The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction
Figures
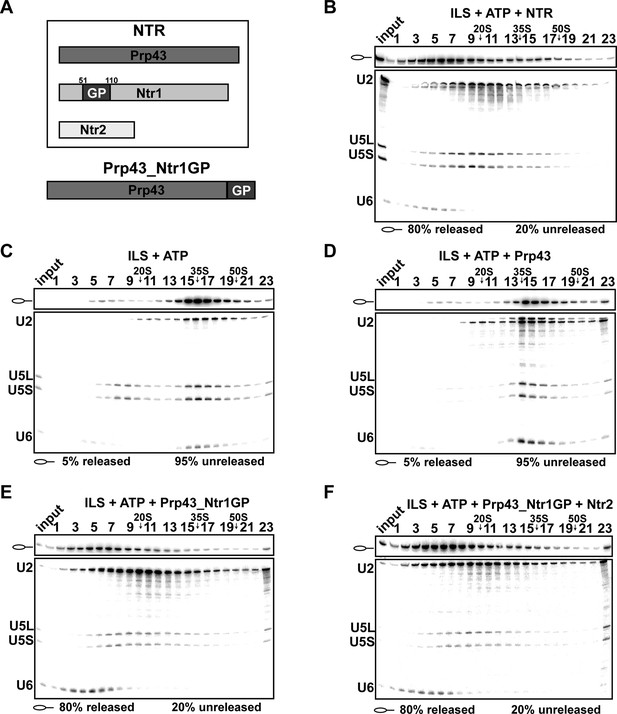
Prp43 fused to the G-patch motif of Ntr1 is sufficient to promote dissociation of the ILS.
(A) Upper panel, schematic representation of the NTR complex, composed of Prp43, Ntr1 and Ntr2; lower panel, Prp43_Ntr1GP in which the G-patch of Ntr1 is fused to the C-terminal domain of Prp43. (B) 10–30% glycerol gradient sedimentation of purified ILS incubated in solution with ATP plus NTR, (C) no recombinant protein, (D) Prp43 (E) Prp43 fused to Ntr1GP (Prp43_Ntr1GP), or (F) Prp43_Ntr1GP and Ntr2. U2, U5 and U6 snRNAs were visualized by Northern blotting followed by autoradiography. RNA identities are indicated on the left. Quantifications were performed with ImageQuant software (Molecular Dynamics, Pittsburg, PA). Numbers represent the percentage of intron-lariat RNA released in the top fractions (sum of fractions 1–11) or associated with the ILS (unreleased, sum of fractions 12–23) relative to the intron-lariat RNA distributed in all 23 fractions, the sum of which was set to 100%.
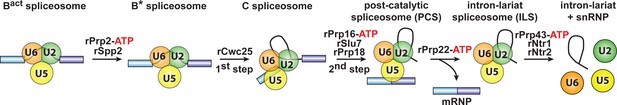
Isolation of intron-lariat spliceosomes (ILSs).
We first purified activated spliceosomes (Bact ΔPrp2), which were assembled on wild-type actin7 pre-mRNA in heat-inactivated splicing extracts from a prp2-1 yeast strain expressing a temperature-sensitive Prp2 mutant. Purified Bact ΔPrp2 complexes were then incubated with recombinant Prp2 and Spp2, generating the B* spliceosome, and then Cwc25 was added to promote catalysis of step 1 of splicing and the formation of complex C. For catalysis of step 2, which generates post-catalytic spliceosomes (PCS), recombinant Prp16, Slu7 and Prp18 were added. Finally, for the purification of the ILS, the spliced mRNA was dissociated from the ILS by incubation of the PCS with Prp22 and ATP. Addition of ATP and recombinant Prp43, Ntr1 and Ntr2, leads to disassembly of the ILS into the intron-lariat, 20S U2 snRNP, 18S U5 snRNP and free U6 snRNA (Fourmann et al., 2013).
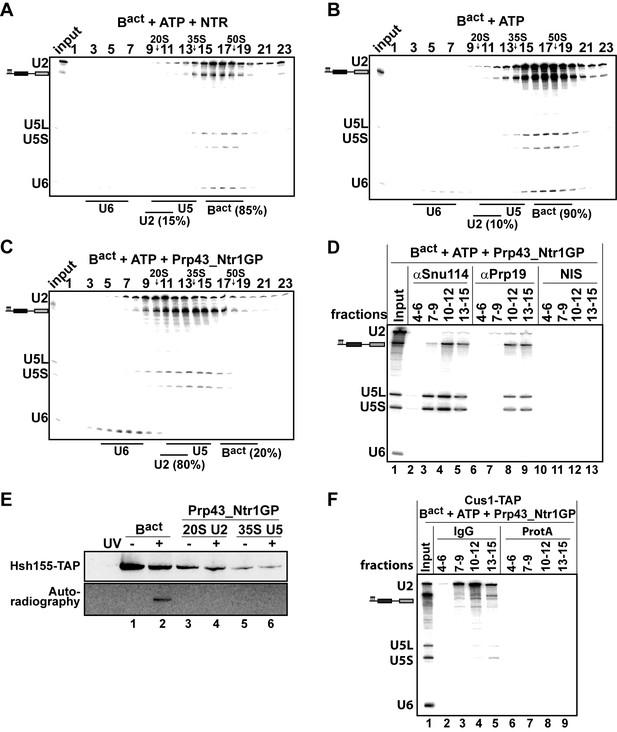
Prp43_Ntr1GP but not NTR disassembles Bact complexes formed on wild-type actin7 pre-mRNA.
10–30% glycerol gradient sedimentation of purified Bact ΔPrp2 complexes incubated with ATP and (A) NTR, (B) no recombinant protein, or (C) Prp43_Ntr1GP. Samples were analyzed as described in Figure 1. Quantifications were performed with ImageQuant software (Molecular Dynamics). Numbers represent the percentage of U2 snRNA released in the top fractions (sum of fractions 1–11) or associated with the Bact ΔPrp2 complex (unreleased, sum of fractions 12–23) relative to the U2 snRNA distributed in all 23 fractions, the sum of which was set to 100%. (D) Bact ΔPrp2 spliceosomes were incubated with ATP and Prp43_Ntr1GP. Five% of the input was withdrawn (lane 1). Samples were separated on 10–30% glycerol gradients. Every three fractions from 4 to 15 (4–6, 7–9, etc.) were combined and immunoprecipitated with anti-Snu114, anti-Prp19 antibodies or with non-immune serum (NIS). Co-precipitated RNAs were analysed by Northern blotting using probes that hybridize to U2, U5 and U6 RNA. (E) Bact ΔPrp2 spliceosomes harboring Hsh155-TAP were assembled on uniformly radiolabeled pre-mRNA. One half was left untreated and the other half was incubated with Prp43_Ntr1GP and ATP, and then analysed on separate glycerol gradients. Peak fractions corresponding to intact Bact ΔPrp2 (15–18), 20S U2 (9–11) and 35S U5 (13–15) were UV-irradiated or untreated. Proteins and protein-RNA crosslinks were separated by SDS-PAGE and transferred to a membrane. Western blot analysis was performed using PAP complex antibodies. Protein crosslinked to radiolabeled pre-mRNA nucleotides was visualized by autoradiography of the same membrane. (F) Bact ΔPrp2 complexes containing the U2 protein Cus1 tagged with the TAP tag (Cus1-TAP) were incubated with ATP and Prp43_Ntr1GP and separated on a glycerol gradient. The indicated three gradient fractions were combined and immunoprecipitated with IgG Sepharose beads (IgG) allowing the selective immunoprecipitation of 20S U2 snRNP (lanes 3,4). The same fractions were also precipitated with Protein A–Sepharose beads (ProtA) (lanes 6–9). Co-precipitated snRNAs were analyzed on an 8% urea-polyacrylamide gel and identified by Northern blotting as above.
-
Figure 2—source data 1
Protein composition of Bact ΔPrp2 complexes disassembled by Prp43_Ntr1GP and ATP.
Every two fractions from the glycerol gradient shown in Figure 2C were pooled, and proteins were recovered and separated by PAGE. Proteins were cut out of the gel, digested in-gel with trypsin and extracted. The extracted peptides were analyzed in a liquid-chromatography-coupled electrospray ionization orbitrap mass spectrometer (LTQ Orbitrap XL) under standard conditions. Proteins were identified by searching fragment spectra against the yeast S. cerevisiae Genomic Database (SGD), using Mascot as a search engine. Numbers represent the absolute number of peptides sequenced for a protein found in a particular preparation. The table contains information about the S. cerevisiae protein and the calculated molecular weight in kilodaltons. Proteins are grouped in organizational and/or functional subgroups. The peaks in the gradient of the U2 and 35S U5 snRNPs and their associated proteins are marked in green and orange, respectively.
- https://doi.org/10.7554/eLife.15564.005
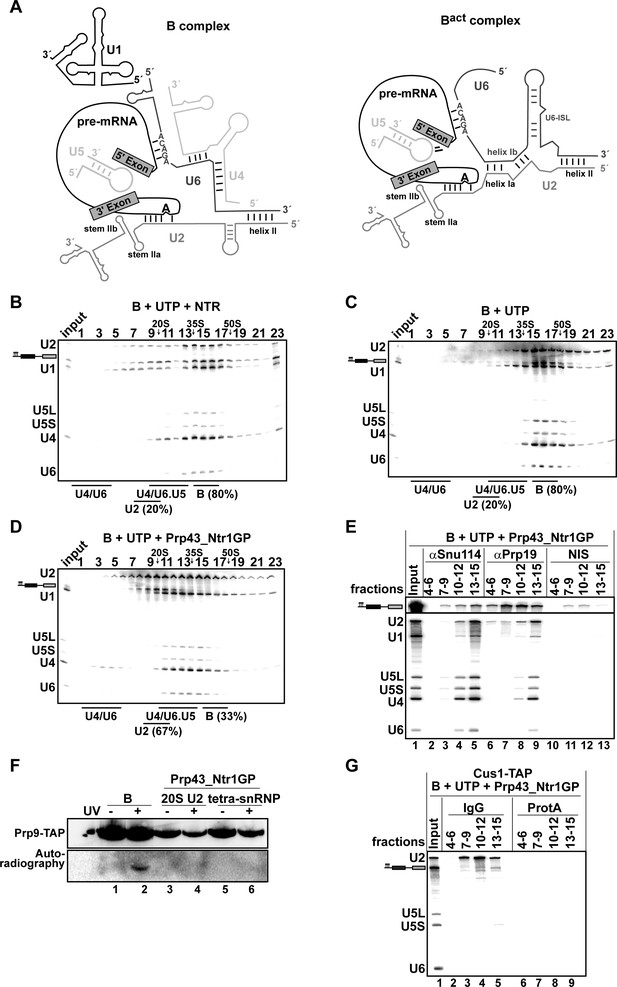
Prp43_Ntr1GP disassembles B complexes.
(A) Scheme of the RNA network in B and Bact complexes (Madhani and Guthrie, 1992). The U6 ACAGA box and U2/U6 helixes I and II are indicated. 10–30% glycerol gradient sedimentation of purified B complexes incubated with UTP and (B) NTR or (C) no recombinant protein or (D) Prp43_Ntr1GP. Samples were analyzed as described in Figure 1. Quantification of U2 snRNA released or associated with the B complex (unreleased) was performed as described in Figure 2. (E) B complexes were incubated with UTP and Prp43_Ntr1GP and separated on a glycerol gradient. The indicated three gradient fractions were combined and immunoprecipitated with anti-Snu114, anti-Prp19 antibodies or with non-immune serum (NIS). Co-precipitated snRNAs were analyzed on an 8% urea-polyacrylamide gel and identified by Northern blotting using probes that hybridize to U2, U1, U5, U4 and U6 RNA (E) B complexes harboring Prp9-TAP were assembled on uniformly radiolabeled pre-mRNA. One half was left untreated and the other half was disassembled with Prp43_Ntr1GP and UTP, and then analysed on separate glycerol gradients. Peak fractions corresponding to intact B complex (15–18), 20S U2 (9–11) and tetra-snRNP (13–15) were UV-irradiated or untreated. Proteins and protein-RNA crosslinks were analysed as in Figure 2. (G) B complexes containing the U2 protein Cus1 tagged with the TAP tag (Cus1-TAP) were incubated with UTP and Prp43_Ntr1GP and separated on a glycerol gradient. The indicated three gradient fractions were combined and immunoprecipitated with IgG Sepharose beads (IgG, lanes 2–5) allowing the selective immunoprecipitation of 20S U2 snRNP (lanes 3,4) or tetra-snRNP (lane 5). The same fractions were also precipitated with Protein A–Sepharose beads (ProtA) (lanes 6–9). Co-precipitated snRNAs were analyzed as above and identified by Northern blotting using probes that hybridize to U2, U5 and U6 RNA.
-
Figure 3—source data 1
Protein composition of B complexes disassembled by Prp43_Ntr1GP and UTP.
Every three fractions from the glycerol gradient shown in Figure 3B were pooled, and proteins were recovered and separated by PAGE. Proteins were identified by mass spectrometry as above. Numbers represent the absolute number of peptides sequenced for a protein found in a particular preparation. The table contains information about the Saccharomyces cerevisiae protein and the calculated molecular weight in kilodaltons. Proteins are grouped in organizational and/or functional subgroups. The peaks in the gradient of the U2, U1 and tetra snRNPs and their associated proteins are marked in green, blue and yellow, respectively.
- https://doi.org/10.7554/eLife.15564.007
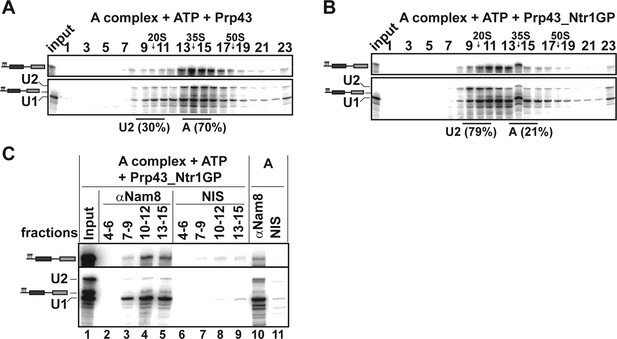
Prp43_Ntr1GP disassembles A complexes.
10–30% glycerol gradient sedimentation of purified A complexes incubated with ATP and (A) Prp43 or (B) Prp43_Ntr1GP. Samples were analyzed as described in Figure 1 and quantifications were performed as described in Figure 2. (C) A complexes were incubated with ATP and Prp43_Ntr1GP. Five% of the input was withdrawn (lane 1). Samples were separated on 10–30% glycerol gradients. Every three fractions from 4 to 15 (4–6, 7–9, etc.; panel b) were combined and immunoprecipitated with anti-Nam8 antibodies or with non-immune serum (NIS). In lanes 10–11, immunoprecipitations were performed with purified A complexes.
-
Figure 4—source data 1
Protein composition of A complexes disassembled by Prp43_Ntr1GP and ATP.
Every three fractions from the glycerol gradient shown in Figure 4B were pooled, and proteins were recovered and separated by PAGE. Proteins were identified by mass spectrometry as above. Numbers represent the absolute number of peptides sequenced for a protein found in a particular preparation. The table contains information about the Saccharomyces cerevisiae protein and the calculated molecular weight in kilodaltons. Proteins are grouped in organizational and/or functional subgroups. The peaks in the gradient of the U2 and U1 snRNPs and their associated proteins are marked in green and blue, respectively.
- https://doi.org/10.7554/eLife.15564.009
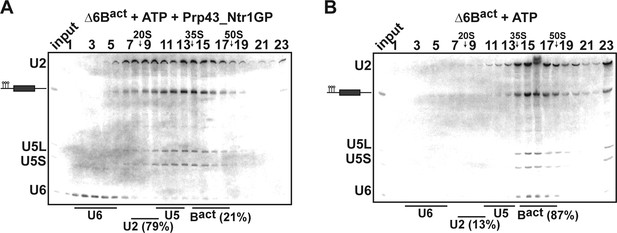
Prp43_Ntr1GP disassembles Δ6Bact complexes.
10–30% glycerol gradient sedimentation of purified ActΔ6Bact spliceosomes assembled on ActinΔ6 pre-mRNAs (Fabrizio et al., 2009) and incubated with (A) ATP and Prp43_Ntr1GP or (B) ATP alone. Samples were further processed and quantified as described in Figures 1 and 2.
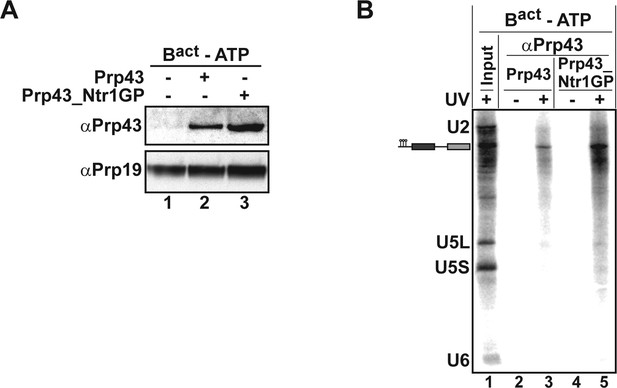
Prp43 and Prp43_Ntr1GP bind to the Bact ΔPrp2 complex and interact with the pre-mRNA.
(A) Purified Bact ΔPrp2 complexes formed on radiolabelled Act7-wt pre-mRNA were incubated without recombinant proteins (lane 1) with Prp43 (lane 2) or Prp43_Ntr1GP (lane 3) in the absence of ATP. Samples were loaded on distinct glycerol gradients. Proteins from the peak fraction were recovered and separated by SDS-PAGE on a 4–12% Bis-TrisNuPAGE polyacrylamide gel (Invitrogen) and visualized by Western blotting using anti-Prp43 or anti-Prp19 antibodies (B) Purified Bact ΔPrp2 complexes formed on radiolabelled Act7-wt pre-mRNA were incubated with Prp43 (lanes 2 and 3) or Prp43_Ntr1GP (lanes 4 and 5) in the absence of ATP and the reaction mixture was irradiated with UV light at 254-nm (+) or left untreated (-). Following denaturation, the reaction mixtures were immunoprecipitated with anti-Prp43 antibodies. After proteolytic digestion, coprecipitated snRNAs were identified by Northern blotting using radiolabelled probes that hybridise to U2, U5 or U6 snRNA.
-
Figure 6—source data 1
Summary of inter-molecular and intra-molecular protein–protein crosslinking for Prp43_Ntr1GP bound to the Bact ΔPrp2 spliceosome in the absence of ATP.
We used BS3, a chemical crosslinker with a spacer length of 11 Å, which is specific for lysines and N-termini. Data were analyzed by mass spectrometry and the spectra of Prp43_Ntr1GP interactions were selected. Spectra count indicates the number of times that one particular interaction was seen. The score indicates the probability of interaction between two proteins. High scores (i.e. ≥2) signify a higher confidence of crosslink identification. In the case of multiple spectra, only the highest score is reported. Three separate experiments were performed and are shown in Figure 6—source data 1a and b. (a) Proteins crosslinked to Prp43_Ntr1GP in affinity-purified Bact ΔPrp2 spliceosomes. Inter-molecular interactions between residues of Prp43_Ntr1GP and residues of the spliceosomal proteins indicated (i.e. Protein 2). (b) Intra-molecular interactions between residues (i.e. residue 1 and 2) of Prp43_Ntr1GP.
- https://doi.org/10.7554/eLife.15564.012
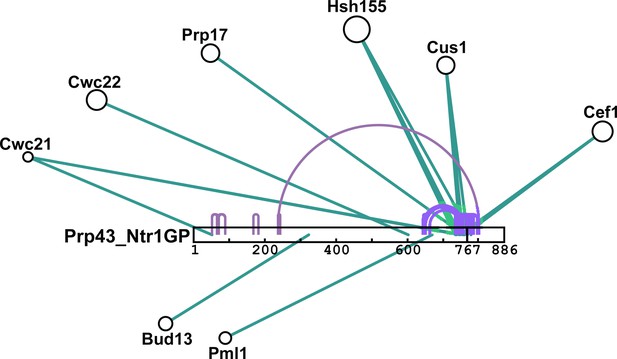
Schematic summary of BS3 crosslinking of Prp43_Ntr1GP with proteins of the Bact ΔPrp2 spliceosome.
Detailed BS3 crosslinking data are shown in Figure 6—source data 1a and b. Prp43_Ntr1GP is shown schematically as a white bar, where the amino acid positions are indicated. Intermolecular crosslinks between Prp43_Ntr1GP and proteins of the Bact ΔPrp2 complex are indicated by green lines. Intramolecular crosslinking sites for Prp43_Ntr1GP are indicated by purple lines. The cartoon was prepared with xiNET (Combe et al., 2015).





