentin (p-VIM) in BOrs at day (D) 30. Scale bar, 50 μm. (C) Immunostaining for young neuron marker DCX, proliferation marker KI67, intermediate progenitor marker TBR2, and neuron marker TUJ1 in D50 BOrs. Scale bar, 50 μm. (D, E) Immunostaining for cortical layer markers TBR1, CTIP2 (D) and REELIN, SATB2 (E) in D60 BOrs. Scale bar, 20 μm. (F) Immunostaining for EC marker CD31 in BOrs at D25 and D45. Scale bar, 200 μm.
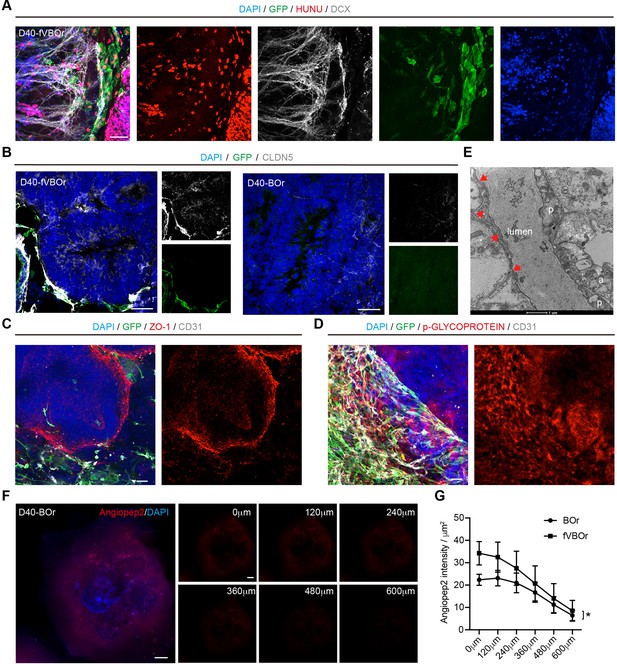
Blood–brain barrier (BBB)-like structures in fused vasculature and brain organoids (fVBOrs).
(A) fVBOrs immunostaining for Human-Nuclei (HUNU) showing human cell identity, DCX for neural cells, and GFP for vessel structures. Scale bar, 50 μm. (B) Fluorescence image showing the expression of CLDN5 in fVBOrs and BOrs, respectively. Scale bar, 50 μm. (C) Fluorescence image showing the expression of tight junctions marker ZO-1 in fVBOrs at day (D) 40. Scale bar, 50 μm. (D) Fluorescence image showing the expression of the efflux transporter p-glycoprotein in fVBOrs at D40. Scale bar, 50 μm. (E) Transmission electronic microscopy (TEM) of vascular and BBB structure in D80-fVBOr. Arrows: basement membrane; p: pericyte; a: end-foot of astrocyte. Scale bar, 1 μm. (F) Confocal z-stack images of rhodamine-labeled angiopep-2 (Angiopep-2–Rhod) in D40 BOrs. (G) Quantification for the intensity of Angiopep-2–Rhod across different z-stack images of fVBOrs compared with BOrs. Data are presented as mean ± SEM (n = 3 organoids in each sample). Errors indicate SEM. *p<0.05, paired t-test.
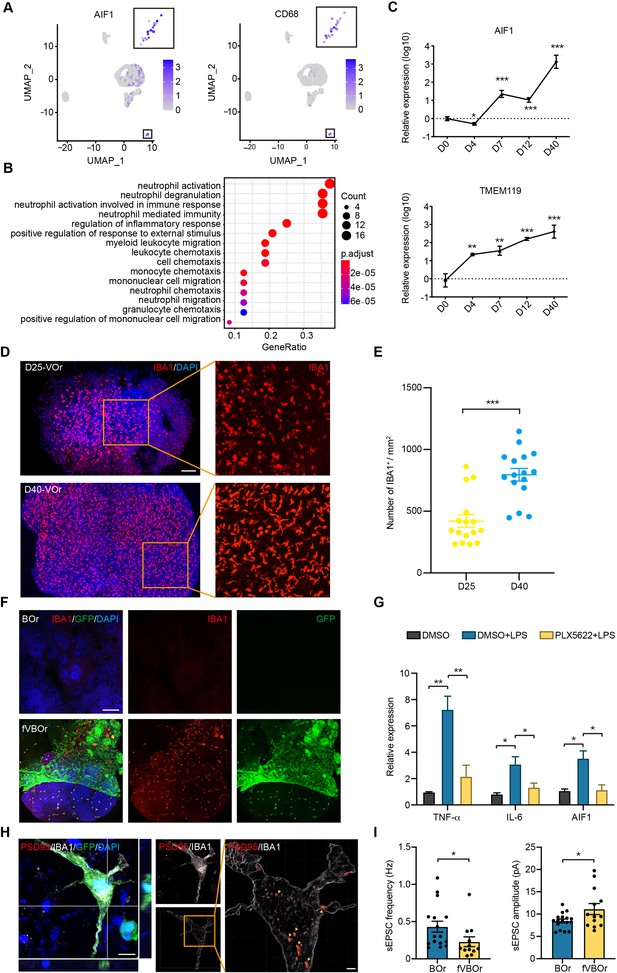
Microglial cells in fused vasculature and brain organoids (fVBOrs).
(A) UMAP plot showing single-cell expression pattern of microglial-specific markers in vessel organoids (VOr). Relative expression level is plotted from gray to blue colors. (B) Gene Ontology (GO) analysis of microglial cell marker genes (p-value<0.1 and false discovery rate [FDR] < 0.05). (C) qPCR analysis for expression of microglial markers AIF1 and TMEM119 in developing VOrs. Data are presented as mean ± SEM (n = 3 independent experiments with 6–7 organoids in each group at indicated time point). Error bars indicate SEM. **p<0.01, ***p<0.001. (D) Immunostaining of IBA1 for labeling microglial cells in day (D) 25 and D40 VOrs. Scale bar, 200 μm. (E) Quantification of the IBA1+ cell number in D25 and D40 VOrs. n = 16. Error bars indicate SEM. Student’s t-test, ***p<0.001. (F) Immunostaining of IBA1 for labeling microglial cells in BOrs and fVBOrs, respectively. Scale bar, 200 μm. (G) qPCR analysis for the expression of indicated genes in D40 fVBOrs treated with lipopolysaccharide (LPS) (500 ng/ml, MCE, HY-D1056) without or with PLX5622 2 μM (MCE, HY-11415) using DMSO as vehicle control. Relative expression was normalized to GAPDH. n = 3 independent experiments with 8–10 organoids in each group. Error bars indicate SEM. One-way ANOVA, *p<0.05, **p<0.01. (H) Double immunostaining and orthogonal view of IBA1 and PSD95 signals within microglia (MG)-like cells (left) and 3D-surface-reconstructed image (right). Arrows indicate synapse puncta engulfed in MG-like cells. Scale bar, 10 μm (left); 2 μm (right). (I) Quantification of the spontaneous excitatory post-synaptic current (sEPSC) frequency (left) and amplitude (right) in neurons of D70 BOrs and fVBOrs. Data are presented as mean ± SEM (BOrs: n = 16 neurons from six organoids; fVBOrs: n = 13 neurons from six organoids). Error bars indicate SEM. Two-tailed Student’s t-test. *p<0.05.
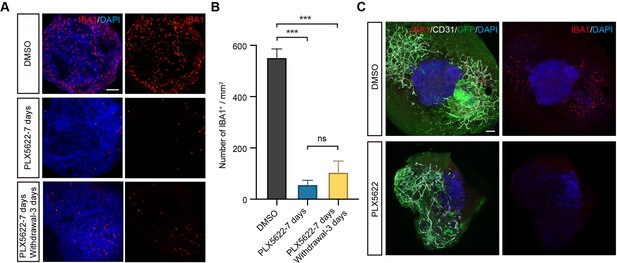
PLX5622 ablates microglias (MGs) in vessel organoids (VOrs).
(A) IBA1 staining in VOrs showing ablation of microglial cells after treatment with 2 μM PLX5622 for 7 days, and drug withdrawal for 3 days. Scale bar, 200 μm. (B) Quantification of microglial cell numbers in VOrs with indicated treatments. Data are shown as mean ± SEM (n = 3 independent experiments with 8–10 organoids in each group). Error bars indicate SEM. One-way ANOVA, ***p<0.001, ns, no significant difference. (C) Ablation of microglia in fused vasculature and brain organoids (fVBOrs) treated with 2 μM PLX5622 for 7 days. Scale bar, 200 μm.
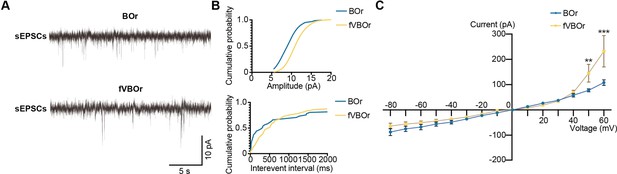
Whole-cell patch-clamp recoding of neurons in brain organoids (BOrs) and fused vasculature and brain organoids (fVBOrs).
(A) Representative spontaneous excitatory post-synaptic current (sEPSC) traces of BOrs and fVBOrs. (B) Representative cumulative distribution curves for amplitude (top) and inter-event interval (bottom) of sEPSCs in the neurons of BOrs and fVBOrs. (C) The amplitudes of inward currents of cells in BOrs and fVBOrs at day (D)70 by injection voltage ranging from –80 mV to 60 mV (step 10 mV). Data are presented as mean ± SEM (BOrs: n = 10 neurons from six organoids; fVBOrs: n = 6 neurons from six organoids). Error bars indicate SEM. Two-way ANOVA. **p<0.01, ***p<0.001.
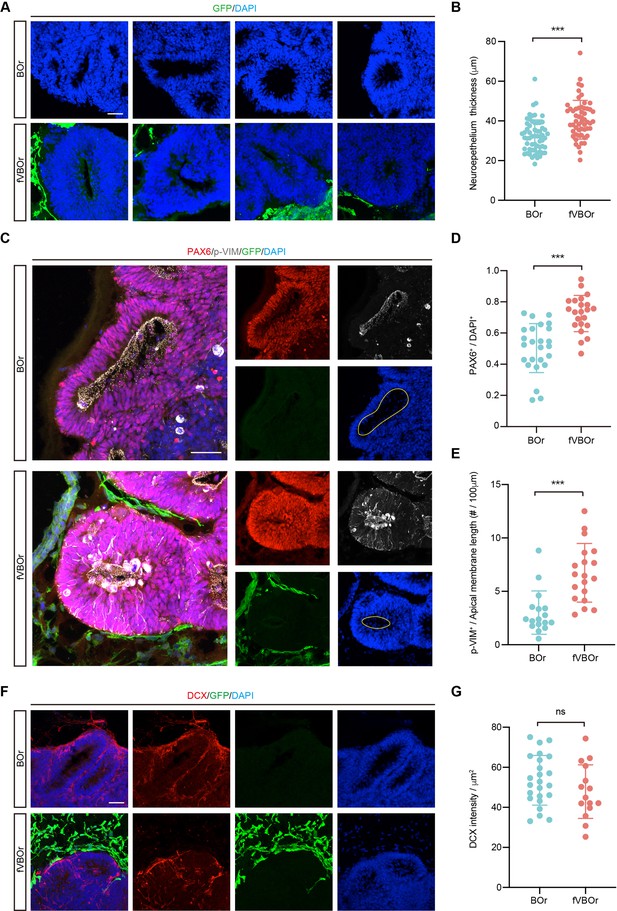
Increased neurogenesis in fused vasculature and brain organoids (fVBOrs).
(A) Immunostaining for DAPI showing the neuroepithelium rosettes of brain organoids (BOrs) and fVBOrs at D25. Scale bar, 50 μm. (B) Quantification of neuroepithelium thickness of BOrs and fVBOrs. Data are presented as mean ± SEM (BOrs: n = 60 rosettes from seven organoids; fVBOrs: n = 55 rosettes from six organoids). Error bars indicate SEM. Two-tailed Student’s t-test. ***p<0.001. (C) Immunostaining for PAX6 and phospho-vimentin (p-VIM) in VZ-like area of BOrs and fVBOrs at day (D) 25. Scale bar, 50 μm. Apical membrane is shown in yellow circle. (D, E) Quantification of the density of PAX6+ (D) and the density of p-VIM+ cells per 100 μM apical membrane length (E) in BOrs and fVBOrs. Data are presented as mean ± SEM (PAX6: n = 25 rosettes from four organoids; p-VIM: n = 23 rosettes from four organoids). Error bars indicate SEM. Two-tailed Student’s t-test. ***p<0.001. (F) Immunostaining for DCX in BOrs and fVBOrs at D25. Scale bar, 50 μm. (G) Quantification of the intensity of DCX in BOrs and fVBOrs. Data are presented as mean ± SEM (BOrs: n = 24 rosettes from three organoids, fVBOrs: n = 15 rosettes from four organoids). Error bars indicate SEM. ns, no significant difference (p=0.308, two-tailed Student’s t-test).
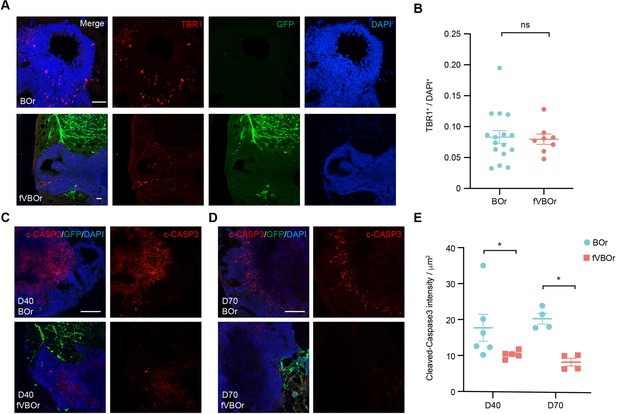
Reduced apoptotic areas in fused vasculature and brain organoids (fVBOrs).
(A) Immunostaining for early born deep-layer neuronal marker TBR1 in brain organoids (BOrs) and fVBOrs at day (D) 40. Scale bar, 50 μm. (B) Quantification of TBR1+ cells in BOrs and fVBOrs (BOr: n = 16 rosettes from six organoids, fVBOr: n = 8 rosettes from three organoids). Data are presented as mean ± SEM (two-tailed Student’s t-test, p=0.8340). (C, D) Staining of cleaved-caspase 3 (c-CASP3) in BOrs or fVBOrs at D40 (C) and D70 (D). Scale bar, 200 μm. (E) Quantification of the cleaved-caspase3 intensity in BOrs and fVBOrs at D40 and D70, respectively. Data are presented as mean ± SEM of chosen fields of five organoids in each sample. Error bars indicate SEM. *p<0.05 (p=0.0303 for D40, p=0.0286 for D70, two-tailed Student’s t-test).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | GFP (chicken polyclonal) | Aves Lab | Cat# GFP-1020 | IF (1:1000) |
| Antibody | CD31 (mouse monoclonal) | Abcam | Cat# ab9498 | IF (1:300) |
| Antibody | PDGFRβ (goat polyclonal) | R&D | Cat# AF1042 | IF (1:200) |
| Antibody | αSMA (rabbit monoclonal) | Abcam | Cat# ab124964 | IF (1:500) |
| Antibody | DCX (goat polyclonal) | Santa Cruz | Cat# sc-8066 | IF (1:200) |
| Antibody | CLDN5 (mouse monoclonal) | Abcam | Cat# ab131259 | IF (1:500) |
| Antibody | IBA1 (rabbit monoclonal) | Wako | Cat# 019-19741 | IF (1:500) |
| Antibody | PAX6 (sheep polyclonal) | R&D | Cat# AF8150 | IF (1:500) |
| Antibody | p-VIM (mouse monoclonal) | MBL | Cat# D076-3 | IF (1:1000) |
| Antibody | DLL4 (rabbit polyclonal) | Abcam | Cat# ab7280 | IF (1:500) |
| Antibody | EPHB4 (rabbit monoclonal) | Cell Signaling | Cat# 14960 | IF (1:500) |
| Antibody | KI67 (mouse monoclonal) | BD | Cat# 550609 | IF (1:1000) |
| Antibody | TBR2 (mouse monoclonal) | R&D | Cat# AF6166 | IF (1:400) |
| Antibody | TUJ1 (chicken polyclonal) | Abcam | Cat# ab41489 | IF (1:1000) |
| Antibody | TBR1 (rabbit polyclonal) | Abcam | Cat# ab31940 | IF (1:500) |
| Antibody | CTIP2 (rat monoclonal) | Abcam | Cat# ab18465 | IF (1:500) |
| Antibody | SATB2 (rabbit polyclonal) | Abcam | Cat# ab69995 | IF (1:400) |
| Antibody | PSD95 (rabbit monoclonal) | Cell Signaling | Cat# 3450S | IF (1:500) |
| Antibody | ZO-1 (rabbit monoclonal) | Abcam | Cat# ab221547 | IF (1:500) |
| Antibody | GLUT1 (rabbit monoclonal) | Abcam | Cat# ab115730 | IF (1:500) |
| Antibody | p-Glycoprotein (rabbit monoclonal) | Abcam | Cat# ab170904 | IF (1:500) |
| Antibody | Cleaved-CASPASE3 (rabbit polyclonal) | Cell Signaling | Cat# 9661L | IF (1:500) |
| Antibody | TMEM119 (rabbit polyclonal) | Abcam | Cat# ab185333 | IF (1:500) |
| Antibody | TREM2 (rabbit monoclonal) | Abcam | Cat# ab209814 | IF (1:500) |
| Antibody | Human-Nuclei (mouse monoclonal) | Millipore | Cat# MAB1281 | IF (1:500) |
| Antibody | Anti-Hu CD31 Alexa 647 WM59 50Tst | BD | Cat# 561654 | Flow cytometry (1:1000) |
| Peptide, recombinant protein | Hu Recom bFGF | STEMCELL | Cat# 78003 | |
| Peptide, recombinant protein | Hu Recom VEGF | STEMCELL | Cat# 78073 | |
| Peptide, recombinant protein | Hu Recom BMP4 | R&D | Cat# 314BP | |
| Peptide, recombinant protein | Insulin | Sigma-Aldrich | Cat# I9278 | |
| Peptide, recombinant protein | Human Dil-acetylated low-density lipoprotein | Yeasen | Cat# 20606ES76 | |
| Chemical compound, drug | Y27632 | STEMCELL | Cat# 72304 | |
| Chemical compound, drug | Dorsomorphine | Tocris | Cat# 3093/10 | |
| Chemical compound, drug | A83-01 | Tocris | Cat# 2939/10 | |
| Chemical compound, drug | CHIR99021 | Selleck | Cat# S1263 | |
| Chemical compound, drug | LDN-193189 2HCL | Selleck | Cat# S7507 | |
| Chemical compound, drug | SB431542 | Selleck | Cat# S1067 | |
| Chemical compound, drug | TTX | Tocris | Cat# 1069 | |
| Chemical compound, drug | DNQX | Tocris | Cat# 0189 | |
| Chemical compound, drug | DL-AP5 | Tocris | Cat# 3693 | |
| Chemical compound, drug | Trypsin inhibitor | Sigma-Aldrich | Cat# T6522 | |
| Chemical compound, drug | PLX5622 | MCE | Cat# HY-11415 | |
| Chemical compound, drug | LPS | MCE | Cat# HY-D1056 | |
| Commercial assay, kit | SYBR Green PCR mix | Bimake | Cat# B21702 | |
| Commercial assay, kit | RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 | |
| Commercial assay, kit | GoScript Reverse Transcription Kit | Promega | Cat# A5001 | |
| Commercial assay, kit | Pon812 812 kit | SPI | Cat# GS02660 | |
| Commercial assay, kit | Toluidine blue | Sinopharm | Cat# XW65860453 | |
| Commercial assay, kit | Uranyl acetate | SPI | Cat# GS02624 | |
| Commercial assay, kit | Lead citrate | SPI | Cat# GP19314 | |
| Commercial assay, kit | Single Cell Reagent Kits | 10x Genomics | N/A | |
| Software, algorithm | Cellranger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome | |
| Software, algorithm | Seurat (v3) | Macosko et al., 2015 | https://satijalab.org/seurat/ | |
| Software, algorithm | R (v3.5.2) | N/A | https://www.r-project.org/ | |
| Software, algorithm | clusterProfiler (v3.10.1) | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html | ||
| Software, algorithm | Limma (v3.38.3) | http://bioconductor.org/packages/release/bioc/html/limma.html | ||
| Software, algorithm | Monocle (v2.10.1) | http://cole-trapnell-lab.github.io/monocle-release/ | An analysis toolkit for single-cell RNA-seq. | |
| Software, algorithms | Fiji | N/A | https://fiji.sc | |
| Software, algorithm | Angiotool (v 0.6a) | Zudaire et al., 2011 | http://angiotool.nci.nih.gov | |
| Software, algorithm | Reference Transcriptome for GRCh38 (v1.2.0) | N/A | https://genome.ucsc.edu/ | |
| Other | Heparin | Sigma-Aldrich | Cat# H3393 | Section ‘Generation of human brain organoid’ |
| Other | Lipidure | NOF CORPORATION | Cat# CM5206 | Section ‘Generation of human brain organoid’ |
| Other | Antibiotic-Antimycotic | Gibco | Cat# 15240096 | Section ‘Generation of human brain organoid’ |
| Other | Matrigel hESC-Qualified Matrix | BD-Biocoat | Cat# 354277 | Section ‘hESCs culture’ |
| Other | Matrigel growth factor reduced (GFR) basement membrane matrix | BD-Biocoat | Cat# 354230 | Section ‘Generation of human brain organoid’ |
| Other | STEMdiff APEL2 Medium | STEMCELL | Cat# 05270 | Section ‘Generation of human vessel organoid’ |
| Other | Endothelial cell growth medium MV2 | PromoCell | Cat# C-22022 | Section ‘Generation of human vessel organoid’ |
| Other | mTeSR1 | STEMCELL | Cat# 85850 | Section ‘hESCs culture’ |
| Other | ReLeSR | STEMCELL | Cat# 05872 | Section ‘hESCs culture’ |
| Other | Accutase | STEMCELL | Cat# 07920 | Section ‘Generation of human brain organoid’ |
| Other | O.C.T | Sakura | Cat# 4583 | Section ‘Immunofluorescence’ |
| Other | BSA | Sigma-Aldrich | Cat# V900933 | Section ‘Immunofluorescence’ |
| Other | TritonX-1000 | Sigma-Aldrich | Cat# T8787 | Section ‘Immunofluorescence’ |
| Other | Neurobasal | Life/Invitrogen | Cat# 21103049 | Section ‘Generation of human brain organoid’ |
| Other | N2 supplement | Life/Invitrogen | Cat# 17502048 | Section ‘Generation of human brain organoid’ |
| Other | B27 supplement without vitamin A | Life/Invitrogen | Cat# 12587010 | Section ‘Generation of human brain organoid’ |
| Other | B27 supplement | Life/Invitrogen | Cat# 17504044 | Section ‘Generation of human brain organoid’ |
| Other | DPBS | Life/Invitrogen | Cat# 14190144 | Section ‘hESCs culture’ |
| Other | HBSS | Life/Invitrogen | Cat# 14175069 | Section ‘Single-cell dissociation and 10x Genomics chromium library construction’ |
| Other | HEPES | Sigma-Aldrich | Cat# H4034 | Section ‘Single-cell dissociation and 10x genomics chromium library construction’ |
| Other | DNase I | Roche | Cat# 10104159001 | Section ‘Single-cell dissociation and 10x Genomics chromium library construction’ |
| Other | Knockout Serum Replacer | Gibco | Cat# 10828028 | Section ‘Generation of human brain organoid’ |
| Other | MEM-NEAA | Gibco | Cat# 11140050 | Section ‘Generation of human brain organoid’ |
| Other | GlutaMAX | Gibco | Cat# 35050061 | Section ‘Generation of human brain organoid’ |
| Other | β-Mercaptoethanol | Sigma-Aldrich | Cat# M3148 | Section ‘Generation of human brain organoid’ |
| Other | Bovine pancreatic trypsin | Sigma-Aldrich | Cat# 6502 | Section ‘Single-cell dissociation and 10x Genomics chromium library construction’ |
| Other | Leibovitz L-15 medium | Thermo | Cat# 11415064 | Section ‘Single-cell dissociation and 10x Genomics chromium library construction’ |
| Other | DMEM/F12 | Life/Invitrogen | Cat# 10565018 | Section ‘hESCs culture’ |





