Antagonistic role of the BTB-zinc finger transcription factors Chinmo and Broad-Complex in the juvenile/pupal transition and in growth control
Figures
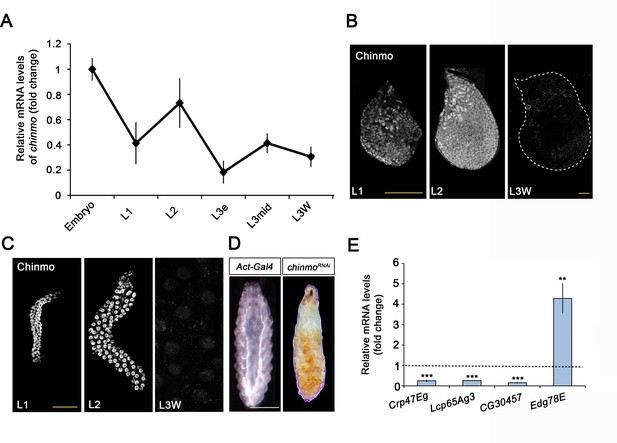
Chinmo is expressed during early larval stages and is essential for proper larval development.
(A) chinmo mRNA levels measured by quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) from embryo to the wandering stage of L3 (L3W). Transcript abundance values were normalised against the Rpl32 transcript. Fold changes were relative to the expression of embryo, arbitrarily set to 1. Error bars indicate the SEM (n = 3). (B–C) Chinmo protein levels in the wing disc (B) and salivary glands (C) of larval L1, L2, and L3W (females) stages. (D) Compared with the control (Act-Gal4), overexpression of UAS chinmoRNAi in the whole body induced developmental arrest at the L1 stage. Scale bars represent 50 µm (B and C) and 0.5 mm (D). (E) Relative expression of larval-specific (Crp47Eg, Lcp65Ag3, and CG30457) and pupal-specific genes (Edg78E) in UAS-chinmoRNAi L1 larvae measured by qRT-PCR. Transcript abundance values were normalised against the Rpl32 transcript. Fold changes were relative to the expression in control larvae, arbitrarily set to 1 (dashed black line). Error bars indicate the SEM (n = 3). Statistical significance was calculated using t test (***p≤0.001; **p≤0.005).
-
Figure 1—source data 1
Numerical data for Figure 1A and E.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig1-data1-v2.xlsx

Chinmo is required for proper growth and function of the salivary glands during larval development.
(A) DAPI staining of salivary glands from control (fkh-Gal4) and UAS-chinmoRNAi larvae at L3W. Scale bar represents 50 µm. (B–D) Comparison of the relative size of salivary glands (n = 10 for each genotype) (B), DAPI intensity (n = 50 for each genotype) (C), and nucleic size of salivary glands (n = 50 for each genotype) (D) between UAS-chinmoRNAi and control larvae at L3W. Error bars indicate the SEM (n = 5–8). (E) Relative expression of ng1-3 and Salivary glands secretion genes (Sgs) in UAS-chinmoRNAi L3W animals measured by quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR). Transcript abundance values were normalised against the Rpl32 transcript. Error bars indicate the SEM (n = 5–8). Statistical significance was calculated using t test (***p≤0.001).
-
Figure 2—source data 1
Numerical data for Figure 2B–E.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig2-data1-v2.xlsx
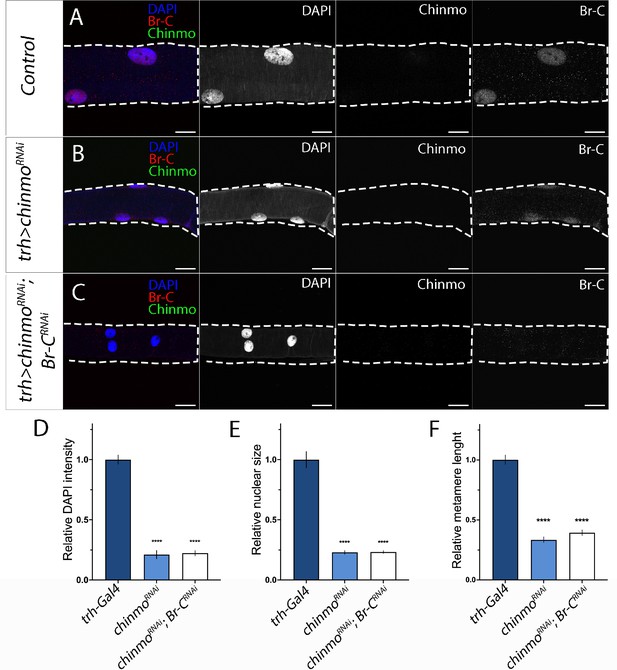
The role of Chinmo in the larval tracheal system.
(A–C) Chinmo (in green), Br-C (in red) and DAPI-stained trachea from control (trh-Gal4) (A), UAS-chinmoRNAi (B) and UAS-Br-CRNAi; UAS-chinmoRNAi (C) mid L3 larvae. Growth defect of tracheal cells induced by the absence of chinmo (B) was not rescued by Br-C depletion (C). (D–F) Comparison of the relative size of the trachea by measuring (n = 10 for each genotype) (D), DAPI intensity (E), and nucleic size of tracheal cells (F), and tracheal metamere length of control, UAS-chinmoRNAi and UAS-Br-CRNAi; UAS-chinmoRNAi mid L3 larvae. Error bars indicates the SEM (n = 5–8). Statistical significance was calculated using t test ( ****p≤0.001). Scale bars represent 50µm.
-
Figure 2—figure supplement 1—source data 1
Numerical data for Figure 2—figure supplement 1D-F.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig2-figsupp1-data1-v2.xlsx

Chinmo is necessary for wing development during the larval period.
Expression of Ct and Wg in wing discs of control (esg-Gal4) and UAS-chinmoRNAi L3W larvae. Wing discs were labelled to visualise the esg domain (GFP in green) and nuclei (DAPI). Ct and Wg were not detected in UAS-chinmoRNAi. Scale bars represent 50µm.
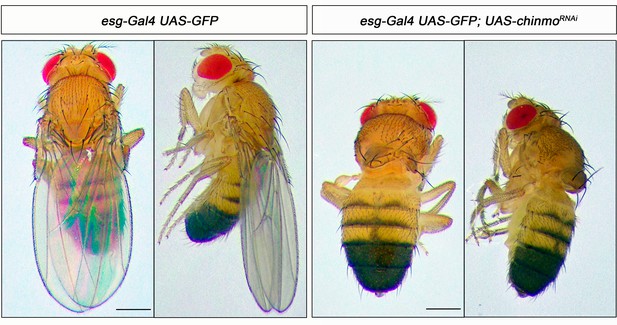
Chinmo is required for wing development during the larval period.
Dorsal and lateral view of control (right panel) and UAS-chinmoRNAi (left panel) adult flies. In the absence of Chinmo, flies emerged without wings. Scale bar represents 1 mm.
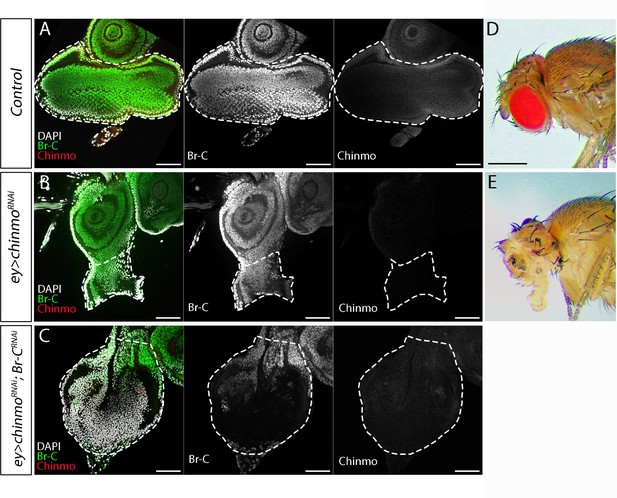
Br-C-dependent requirement of chinmo in the eye disc.
(A–C) Chinmo (in red), Br-C (in green), and DAPI (in grey) staining in eye discs of control (ey-Gal4) (A), UAS-chinmoRNAi (B), and UAS-Br-CRNAi; UAS-chinmoRNAi (C) L3W larvae. The dramatic eye disc reduction induced by the absence of chinmo (B) was rescued by Br-C depletion (C). (D–E) Lateral view of control (D) and UAS-chinmoRNAi (E) adult flies. In the absence of Chinmo, flies emerged without eyes. Scale bars represent 50 µm in A–B and 1 mm in D–E. Scale bars represent 50µmin A-C and 1mm in D-E .
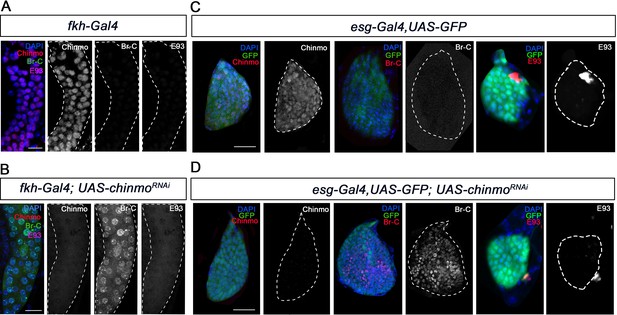
Chinmo represses Br-C in salivary glands and wing discs during early larval development.
(A–B) Expression of Chinmo, Br-C, and E93 in salivary glands of L1 control (fkh-Gal4) (A), and UAS-chinmoRNAi (B). (C–D) Expression of Chinmo, Br-C, and E93 in wing discs of early L2 control (esg-Gal4) (C) and UAS-chinmoRNAi (D). The esg domain is marked with GFP and all cell nucleus with DAPI. In the absence of chinmo only Br-C shows early upregulation in both tissues. Scale bars represent 25 µm.
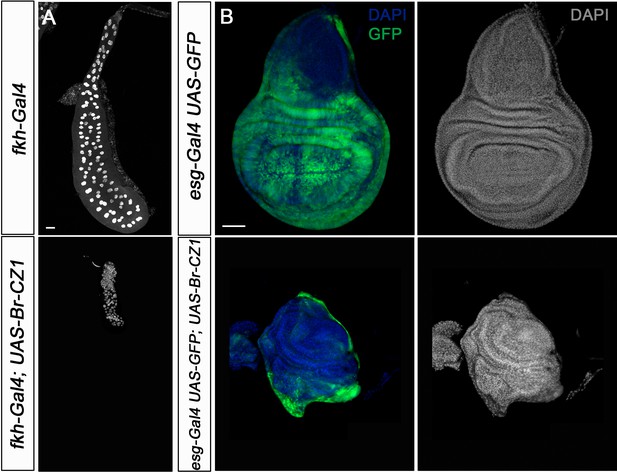
Overexpression of Br-CZ1 phenocopies chinmo loss of function in SGs and wing discs.
(A) DAPI-stained SGs from control (fkh-Gal4) and UASBr-CZ1 in L3W larvae. Overexpression of Br-CZ1 impairs SGs grow. (B) Wing discs of control (esg-Gal4) and UA-SBr-CZ1 L3W larvae. Wing discs were labelled to visualise the esg domain (GFP in green) and nuclei (DAPI). Overexpression of Br-CZ1 in the esg domain abolishes wing development. Scale bar represents 50 µm in all panels.
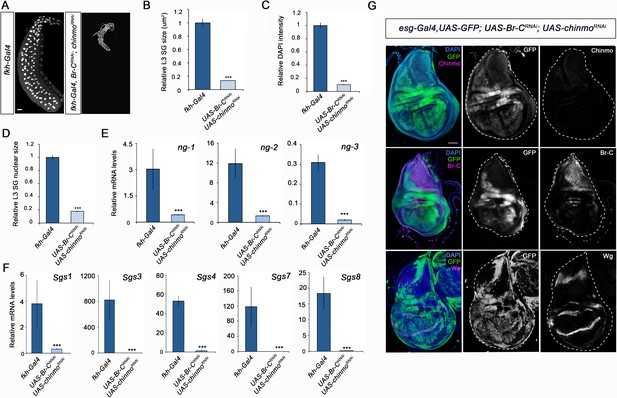
Different requirement of chinmo for the larval growth of salivary glands and wing discs.
(A) DAPI-stained salivary glands from control (fkh-Gal4) and UAS-Br-CRNAi; UAS-chinmoRNAi L3W larvae. In the absence of chinmo and Br-C, salivary glands did not grow. (B–D) Comparison of the relative size of salivary glands (n = 10 for each genotype) (B), DAPI intensity (n = 50 for each genotype) (C), and nucleic size of salivary glands (n = 30 for each genotype) (D) of control and UAS-Br-CRNAi; UAS-chinmoRNAi L3W larvae. (E–F) Relative expression of (E) ng1-3 and (F) Salivary glands secretion genes in control and UAS-Br-CRNAi; UAS-chinmoRNAi L3W larvae measured by quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR). Transcript abundance values were normalised against the Rpl32 transcript. Error bars in B and C indicate the SEM (n = 5–8). Statistical significance was calculated using t test ( ****p≤0.001). (G) Expression of Chinmo, Br-C, and Wg in wing discs of UAS-Br-CRNAi; UAS-chinmoRNAi L3W larvae. Wing discs labelled to visualise the esg domain (GFP in green). In the absence of chinmo and Br-C, wing discs grow normally and express Wg correctly. Scale bars represent 50 µm.
-
Figure 5—source data 1
Numerical data for Figure 5B–F.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig5-data1-v2.xlsx
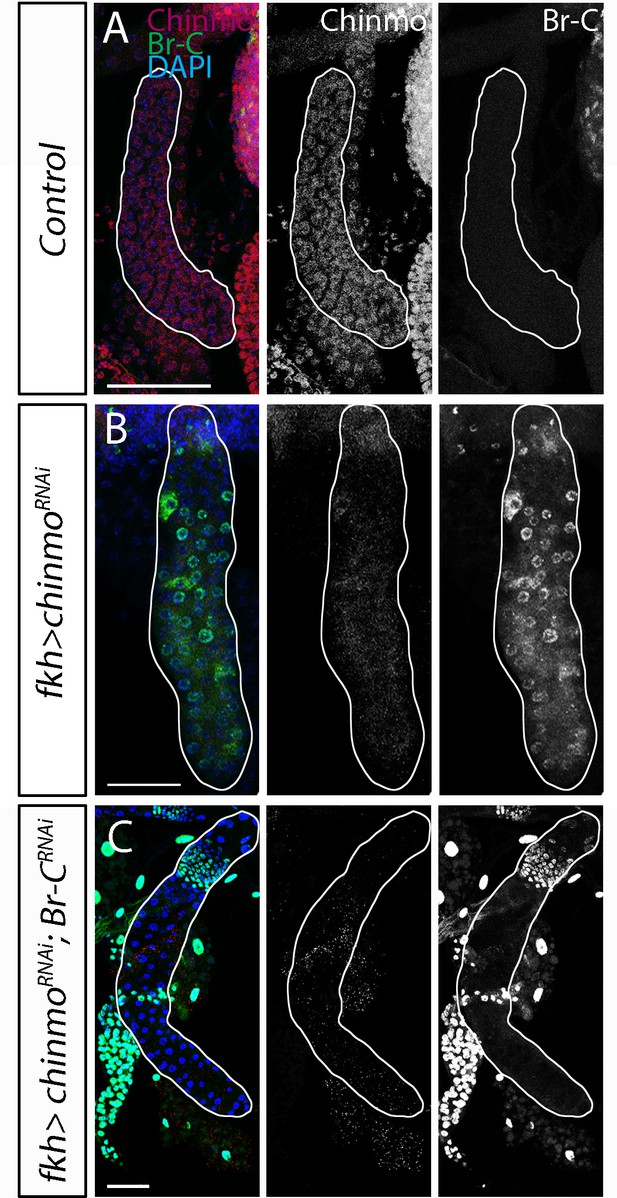
Effectiveness of chinmo and Br-C RNAis in the salivary glands.
(A–C) Expression of Chinmo (in red) and Br-C (in green) in SG of control (fkh-Gal4) (A) UAS-chinmoRNAi and (B) UAS-Br-CRNAi; UAS-chinmoRNAi (C) L2 larvae. Nuclei of SG cells are labelled with DAPI (in blue). Note that the (C) double knockdown of chinmo and Br-c completely abolished the expression of Br-C in the absence of Chinmo (C). Scale bars represent 50 µm.
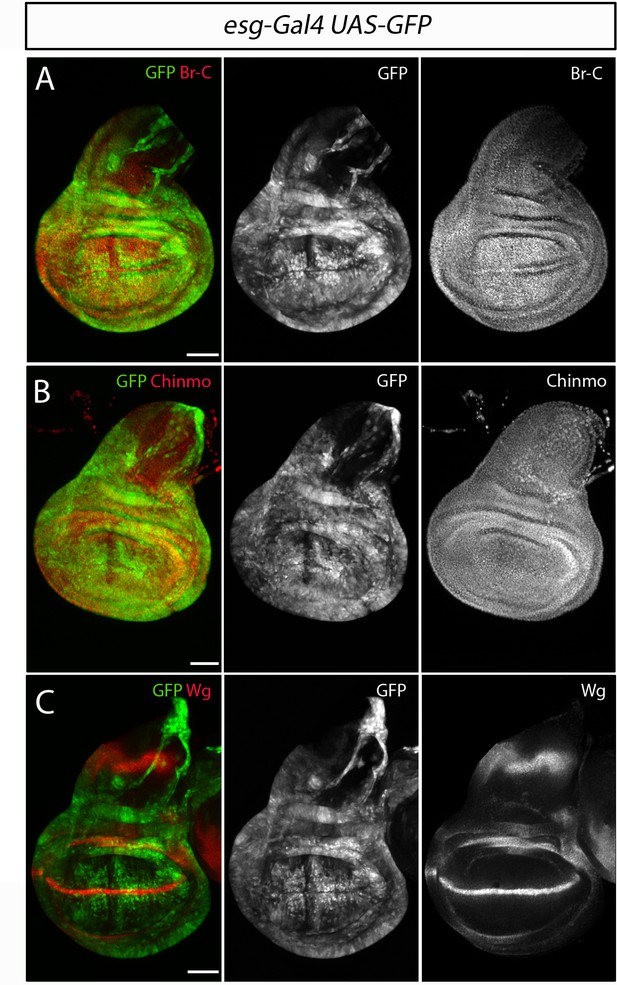
Expression of Chinmo and Br-C in the developing wing disc.
(A–C) Control wing discs, esg-Gal4; UAS-GFP, were labelled to visualise the esg domain in green and in red (A) Br-C expression in L3W larvae, (B) Chinmo expression in early-mid L3 larvae and (C) the morphogenetic marker Wg in L3W larvae. Scale bar represents 50 µm in all panels.
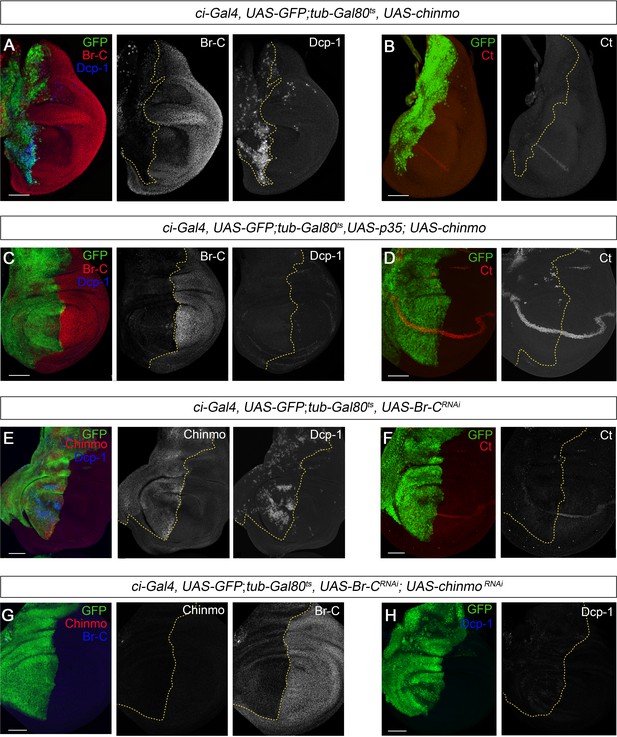
chinmo depletion during late L3 is required for proper larva to pupa transition.
(A–H) Images of wing imaginal discs from L3W larvae. The indicated constructs were expressed under the control of the ci-Gal4 driver. Overexpression or depletion of the transgenes was activated in early L3 larvae and analyzed at the L3W stage. An UAS-GFP construct was used to mark the anterior region of the disc where the transgenes were induced or repressed (green). (A) Overexpression of chinmo repressed Br-C, induced Dcp-1, and (B) abolished Ct. (C) Overexpression of chinmo together with p35 repressed Br-C and blocked Dcp-1, but fails to restore normal expression of Ct (D). (E) Depletion of Br-C induced Chinmo and Dcp-1 and (F) repressed Ct. (G) In double depletion of Br-C and chinmo (H), Dcp-1 was not detected. Scale bars represent 50 µm.
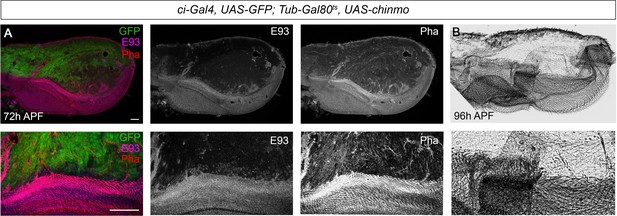
Presence of Chinmo during pupal development blocks adult differentiation.
(A) Overexpression of chinmo in the anterior part of the pupal wing at 72 hr after pupa formation (APF) using ci-Gal4 driver represses E93 expression and produced alterations in phalloidin (Pha) pattern. (B) Cuticle preparation of a pupal wing at 96 hr APF expressing chinmo under the control of the ci-Gal4 driver. Bottom panels are magnifications from upper images. The scale bars represent 50 μm (top panels) and 100 μm (bottom panels).
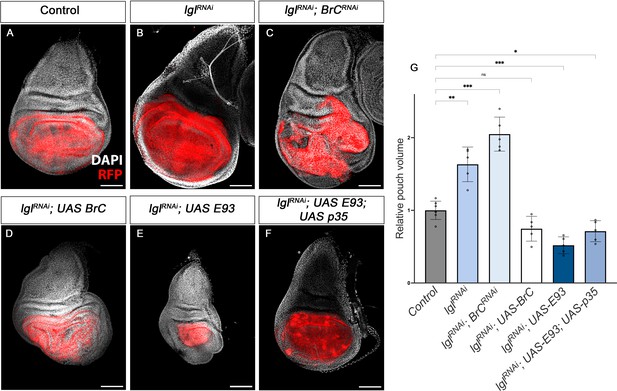
Tumour suppression action of Br-C and E93.
(A–F) Confocal images of L3 wing imaginal discs. The indicated constructs were expressed under the control of the nub-Gal4 driver. An UAS-RFP construct was used to mark the pouch region of the disc where the transgenes were induced (magenta). Nuclei were labelled with DAPI (grey). Scale bars at 100 μm. (G) Volumetric quantification of the RFP-positive area of the wing discs for the indicated groups. The pouch volumes were normalised to the mean of the control. Error bars in G indicate the SEM (n = 10). Welch’s ANOVA (p<0.0001) followed by Dunnett’s T3 post hoc tests (*p<0.05, **p<0.01, ***p<0.001).
-
Figure 8—source data 1
Numerical data for Figure 8G.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig8-data1-v2.xlsx
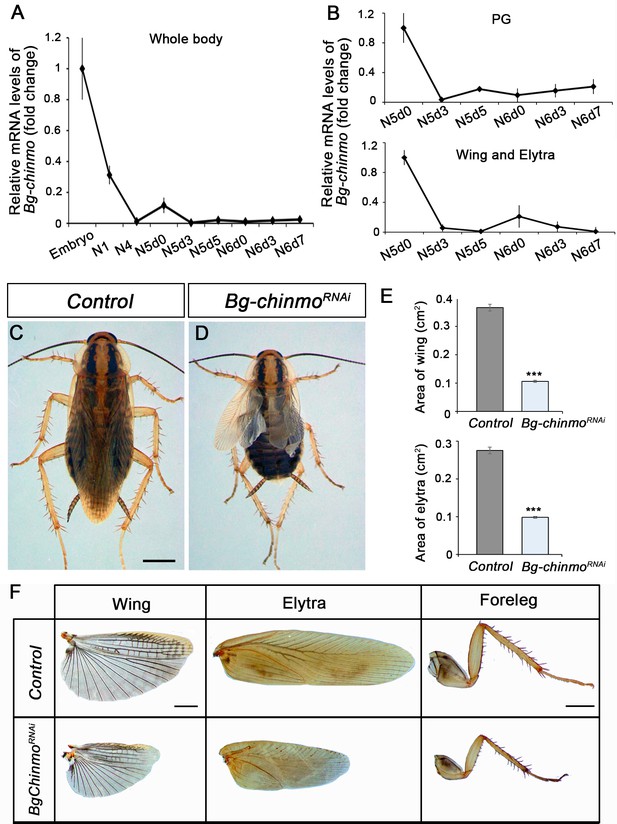
Depletion of chinmo in B. germanica promotes premature adulthood.
(A–B) Bg-chinmo mRNA levels measured by quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) from embryo to the last nymphal stage (N6) in whole body (A), and prothoracic gland (PG), and wings and elytra (B). Transcript abundance values were normalised against the Rpl32 transcript. Fold changes were relative to the expression of embryo (for whole body) or N5d0 (for PG and wings and elytra), arbitrarily set to 1. Error bars indicate the SEM (n = 3–5). (C–D) Newly moulted N4 nymphs of B. germanica were injected with dsMock (Control) or dschinmo (Bg-chinmoRNAi) and left until adulthood. (C) Dorsal view of adult Control, and (B) premature adult Bg-chinmoRNAi. (E) Quantification of wing and elytra areas (cm2) of adult Control and Bg-chinmoRNAi premature adults. Error bars indicate the SEM (n = 4–6). Statistical significance was calculated using t-test (***p≤0.001). (F) Control and Bg-chinmoRNAi wing, elytra and foreleg of newly emerged adult of B. germanica. The scale bar represents 2 mm.
-
Figure 9—source data 1
Numerical data for Figure 9A and D.
- https://cdn.elifesciences.org/articles/84648/elife-84648-fig9-data1-v2.xlsx





