RNA fusion in human retinal development
Figures
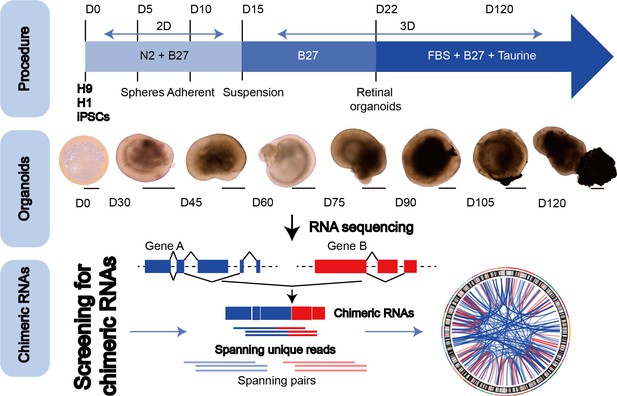
The general scheme of detection of chimeric RNAs.
(Top panel) Procedure for the generation of human retinal organoids (ROs) from pluripotent stem cells. (Middle panel) D0–120 human ROs were used for bulk RNA sequencing. Scale bars: 200 μm (D0) and 400 μm (D30–D120). (Bottom panel) Illustration of chimeric RNA screening process and criteria by FusionCatcher.
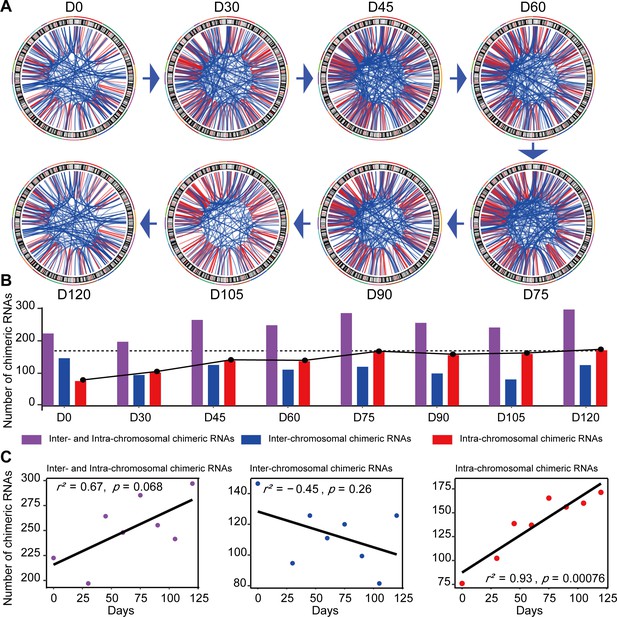
Expression of chimeric RNAs in the developing human retinal organoids.
(A) Circos plots of genomic distribution of chimeric RNA parental genes observed in this work. Red lines indicate parental genes located in the same chromosome. Blue lines indicate parental genes located in different chromosomes. The outermost colored lines of circos plots represent chromosomes. (B) Types of chimeric RNAs based on parental genes’ genomic distribution. (C) Pearson’s correlation analysis of the number of chimeric RNAs and developmental stages.
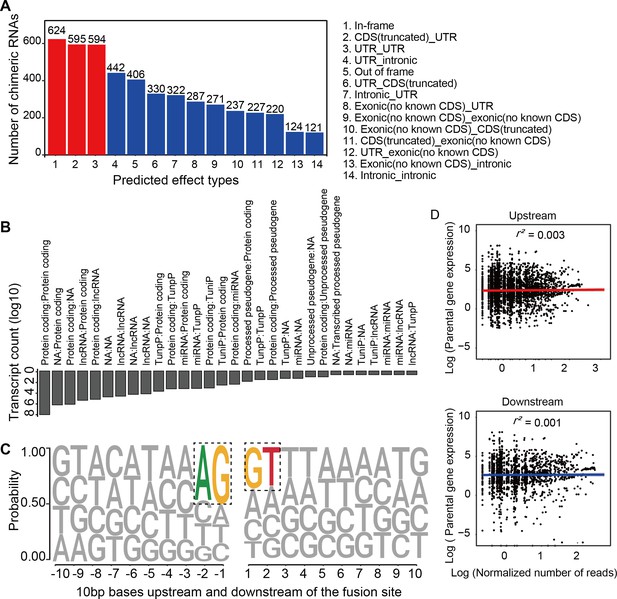
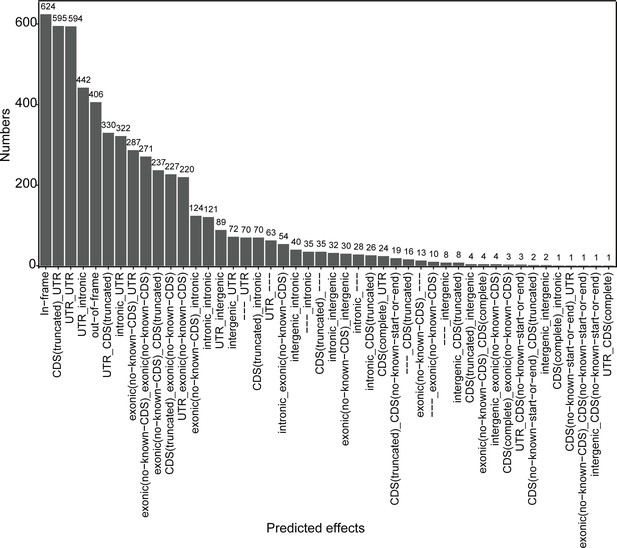
Characterization of chimeric RNAs in the developing human retinal organoids.
(A) Top 14 types and corresponding numbers of chimeric RNAs based on predicted effects. (B) Biotype quantification of parental gene combinations in all samples. TunpP: transcribed unprocessed pseudogene; TuniP: transcribed unitary pseudogene. (C) Motifs consisting of 20 bp DNA sequences around the fusion site. (D) Spearman correlation analysis of expression level of chimeric RNAs and their parental genes, p > 0.05.
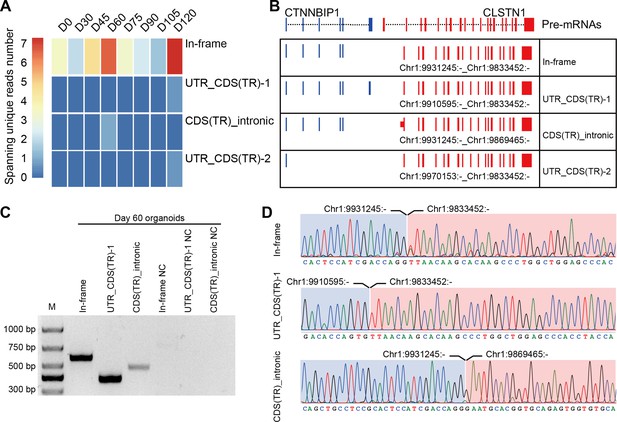
Four isoforms of CTCL are present in the retinal organoids (ROs).
(A) Heatmap of CTCL’s spanning unique reads of each isoform in the indicated stages. UTR_CDS(TR)-1: UTR_CDS(truncated)-1; CDS(TR)Intronic: CDS(truncated)Intronic; UTR_CDS(TR)-2: UTR_CDS(truncated)-2. (B) Schematic diagram of the structures of the four isoforms of CTCL. Blue represents upstream parental gene, red represents downstream parental gene. (C) Four isoforms of CTCL in D60 ROs were validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR). (D) Sanger sequencing to verify three isoforms of CTCL.
-
Figure 4—source data 1
Source data for Figure 4C.
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig4-data1-v3.pdf
-
Figure 4—source data 2
Source data for Figure 4C (labelled).
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig4-data2-v3.pdf
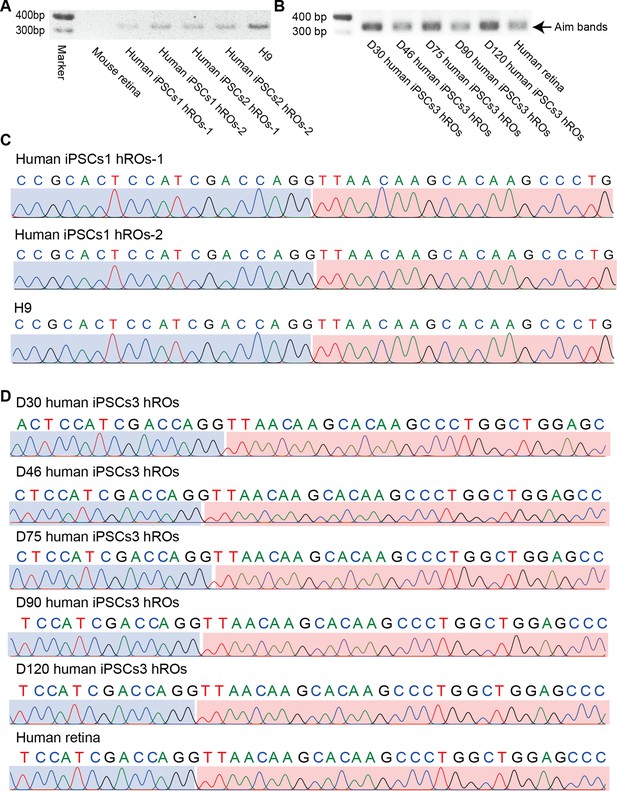
In-frame CTCL is common in human retinas and retinal organoids (ROs).
(A) The qRT-PCR validation of the In-frame CTCL in the mouse retina, iPSCs1/iPSCs2-derived ROs, and H9 cells. (B) The qRT-PCR validation of In-frame CTCL in the iPSCs3-derived ROs and human retina. (C) Verification of the In-frame CTCL in the mouse retina, iPSCs1/iPSCs2-derived ROs and H9 cells by Sanger sequencing. (D) Verification of the In-frame CTCL in the iPSCs3-derived ROs and human retinas by Sanger sequencing.
-
Figure 5—source data 1
Source data for Figure 5A.
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig5-data1-v3.pdf
-
Figure 5—source data 2
Source data for Figure 5A (labelled).
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig5-data2-v3.pdf
-
Figure 5—source data 3
Source data for Figure 5B.
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig5-data3-v3.pdf
-
Figure 5—source data 4
Source data for Figure 5B (labelled).
- https://cdn.elifesciences.org/articles/92523/elife-92523-fig5-data4-v3.pdf
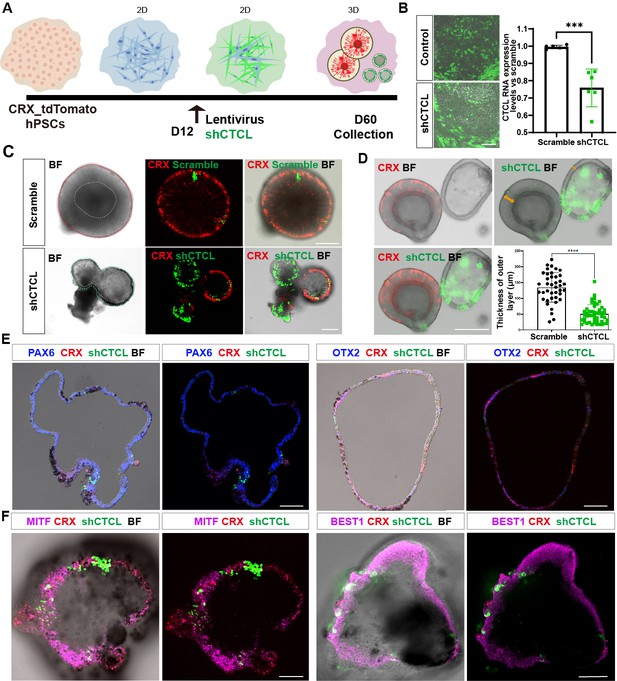
The CTCL knockdown obstructed neural retina’s (NR) cell fates but prompted the retinal pigment epithelial (RPE) differentiation.
(A) A schema illustrated the shRNA experiments. A CRX-tdTomato report line was used in this experiment. The shCTCL or scramble shRNA lentivirus transfected the retinal cells in three independent experiments on D12. All samples for shRNA experiments were collected on day 60, except those for analysis of knockdown efficiency. (B) Samples were obtained 48–72 hr after infection to examine the knockdown efficiency. There were two technical replicates for three independent experiments, t-test, ***, p < 0.001. Data were shown as mean ± standard deviation (SD). Scale bar = 200 μm. (C) D60 scramble shRNA-transfected retinal organoids (ROs) displayed a typical morphology of ROs with the expression of CRX. ShCTCL-transfected organoids showed thinner outer layers with much less CRX expression. BF, bright-field images, same as below.Scale bars = 200 μm. (D) The shCTCL group displayed much thinner outer layers (orange arrows in right panel). The outer layer thickness of 25 organoids from each group was measured for statistics analysis, t-test, ****, p < 0.0001. Data were shown as mean ± SD. Scale bar = 400 μm.(E) The section immunostaining of neural progenitor markers PAX6 and OTX2, and differentiated cell markers CRX, in shCTCL-treated organoids. Scale bars = 100 μm. (F) Whole-mount immunostaining of shCTCL-transfected organoids with CRX, shCTCL, and RPE-specific markers MITF and BEST1. Scale bars = 100 μm.
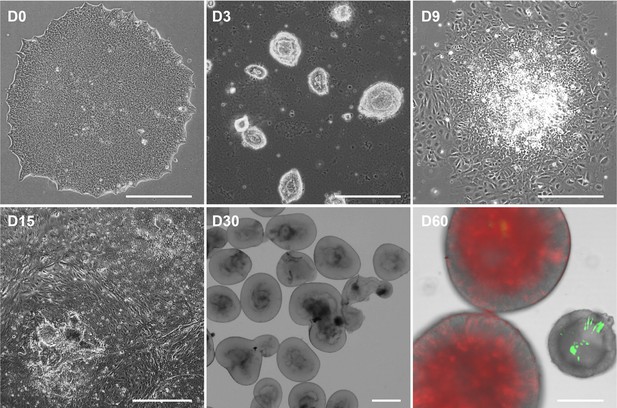
Retinal organoid (RO) differentiation at different timepoints.
This RO differentiation method was a combination of 2D and 3D culture. On D60, CTCL downregulated organoids displayed a thinner outer layer. Live imaging showed that D60 organoids expressed CRX reporter tdTomato (red) and shCTCL reporter GFP (green). Scale bars = 400 μm.
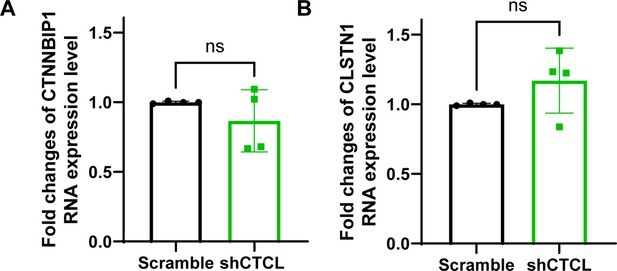
The qRT-PCR quantification of CTCL parental gene expression.
(A) The knockdown experiments showed no effect on the RNA expression levels of CTNNBIP1, n = 4, t-test, p > 0.05. (B) The knockdown experiments showed no effect on the RNA expression levels of CLSTN1, n = 4, t-test, p > 0.05.
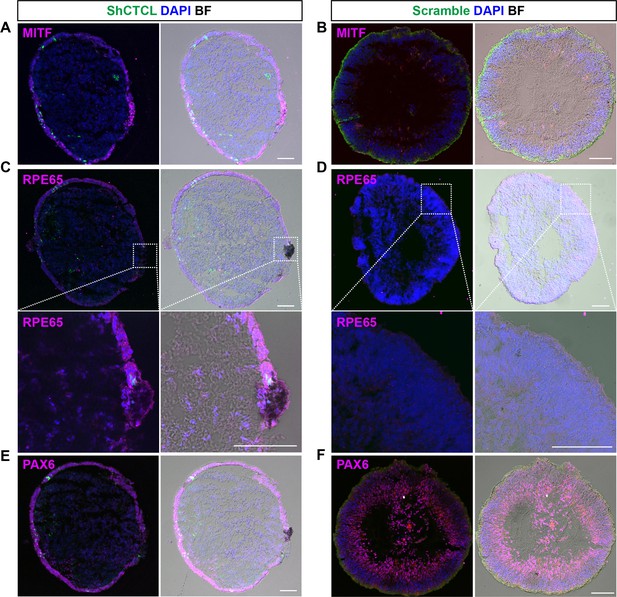
Cryosection immunofluorescence staining of D60 organoids.
Immunostaining of retinal pigment epithelial (RPE)-specific marker MITF on CTCL-knockdown organoids (A) and scramble shRNA organoids (B). Immunostaining of RPE-specific marker RPE65 on CTCL-knockdown organoids (C) and scramble shRNA organoids (D). The lower panel showed the amplified images of the indicated areas. Immunostaining of PAX6 on CTCL-knockdown organoids (E) and scramble shRNA organoids (F). Scale bars = 100 μm. BF, bright-field images.
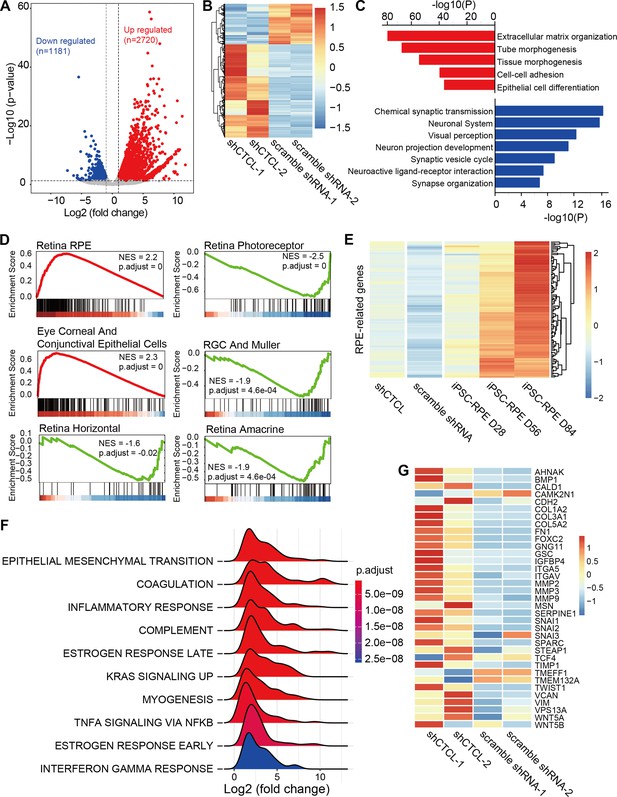
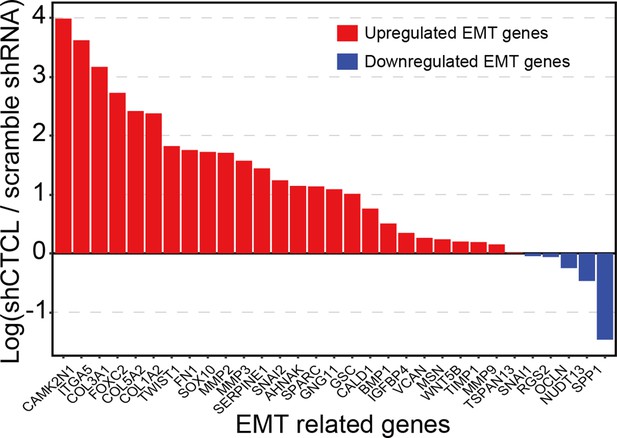
Transcriptomic alterations in CTCL-knockdown retinal organoids.
(A) Volcano plot shows differentially expressed genes (DEGs, p-values <0.05 and |Log2(shCTCL/scramble)| > 1) between shCTCL- and scramble shRNA-treated organoids at D60. (B) Heatmap of DEGs in shCTCL- and scramble shRNA-treated organoids. (C) Functional enrichment analysis of up- and downregulated DEGs. (D) Gene Set Enrichment Analysis (GSEA) results showed the enriched gene sets in shCTCL-treated organoids using the c8 reference gene set. (E) Expression of retinal pigment epithelial (RPE)-related genes in shCTCL- and scramble shRNA-treated organoids. (F) GSEA results showed the enriched gene sets in shCTCL-treated organoids using the hallmark reference gene set. (G) Expression of epithelial–mesenchymal transition (EMT)-related genes in shCTCL- and scramble shRNA-treated organoids.

Expression of CTCL is lower in differentiated retinal pigment epithelial (RPE) cells.
(A) Bright-filed image of mature RPE differentiated from iPSCs. (B) The RT-qPCR results of CTCL RNA expression levels in D60 CTCL-knockdown organoids and the differentiated RPEs at the indicated timepoints. It showed that CTCL was expressed at a lower level in RPEs, even compared with that in the shCTCL-treated retinal organoids, n = 4, t-test, ****, p < 0.0001.
Videos
The 4 days live-cell imaging of retinal organoid differentiation from D3 to D7.
Tables
CTCL is detected in multiple healthy human retinas by RNA-seq.
| Data source | Samples | Gene 1 symbol | Gene 2 symbol | Fusion point for gene 1 | Fusion point for gene 2 |
|---|---|---|---|---|---|
| PMID: 30874468 (Sun et al., 2019) Data published by our lab | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID: 30874468 (Sun et al., 2019) Data published by our lab | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315404 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315407 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315408 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315412 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315419 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315420 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| PMID:35784369 (Zauhar et al., 2022) SRR22315424 | Human retina | CTNNBIP1 | CLSTN1 | 1:9871187:- | 1:9773394:- |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | CRX-tdTomato human ES reporter line-H9 | PMID:32831148 | Available upon reasonable request. | |
| Cell line (Homo sapiens) | iPSCs1 | PMID:36714839 | Available upon reasonable request. | |
| Cell line (Homo sapiens) | iPSCs2 | PMID:35451725 | Available upon reasonable request. | |
| Cell line (Homo sapiens) | iPSCs3 | PMID:33970142 | Available upon reasonable request. | |
| Antibody | anti-Pax6 (rabbit polyclonal) | Biolegend | Cat.# 862002 RRID: AB_3076431 | Dilution: 1:200 |
| Antibody | anti-Ki67 (rabbit polyclonal) | Abcam | Cat.# ab15580 RRID:AB_443209 | Dilution: 1:200 |
| Antibody | anti-OTX2 (rabbit monoclonal) | Abcam | Cat.# ab183951 RRID: AB_3076432 | Dilution: 1:200 |
| Antibody | anti-HuC/D (mouse monoclonal) | Invitrogen | Cat.# A21271 RRID:AB_221448 | Dilution: 1:100 |
| Antibody | anti-SOX2 (mouse monoclonal) | Santa Cruz | Cat.# sc-365823 RRID:AB_10842165 | Dilution: 1:200 |
| Antibody | anti-MITF (mouse monoclonal) | Abcam | Cat.# ab3201 RRID:AB_303601 | Dilution: 1:100 |
| Antibody | anti-GFAP (mouse monoclonal) | Santa Cruz | Cat.# sc-33673 RRID:AB_627673 | Dilution: 1:200 |
| Antibody | anti-Sox9 (rabbit mono/oligo-colonal ) | Invitrogen | Cat.# 711048 RRID:AB_2633109 | Dilution: 1:200 |
| Antibody | anti- RxRγ (mouse monoclonal) | Santa Cruz | Cat.# sc-365252 RRID:AB_10850062 | Dilution: 1:100 |
| Recombinant DNA reagent | pLenti-U6-shRNA-EF1a-EGFP-T2A-Puro-WPRE | This paper | Available upon reasonable request. | |
| Sequence-based reagent | UTR_CDS(truncated)-1 CTCL-forward | Tsingke Biotechnology Co., Ltd | TGCAAAGCCCTTGGAACA | |
| Sequence-based reagent | UTR_CDS(truncated)-1 CTCL-reverse | Tsingke Biotechnology Co., Ltd | TCCACTACCACTGCATCAAAG | |
| Sequence-based reagent | in-frame CTCL-forward | Tsingke Biotechnology Co., Ltd | TTCCTACTTCTGCCCAGCC | |
| Sequence-based reagent | in-frame CTCL-reverse | Tsingke Biotechnology Co., Ltd | AGGCCTGGATGGTGAATGAAT | |
| Sequence-based reagent | CDS(truncated)_intronic CTCL-forward | Tsingke Biotechnology Co., Ltd | GAAGAGTCCGGAGGAGATGTA | |
| Sequence-based reagent | CDS(truncated)_intronic CTCL-reverse | Tsingke Biotechnology Co., Ltd | ATTCTCTGCCAAGACTTACACC | |
| Sequence-based reagent | UTR_CDS(truncated)-2 CTCL-forward | Tsingke Biotechnology Co., Ltd | CTCCTGCTGCTGCTACTG | |
| Sequence-based reagent | UTR_CDS(truncated)-2 CTCL-reverse | Tsingke Biotechnology Co., Ltd | GGTCCCTTCCCACAATCATAG | |
| Sequence-based reagent | GAPDH forward | Tsingke Biotechnology Co., Ltd | CTCTGACTTCAACAGCGACA | |
| Sequence-based reagent | GAPDH reverse | Tsingke Biotechnology Co., Ltd | GTAGCCAAATTCGTTGTCATACC | |
| Sequence-based reagent | ShCTCL | Tsingke Biotechnology Co., Ltd | TGCTTGTTAACCTGGTCGA | |
| Sequence-based reagent | Scramble | Tsingke Biotechnology Co, Ltd | GCCTAAGGTTAAGTCGCCCTCG | |
| Software, algorithm | R (v4.0.3) | R Development Core Team, 2020 | RRID:SCR_001905 | https://www.r-project.org/ |
| Software, algorithm | FusionCatcher | Nicorici, 2023 | https://github.com/ndaniel/fusioncatcher | |
| Software, algorithm | Hisat2 | Kim and Park, 2022 | https://github.com/DaehwanKimLab/hisat2 | |
| Software, algorithm | samtools | Danecek et al., 2023 | https://github.com/samtools/samtools | |
| Software, algorithm | featureCounts | Liao et al., 2021 | http://subread.sourceforge.net/featureCounts.html | |
| Software, algorithm | ggplot2 | Pedersen, 2022 | https://rdocumentation.org/packages/ggplot2/versions/3.3.6 | |
| Software, algorithm | ggseqlogo | Wagih, 2017 | https://github.com/omarwagih/ggseqlogo | |
| Software, algorithm | chimeraviz | Lågstad, 2023 | https://www.bioconductor.org/packages/release/bioc/html/chimeraviz.html | |
| Software, algorithm | pheatmap | Kolde, 2019 | https://rdocumentation.org/search?q=pheatmap | |
| Software, algorithm | clusterProfiler | Yu, 2012 | https://rdocumentation.org/search?q=clusterProfiler |