Free spermidine evokes superoxide radicals that manifest toxicity
Figures
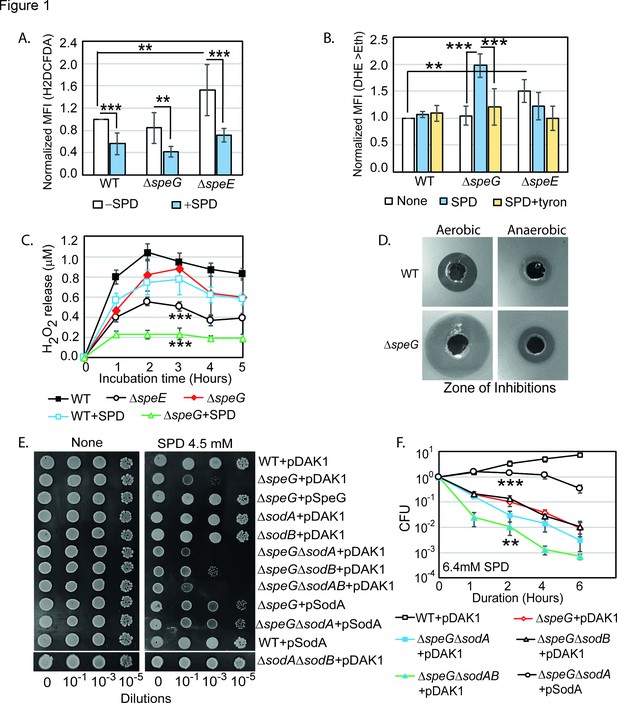
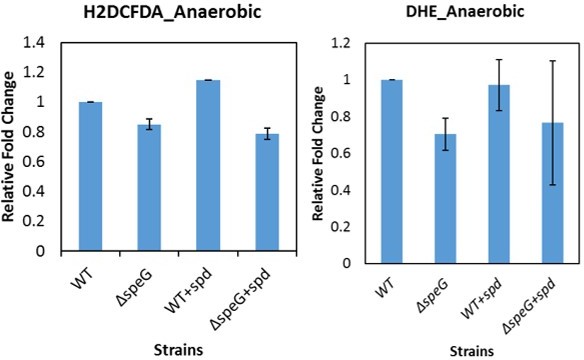
Spermidine (SPD) stress and intracellular reactive oxygen species (ROS) in Escherichia coli.
(A) The relative mean fluorescence intensity (MFI) values for the 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA), which is an indicator of •OH radical production, obtained by flow cytometry analyses are plotted. (B) The relative MFI values of dihydroethidium (DHE) probe, which is an indicator of O2- radical production, obtained by flow cytometry analyses are plotted. (C) The absolute H2O2 production for a span of 5 hr from the different E. coli strains are shown. *** are p-values generated comparing with WT value. (D) Zone of inhibitions (ZOIs) surrounding SPD well on the agar plates were shown for the WT and ΔspeG strains of E. coli under aerobic and anaerobic conditions. (E) Serially diluted E. coli cells were spotted on LB-agar plates to show their sensitivity to SPD. (F) Viability of different knockout strains were plotted from the CFU counts in different time intervals after treatment with lethal dose of SPD. ** and *** are p-values generated comparing with the values of ΔspeG and ΔspeGΔsodA, respectively. Error bars in the panels are mean ± SD from the three independent experiments. Whenever mentioned, *** and ** are <0.001 and <0.01, respectively; unpaired t test. See also Figure 1—figure supplement 1, and Figure 1—source data 1, Figure 1—source data 2, Figure 1—source data 3, Figure 1—source data 4.
-
Figure 1—source data 1
Figure 1A Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Figure 1B Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-data2-v3.xlsx
-
Figure 1—source data 3
Figure 1C Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-data3-v3.xlsx
-
Figure 1—source data 4
Figure 1F Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-data4-v3.xlsx
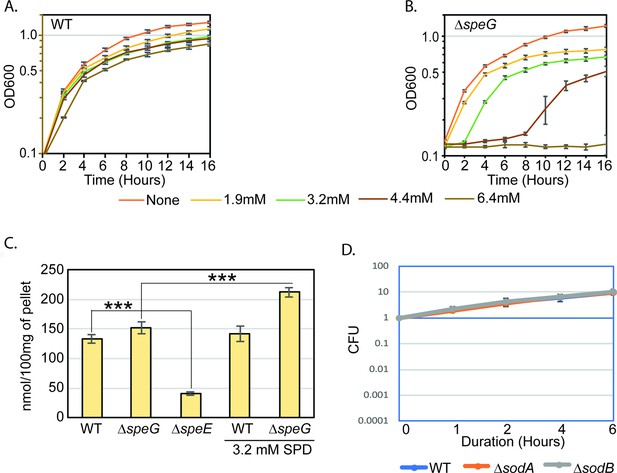
Spermidine-mediated O2- production is apparently toxic in the absence of SpeG function.
(A) and (B) represent the growth curves of Escherichia coli WT and ΔspeG strains, respectively, in the presence of varying concentrations of spermidine. (C) Bar diagram represents intracellular spermidine levels in different strains in the presence or absence of spermidine. (D) Cell viability assays show that single ΔsodA and ΔsodB mutants are not affected by spermidine (SPD). See also Figure 1—figure supplement 1—source data 1 and Figure 1—figure supplement 1—source data 2.
-
Figure 1—figure supplement 1—source data 1
Figure 1—figure supplement 1C raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-figsupp1-data1-v3.xlsx
-
Figure 1—figure supplement 1—source data 2
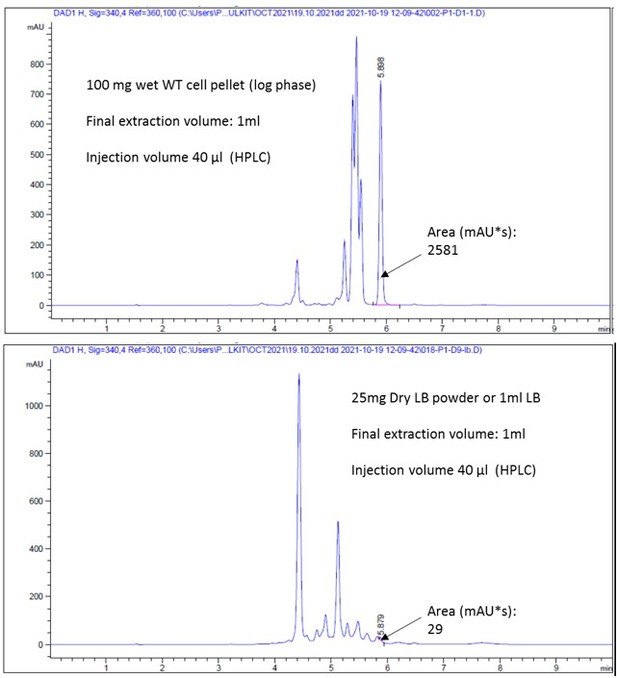
Figure 1—figure supplement 1C raw HPLC peak profiles.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig1-figsupp1-data2-v3.pdf
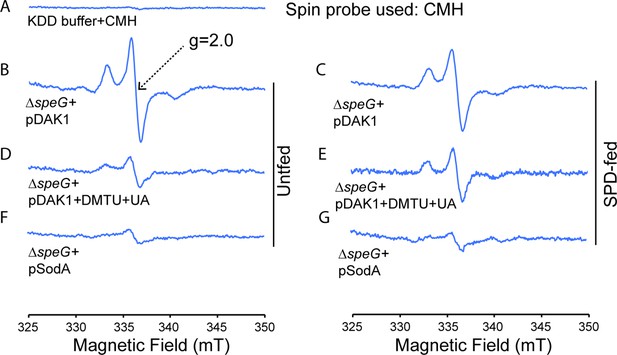
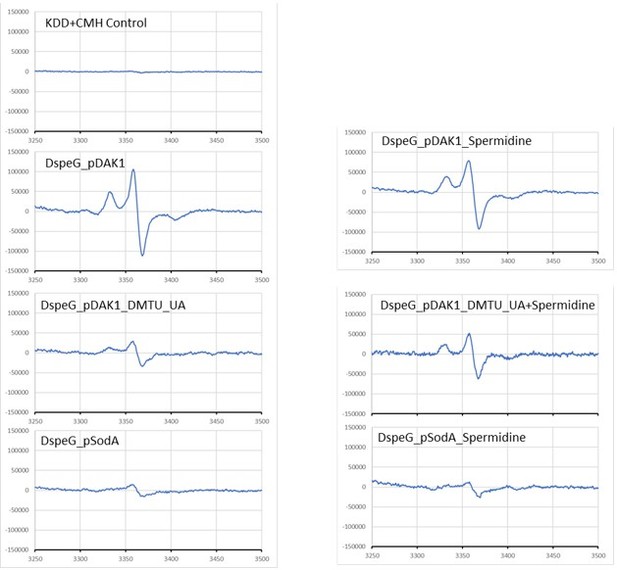
Spermidine stress generates O2- radical production in ΔspeG strain.
(A) 1-Hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidine (CMH) probe incubated with KDD buffer before electron paramagnetic resonance (EPR) analysis. (B, C, D) EPR spectra ΔspeG strain with the plasmids pDAK1 (empty vector) or pSodA were grown without spermidine and performed EPR adding CMH probe. (E, F, G) ΔspeG strain with the plasmids pDAK1 (empty vector) or pSodA were grown with spermidine and performed EPR adding CMH spin probe, as mentioned in the Materials and methods. See also Figure 2—source data 1.
-
Figure 2—source data 1
Figure 2 Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig2-data1-v3.xlsx
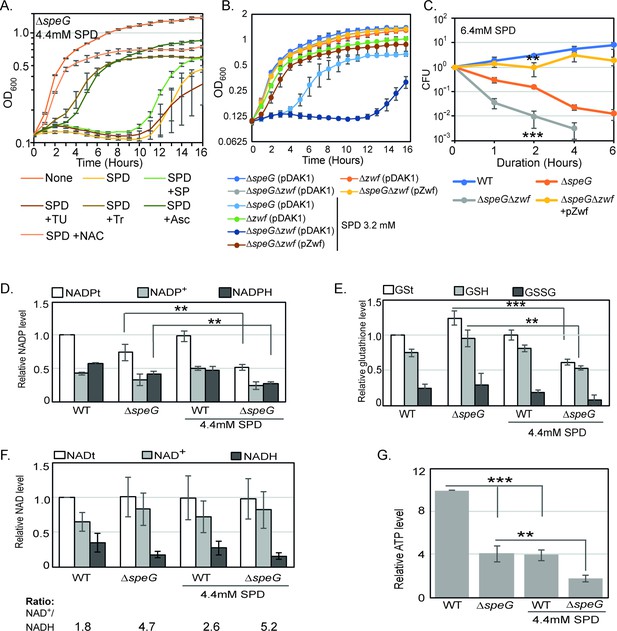
O2- radical production affects redox balance in the spermidine-fed ΔspeG strain.
(A) Growth curves show that Tyron (Tr), ascorbate (Asc), and N-acetyl cysteine (NAC) can overcome spermidine (SPD) stress while sodium pyruvate (SP) and thiourea (TU) fail to do so. (B) Growth curves show that ΔspeGΔzwf strain is hypersensitive to SPD in comparison to ΔspeG strain. Complementation of ΔspeGΔzwf strain with pZwf plasmid overcomes this SPD hypersensitivity. (C) CFUs were obtained for different Escherichia coli strains pretreated with SPD for desired time points and plotted to show the reduced viability of ΔspeGΔzwf strain in comparison to theΔspeG strain. (D) Relative levels of NADPt and reduced nicotinamide adenine dinucleotide phosphate (NADPH) were significantly decreased in the ΔspeG strain under SPD stress. (E) Relative levels of GSt, GSH, and GSSG were significantly decreased in the SPD-fed ΔspeG strain. (F) No significant change in the relative total NAD (NADt), NAD+, and NADH levels were recorded. However, NAD+ to NADH ratio was significantly increased in the ΔspeG strain compared to WT cells. No further increase of the ratio was observed by adding SPD in the growth medium of WT and ΔspeG strain. (G) The relative level of ATP was declined in ΔspeG strain and spermidine-fed WT cells in comparison to the unfed WT. SPD supplementation decreased the ATP level further in the SPD-fed ΔspeG strain. Error bars in the panels are mean ± SD from the three independent experiments. Whenever mentioned, the *** and ** denote p-values < 0.001 and < 0.01, respectively; unpaired t test. See also Figure 3—source data 2, Figure 3—source data 3, Figure 3—source data 4, Figure 3—source data 5.
-
Figure 3—source data 1
Figure 3A Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Figure 3B Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig3-data2-v3.xls
-
Figure 3—source data 3
Figure 3C Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig3-data3-v3.xlsx
-
Figure 3—source data 4
Figure 3D, E and F Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig3-data4-v3.xlsx
-
Figure 3—source data 5
Figure 3G Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig3-data5-v3.xlsx
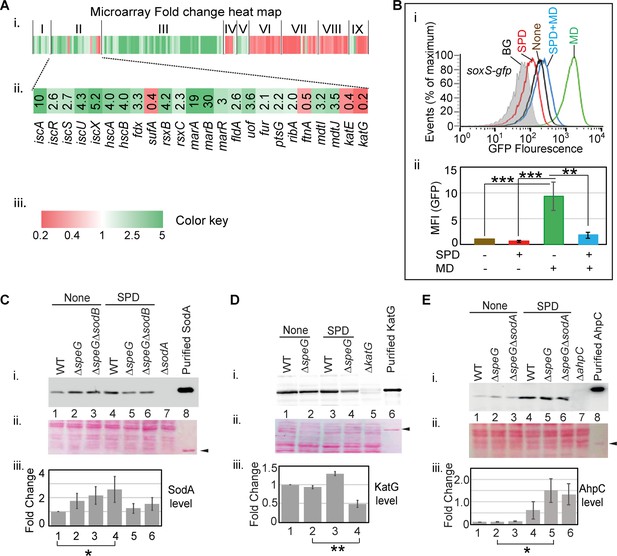
Spermidine blocks the activation of superoxide defense circuit.
(A) (i) Microarray heat map showing various categories of genes (I: Replication and transcription associated genes, II: Iron homeostasis, ROS, multidrug resistance and sugar metabolism genes, III: Ribosomal and ribosome biogenesis-associated genes, IV: Oxidoreductase and ATP synthesis genes, V: Fatty acid metabolism-related genes, VI: Flagellar biogenesis-related genes, VII: Acid resistance and chaperone genes, VIII: Hydrogenase and nitrogen metabolizing genes, IX: Amino acid metabolizing genes; see Supplementary file 1) that were differentially expressed under spermidine stress. (ii) Zoomed in heat map of the category II genes responsible for iron metabolism and reactive oxygen species (ROS) regulation. (iii) Color key represents the expression fold-change (FC) of the genes. (B) The subpanel (i) represents a flow cytometry experiment to demonstrate that spermidine (SPD) stress inhibits menadione (MD)-induced PsoxS-gfpmut2 reporter fluorescence. The subpanel (ii) represents relative mean fluorescence intensities (MFIs) in the presence or absence of SPD and MD calculated from three different flow cytometry experiments. (C, D, E) Western blotting experiments show SodA, KatG, and AhpC levels in the various strains in the presence or absence of SPD: (i) developed blot, (ii) ponceau S-stained counterpart of the same blot, (iii) the bar diagrams represent relative FC of the proteins under SPD stress. The relative FC values were calculated from the band intensity values obtained from three independent blots in comparison to the untreated WT counterparts. Purified 6X His-tagged SodA, KatG, and AhpC proteins were loaded as positive controls. The cellular protein extracts from ΔsodA, ΔkatG, and ΔahpC strains were used for negative controls. Whenever mentioned, the *** and ** denote p-values < 0.001, < 0.01, respectively; unpaired t test.Figure 4—source data 2., Figure 4—source data 3, Figure 4—source data 4, Figure 4—source data 5, Figure 4—source data 6, Figure 4—source data 7, Figure 4—source data 8, Figure 4—source data 9, Figure 4—source data 10, Figure 4—source data 11, Figure 4—source data 12, Figure 4—source data 13 and Figure 4—source data 14.
-
Figure 4—source data 1
Figure 4B–ii Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data1-v3.xlsx
-
Figure 4—source data 2
Figure 4C–i Raw unedited image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data2-v3.zip
-
Figure 4—source data 3
Figure 4C–i Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data3-v3.zip
-
Figure 4—source data 4
Figure 4C–ii Raw full image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data4-v3.zip
-
Figure 4—source data 5
Figure 4C–ii Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data5-v3.zip
-
Figure 4—source data 6
Figure 4D–i Raw full image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data6-v3.zip
-
Figure 4—source data 7
Figure 4D–i Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data7-v3.zip
-
Figure 4—source data 8
Figure 4D–ii Raw full image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data8-v3.zip
-
Figure 4—source data 9
Figure 4D–ii Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data9-v3.zip
-
Figure 4—source data 10
Figure 4E–i Raw full image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data10-v3.zip
-
Figure 4—source data 11
Figure 4E–i Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data11-v3.zip
-
Figure 4—source data 12
Figure 4E–ii Raw full image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data12-v3.zip
-
Figure 4—source data 13
Figure 4E–ii Raw uncropped and labeled image.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data13-v3.zip
-
Figure 4—source data 14
Figure 4C, D and E Fold change values of the western blots.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig4-data14-v3.xlsx
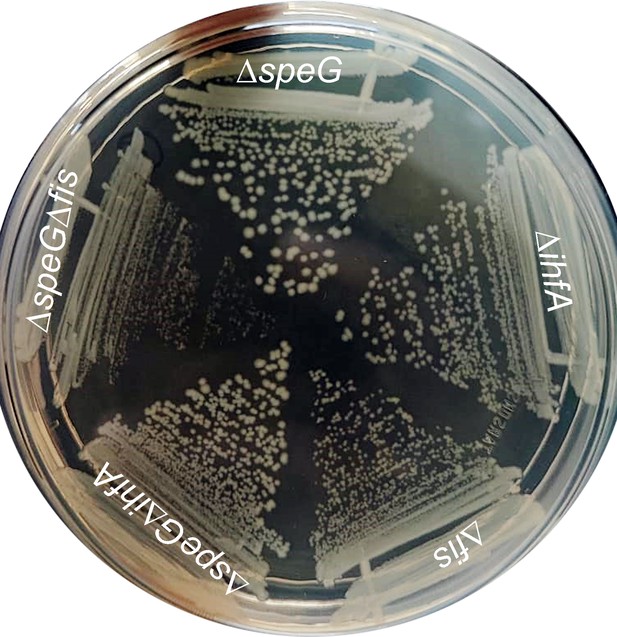
Indication of the importance of Fis regulator under spermidine stress.
The colonies of different mutant Escherichia coli strains were steaked on the LB-agar surface to show that ΔspeGΔfis slow growing compared to ΔspeG and ΔspeGΔihfA strains.
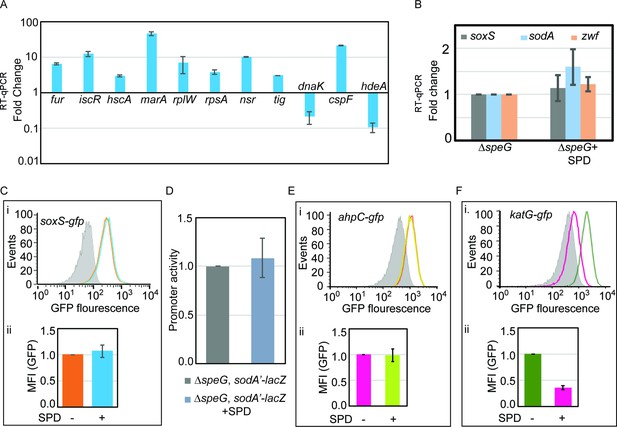
Validation of microarray data.
(A) Microarray data was validated by performing quantitative real-time PCR (RT-qPCR) against some of the genes that were up- or downregulated in the microarray. (B) RT-qPCR data to show that the spermidine stress does not alter the expression of soxS, sodA, and zwf genes in the ΔspeG strain. (C) Flow cytometry experiments determined that spermidine stress does not upregulate PsoxS-gfpmut2 reporter in the ΔspeG strain. The upper portion of the panel (i) represents a flow cytometry histogram. Gray area is background fluorescence of the ΔspeG cells. The areas within saffron line and blue line represent GFP fluorescence in the absence and presence of spermidine, respectively. The bar diagram in the lower part of the panel (ii) exhibits relative mean fluorescence intensity (MFI) values calculated from three independent flow cytometry experiments. (D) β-Galactosidase reporter assay shows that the promoter activity of sodA gene remains unchanged under spermidine stress. (E) Flow cytometry experiments were performed to show that spermidine stress does not upregulate PahpC-gfp reporter in the ΔspeG strain. The subpanel (i) represents a flow cytometry histogram of background fluorescence of the ΔspeG cells (gray area). The pink-lined and green-lined areas of histogram represent fluorescence in the absence and presence of spermidine, respectively. The bar diagram in the subpanel (ii) exhibits relative MFI values calculated from three independent flow cytometry experiments. (F) Flow cytometry experiments show that spermidine stress downregulates PkatG-gfp reporter expression in the ΔspeG strain. The subpanel (i) represents histogram of background fluorescence of the ΔspeG cells (gray area). The green-lined and pink-lined areas of histogram represent GFP fluorescence in the absence and presence of spermidine, respectively. The bar diagram in the lower part of the panel (ii) exhibits relative MFI values in the presence or absence of spermidine calculated from three independent flow cytometry experiments. *** denotes p-value < 0.001; unpaired t test.
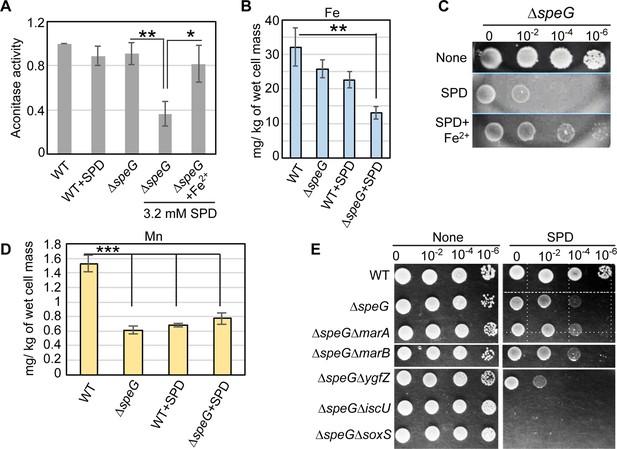
Spermidine-mediated O2- radical production affects iron metabolism.
(A) The bar diagram represents relative aconitase activity in the Escherichia coli WT and ΔspeG strains in the presence and absence of spermidine (SPD). (B) Intracellular levels of Fe in the E. coli strains determined in the presence or absence of SPD stress were plotted. (C) Spot assay using serially diluted ΔspeG cells demonstrated that Fe2+ can rescue SPD stress. (D) Intracellular levels of Mn levels in the E. coli strains determined in the presence or absence of SPD stress were plotted. (E) Spot assay shows the relative sensitivity of various double mutants, ΔspeGΔygfZ, ΔspeGΔiscU, and ΔspeGΔsoxS strains to SPD. Error bars in the panels are mean ± SD from the three independent experiments. Whenever mentioned, the ***, **, and * denote p-values < 0.001, < 0.01, and < 0.1 respectively; unpaired t test. See also Figure 5—figure supplement 1, and Figure 5—source data 1, Figure 5—source data 2, Figure 5—source data 3.
-
Figure 5—source data 1
Figure 5A Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Figure 5B Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig5-data2-v3.xlsx
-
Figure 5—source data 3
Figure 5D Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig5-data3-v3.xlsx
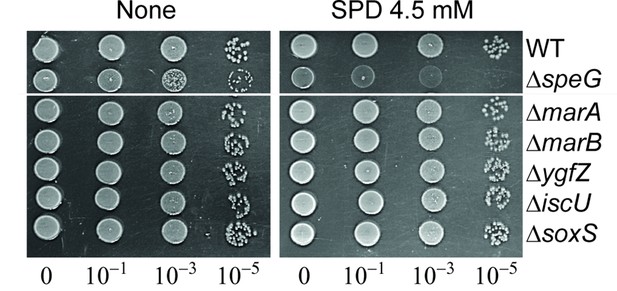
Spermidine (SPD) sensitivity of the single mutants.
The spot assay shows that single mutants are not affected by SPD.
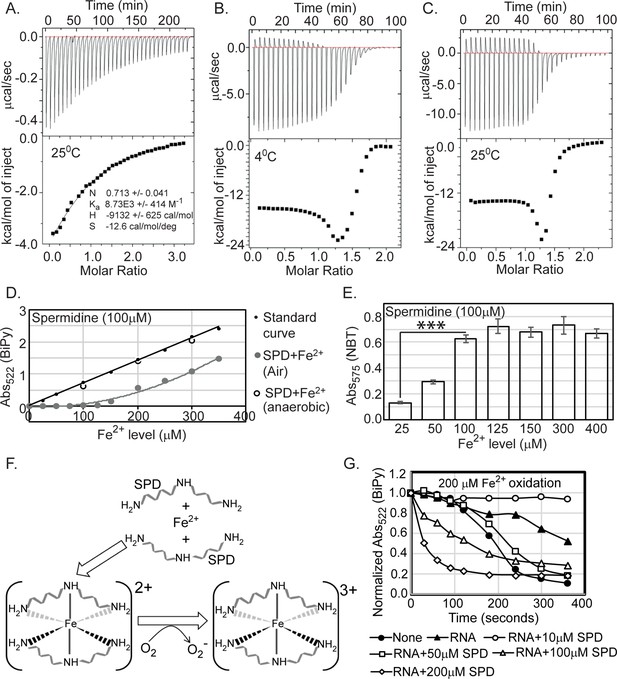
Spermidine oxidizes Fe2+ generating O2- radical in aerobic condition.
(A) Isothermal titration calorimetry (ITC) data demonstrates the interaction of spermidine with Fe3+. (B) and (C) ITC data shows the interaction of spermidine with Fe2+ ion at 4°C and 25°C, respectively. (D) 100 µM spermidine was incubated with different concentrations of Fe2+ followed by estimation of Fe2+ levels by bipyridyl chelator. The color formation was recorded at 522 nm and plotted them along with standard curve. The panel depicts that the incubations of 100 µM spermidine with 100, 200, and 300 µM of Fe2+ in the anaerobic condition do not lead to the loss of Fe2+ ions detected by bipyridyl chelator. However, when 100 µM spermidine was incubated with the different concentrations of Fe2+ (25–350 µM) in the aerobic condition, the bipyridyl-mediated color formation was observed when Fe2+ level was between above 125 µM and 150 µM (i.e., till spermidine to Fe2+ ratio reaches approximately 1.3). The mean values from the three independent experiments were plotted. SD is negligible and is not shown for clarity. (E) Nitro blue tetrazolium (NBT) assay was performed to determine that spermidine and Fe2+ interaction yields O2- radical. The colorimetry at 575 nm suggests that 100 µM of spermidine interacts with approximately 125 µM of Fe2+ (ratio 1:1.3) to generate saturated color. Error bars in the panel are mean ± SD from the three independent experiments. *** denotes p-value < 0.001; unpaired t test. (F) Model to show final coordination complex formation. An Fe2+ interacts with two spermidine molecules forming hexadentate coordination complex. This interaction oxidizes Fe2+ liberating one electron to reduce oxygen molecule. Finally, two spermidine coordinates one Fe3+ with an octahedral geometry. (G) The curves represent the Escherichia coli total RNA inhibits iron oxidation. Spermidine further reduces the RNA-mediated iron oxidation at concentration 10 µM but higher concentrations of spermidine increase the iron oxidation despite the presence of RNA. The mean values are derived from the three independent experiments and plotted. SD is negligible and is not shown for clarity. See also Figure 6—source data 1, Figure 6—source data 2, Figure 6—source data 3.
-
Figure 6—source data 1
Figure 6D Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig6-data1-v3.xlsx
-
Figure 6—source data 2
Figure 6E Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig6-data2-v3.xlsx
-
Figure 6—source data 3
Figure 6G Raw data.
- https://cdn.elifesciences.org/articles/77704/elife-77704-fig6-data3-v3.xlsx
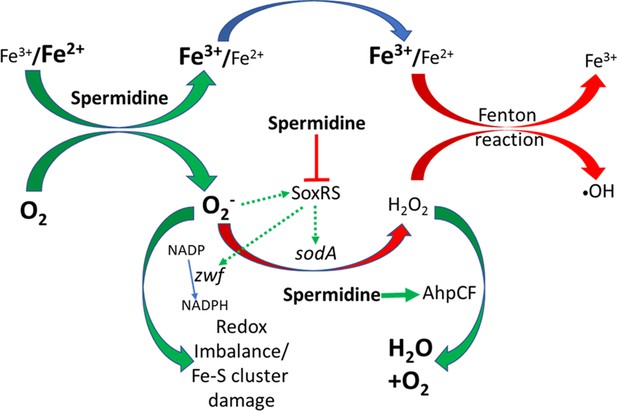
Flowchart explaining the reactive oxygen species (ROS) generation under spermidine stress.
The model describes that the spermidine administration in the cell interacts with free iron and oxygen to generate O2- radical, increasing Fe3+/Fe2+ ratio. Spermidine also blocks O2- radical-mediated activation of SoxRS that upregulates zwf and sodA. Consequently, reduced nicotinamide adenine dinucleotide phosphate (NADPH) production and dismutation of O2- radical to H2O2 were not accelerated, leading to redox imbalance and O2- -mediated damage to the iron-sulfur clusters, respectively. Additionally, spermidine translationally upregulated alkyl hydroperoxidase (AhpCF) that lowers the level of H2O2. Declined cellular Fe2+ and H2O2 levels weaken Fenton reaction to produce •OH radical.
Tables
The list of strains and plasmids used in this work.
| Strains and plasmids | Genotype/features | References |
|---|---|---|
| Strains | ||
| BW25113 | Escherichia coli; rrnB3 ΔlacZ4787 hsdR514Δ(araBAD) 567 Δ(rhaBAD)568 rph-1 | Baba et al., 2006 |
| ΔspeG | BW25113, ΔspeG::kanR | Baba et al., 2006 |
| ΔsodA | BW25113, ΔsodA::kanR | Baba et al., 2006 |
| ΔsodB | BW25113, ΔsodB::kanR | Baba et al., 2006 |
| Δzwf | BW25113, ΔsodC::kanR | Baba et al., 2006 |
| Δfis | BW25113, Δfis::kanR | Baba et al., 2006 |
| ΔihfA | BW25113, ΔihfA::kanR | Baba et al., 2006 |
| ΔiscU | BW25113, ΔiscU::kanR | Baba et al., 2006 |
| ΔygfZ | BW25113, ΔygfZ::kanR | Baba et al., 2006 |
| ΔsoxS | BW25113, ΔsoxS::kanR | Baba et al., 2006 |
| ΔmarA | BW25113, ΔmarA::kanR | Baba et al., 2006 |
| ΔmarB | BW25113, ΔmarB::kanR | Baba et al., 2006 |
| ΔahpC | BW25113, ΔahpC::kanR | Baba et al., 2006 |
| ΔkatG | BW25113, ΔkatG::kanR | Baba et al., 2006 |
| ΔspeGΔsodA | BW25113, ΔspeG, ΔsodA::kanR | This study |
| ΔspeGΔsodB | BW25113, ΔspeG, ΔsodB::kanR | This study |
| ΔspeGΔsodAΔsodB | BW25113, ΔspeG, ΔsodA, ΔsodB::kanR | This study |
| ΔspeGΔzwf | BW25113, ΔspeG, Δzwf::kanR | This study |
| ΔspeGΔsoxS | BW25113, ΔspeG, ΔsoxS::kanR | This study |
| ΔspeGΔfis | BW25113, ΔspeG, Δfis::kanR | This study |
| ΔspeGΔihfA | BW25113, ΔspeG, ΔihfA::kanR | This study |
| ΔspeGΔiscU | BW25113, ΔspeG, ΔiscU::kanR | This study |
| ΔspeGΔygfZ | BW25113, ΔspeG, ΔygfZ::kanR | This study |
| ΔspeGΔmarA | BW25113, ΔspeG, ΔmarA::kanR | This study |
| ΔspeGΔmarB | BW25113, ΔspeG, ΔmarB::kanR | This study |
| JRG3533 | MC4100 ф(sodA-lacZ)49, cmR | Tang et al., 2002 |
| RKM1 | BW25113, ΔspeG, sodA-lacZ:cmR | This study |
| Plasmids | ||
| pET28a (+) | kanR; T7-promoter; IPTG inducible | Novagen |
| pBAD/Myc-His A | ampR; pBAD-promoter; Ara inducible | ThermoFisher |
| pDAK1 | pBAD/Myc-His A; Two NdeI sites were mutated and NcoI site was replaced by NdeI | Lab resource |
| pZwf | zwf cloned in pDAK1 NdeI and HindIII sites | This study |
| pSodA | sodA cloned in pDAK1 vector | This study |
| pET-sodA | sodA cloned in pET28a (+) vector | This study |
| pET-ahpC | ahpC cloned in pET28a (+) vector | This study |
| pET-katG | katG cloned in pET28a (+) vector | This study |
| pSpeG | speG cloned in pDAK1 vector | This study |
| pUA66_soxS | kanR; soxS promoter cloned upstream of gfpmut2 reporter in pUA66 | Zaslaver et al., 2006 |
| pUA66_ahpC | kanR; ahpC promoter cloned upstream of gfpmut2 reporter in pUA66 | Zaslaver et al., 2006 |
| pUA66_katG | kanR; katG promoter cloned upstream of gfpmut2 reporter in pUA66 | Zaslaver et al., 2006 |
| Note: kanR, kanamycin resistance; ampR, ampicillin resistance, and cmR, chloramphenicol resistance. | ||
Additional files
-
Supplementary file 1
Table listing microarray data representing upregulated (green) and downregulated (red) genes.
- https://cdn.elifesciences.org/articles/77704/elife-77704-supp1-v3.xlsx
-
Supplementary file 2
Table listing the Fis- and IHF-regulated genes that were upregulated (green) and downregulated (red).
- https://cdn.elifesciences.org/articles/77704/elife-77704-supp2-v3.xlsx
-
Supplementary file 3
Table listing the oligonucleotide primers used in this study.
- https://cdn.elifesciences.org/articles/77704/elife-77704-supp3-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/77704/elife-77704-transrepform1-v3.docx