PAX3-FOXO1 transgenic zebrafish models identify HES3 as a mediator of rhabdomyosarcoma tumorigenesis
Abstract
Alveolar rhabdomyosarcoma is a pediatric soft-tissue sarcoma caused by PAX3/7-FOXO1 fusion oncogenes and is characterized by impaired skeletal muscle development. We developed human PAX3-FOXO1 -driven zebrafish models of tumorigenesis and found that PAX3-FOXO1 exhibits discrete cell lineage susceptibility and transformation. Tumors developed by 1.6–19 months and were primitive neuroectodermal tumors or rhabdomyosarcoma. We applied this PAX3-FOXO1 transgenic zebrafish model to study how PAX3-FOXO1 leverages early developmental pathways for oncogenesis and found that her3 is a unique target. Ectopic expression of the her3 human ortholog, HES3, inhibits myogenesis in zebrafish and mammalian cells, recapitulating the arrested muscle development characteristic of rhabdomyosarcoma. In patients, HES3 is overexpressed in fusion-positive versus fusion-negative tumors. Finally, HES3 overexpression is associated with reduced survival in patients in the context of the fusion. Our novel zebrafish rhabdomyosarcoma model identifies a new PAX3-FOXO1 target, her3/HES3, that contributes to impaired myogenic differentiation and has prognostic significance in human disease.
https://doi.org/10.7554/eLife.33800.001eLife digest
One of the most common cancers in children and adolescents is rhabdomyosarcoma, a cancer of soft tissue such as muscle, tendon or cartilage. The fusion of DNA on two different chromosomes causes the most aggressive form of rhabdomyosarcoma. The fused DNA produces an abnormal protein called PAX3-FOXO1. During normal muscle development, a subset of rapidly growing cells eventually slow down and form mature, working muscle cells. It is still unclear how exactly rhabdomyosarcoma develops, but it is thought that PAX3-FOXO1 stops muscle cells from maturing and the cells that grow out of control result in a tumor. Learning how PAX3-FOXO1 hijacks normal muscle development could lead to new treatments for rhabdomyosarcoma.
One treatment approach is to slow the growth of a tumor and force the cells to mature. Then, young patients might avoid chemotherapy or radiation treatments and their side effects. Efforts to improve treatment for this type of cancer face several obstacles. Currently, only one vertebrate animal model of the disease is available to test drugs, and it is still not known how PAX3-FOXO1 causes healthy cells to become cancerous. It is also hard to turn off PAX3-FOXO1 itself, so scientists must find additional proteins that collaborate with it to target with drugs.
Now, Kendall et al. show that genetically engineered zebrafish with human PAX3-FOXO1 develop rhabdomyosarcoma-like tumors. Experiments on these zebrafish showed that the protein turns on a gene called her3. Humans have a similar gene called HES3. In zebrafish or mouse cells, human HES3 interferes with muscle-cell maturation and allows cells that acquire PAX3-FOXO1 to persist during development instead of dying. It also increases the cell growth and cancerous behavior in human tumor cells. Kendall et al. further looked at HES3 levels in tumors collected from patients with rhabdomyosarcoma and found that having higher levels of HES3 increased the risk of death from the cancer.
Human rhabdomyosarcoma tumors with high HES3 levels were also more likely to have certain cell-growth and cell-differentiation related proteins. Drugs that turn off or modify the activity of these proteins already exist. Testing these drugs that target processes such as cell growth in the zebrafish with rhabdomyosarcoma-like tumors may help scientists determine if they reduce tumor growth. If they do, additional trials could determine if they would help patients with rhabdomyosarcoma.
https://doi.org/10.7554/eLife.33800.002Introduction
Rhabdomyosarcoma (RMS) presents as a solid tumor that displays characteristics of primitive skeletal muscle (Parham and Ellison, 2006). There are two major histological subtypes, embryonal (ERMS) and alveolar rhabdomyosarcoma (ARMS), with ARMS being more aggressive and particularly prone to metastasis (Williamson et al., 2010; Skapek et al., 2013). At the genomic level, ERMS is characterized by frequent alterations affecting RAS signaling, although this still represents a minority of the cases (Stratton et al., 1989; Langenau et al., 2007; Martinelli et al., 2009). The defining oncogenic event in ARMS is a t(1;13) or t(2;13) chromosomal translocation in which the PAX7 or PAX3 DNA-binding domain, respectively, is fused to the FOXO1 transactivation domain to create a PAX3/7-FOXO1 chimeric oncogene (Barr et al., 1993; Galili et al., 1993; Shapiro et al., 1993; Davis et al., 1994). The PAX3-FOXO1 fusion is the most prevalent fusion in the disease, and functions as an aberrant transcription factor that is expressed in the nucleus and deregulates gene expression signatures (del Peso et al., 1999; Fredericks et al., 1995; Barber et al., 2002; Khan et al., 1999). This activity is the predominant cellular insult required for transformation.
The PAX3/7-FOXO1 oncogenes remain intractable to therapeutic targeting, impeding the development of effective precision medicine therapies. Fusions are notoriously difficult to model in animals, hence the limited availability of vertebrate animal models of this disease. Furthermore, there is a narrow understanding of the cellular origin of RMS, making it difficult to define the expression pattern required for tumorigenesis (Hettmer and Wagers, 2010). Zebrafish are a complementary model system that can address these genetic and cellular issues. Advantages of zebrafish systems are two-fold: (1) they provide insight into the underlying biology of how cancer genes behave in a complex environment and (2) provide a platform for translational drug discovery efforts. Such strengths are intrinsically important for translational models of pediatric disease.
Here, we describe human PAX3-FOXO1-driven zebrafish models. We implemented our zebrafish models to provide the appropriate context to understand the behavior of PAX3-FOXO1 during development and tumorigenesis. The tumor presentation spectrum identified three distinct cellular contexts that are susceptible to transformation, generating insight into basic mechanisms of PAX3-FOXO1 tumorigenesis and human rhabdomyosarcoma. By applying our zebrafish RMS model, we found a novel PAX3-FOXO1 target, her3/HES3. HES3 is a member of the HES family of basic helix-loop-helix transcription factors, which function as direct or indirect transcriptional activators or repressors, thus regulating gene expression and epigenetic identity (Kageyama et al., 2007). HES3 is expressed in the developing brain and inhibits differentiation of neural stem cells (Hatakeyama et al., 2004). In cancer, HES3 is expressed in glioblastoma cell culture, and co-localizes with additional markers of stemness in the mouse brain (Park et al., 2013; Poser et al., 2013; Katoh and Katoh, 2007). However, its role as a cooperating gene in PAX3/7-FOXO1 fusion-positive rhabdomyosarcoma has never been described. Taken together, this model represents a novel strategy to identify new targets and biomarkers in the context of human disease and contributes to our understanding of RMS biology by defining the earliest tumor initiation events.
Results
A transgenic zebrafish model of human PAX3-FOXO1 driven tumorigenesis
To develop a new vertebrate model of PAX3-FOXO1-dependent tumorigenesis, we performed a survey of promoters driving PAX3-FOXO1 expression. These promoters represent ubiquitous (beta actin, CMV, ubiquitin), hematologic (fli1), muscle (unc503), neural crest (mitfa) expression, and a gene trap approach. Selected promoters were chosen because of their relevance in the disease as implicated lineages for the cell of origin or for their capacity to drive PAX3-FOXO1 at high levels of expression. Further, all promoters had been previously validated as functional in zebrafish, with data from our group underscoring the beta actin promoter as a successful expression system for EWS-FLI1 transgenic models of Ewing sarcoma (Leacock et al., 2012). Human PAX3-FOXO1 was integrated into the zebrafish genome utilizing the Tol2 transposon-based system and microinjection in a stable mosaic manner. Genomic integration and transgene expression were tracked using a GFP or mCherry fluorescent protein linked to the coding sequence of PAX3-FOXO1 with a viral 2A sequence (Figure 1A–D). This results in equimolar expression of both genes on the same mRNA transcript, yet translation as independent proteins. Zebrafish were monitored for up to 19 months. Using this strategy, we identified fusion-oncogene driven tumors first based on gross morphology and then by screening suspected tumors for fluorescent signal.
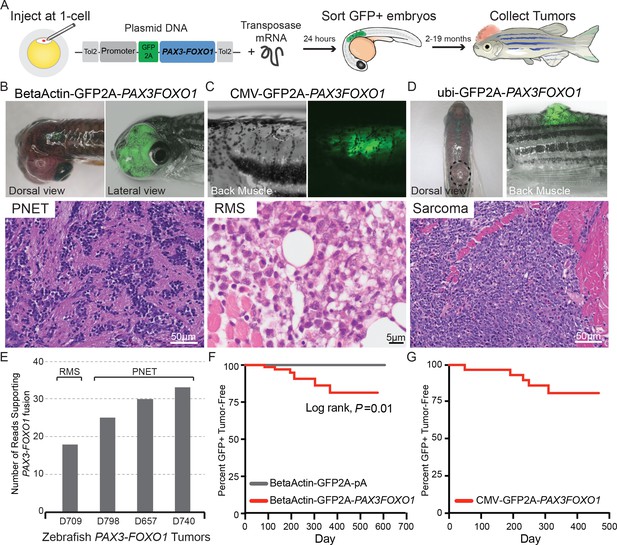
Zebrafish models of human PAX3-FOXO1 tumorigenesis.
(A) Zebrafish were injected at the single-cell stage with mosaic GFP2A-tagged human PAX3-FOXO1 under the control of various promoters. At 24 hr old, embryos were sorted for GFP expression indicating successful injections (typically 99% GFP+) and were allowed to grow and monitored for up to 19 months to develop tumors. (B) Beta-actin-driven PAX3-FOXO1 primarily produced primitive neuroectodermal tumors in a wild-type genetic background. Shown for all tumors are representative examples with the presentation of gross morphology and GFP expression patterns coupled with a hematoxylin and eosin stain. (C) CMV-driven PAX3-FOXO1 produced rhabdomyosarcoma in the tp53M214K/M214K-sensitized genetic background. (D) Ubiquitin-driven PAX3-FOXO1 produced an undifferentiated sarcoma in a wild-type genetic background. (E) RNAseq data from zebrafish PAX3-FOXO1 fluorescent tumors showing the number of reads supporting the presence of the human fusion-oncogene. (F) Tumor incidence of GFP + tumors detected in BetaActin-GFP2A-PAX3FOXO1 (n = 74) injected zebrafish versus BetaActin-GFP (n = 147) injected controls in a wildtype genetic background. (G) Tumor incidence of GFP + tumors detected in CMV-GFP2A-PAX3FOXO1 (n = 31) injected zebrafish in a tp53M214K-sensitizing genetic background.
Human PAX3-FOXO1 under the control of different promoters had unique survival and transforming properties suggesting cell-of-origin specificity to oncogenesis. Tested promoters had a negative impact on early survival (up to 30 days of life), but to different extents (Figure 1—figure supplement 1). Only a subset of the promoters that were tested induced tumor formation, including the beta actin (9% of injected zebrafish), CMV (16% of injected zebrafish), and ubiquitin (1.8% of injected zebrafish) promoters (Supplementary file 1). The fli1, unc503, mitfa, and gene trap approach driving PAX3-FOXO1 were not transforming (Supplementary file 2).
The three promoters (beta actin, CMV, ubiquitin) that induced PAX3-FOXO1 transformation in zebrafish had varied requirements for tp53M214K as a sensitizing mutation. The beta actin promoter driving the expression of PAX3-FOXO1 is tumorigenic in zebrafish, both in a wildtype and a p53-deficient genetic background. In tp53 wild-type zebrafish, BetaActin-PAX3FOXO1 tumors began developing at 3 months of age in 5% of injected zebrafish in the head. Diagnosis of tumors was made by hematoxylin and eosin staining and evaluation by light microscopy. Based on this analysis, the majority of beta-actin-driven PAX3-FOXO1 zebrafish tumors were consistent with primitive neuroectodermal tumors (Figure 1B). However, beta-actin-driven PAX3-FOXO1 injected into the tp53M214K/M214K mutant resulted in one undifferentiated sarcoma after 412 days.
The CMV promoter restricting PAX3-FOXO1 expression results in RMS tumors that present in the skeletal muscle of the back. Histological analysis of the zebrafish tumors was consistent with human RMS (Figure 1C). This was true only in the context of the tp53M214K mutation, indicating that the tp53 mutation is sensitizing to and favors RMS. These findings are concordant with the Pax3-Foxo1 RMS mouse model, in which Tp53 deletion, or other secondary cooperating mutations are required for RMS development (Keller et al., 2004). Further, patients with Li Fraumeni syndrome develop RMS as part of the spectrum of the human disease (Li and Fraumeni, 1969).
The ubiquitin promoter driving PAX3-FOXO1 generated one undifferentiated sarcoma by 378 days of age in a wild-type genetic background (Figure 1D). We performed RNA-seq on zebrafish tumors derived from all three promoters (beta actin, CMV, ubiquitin), and detected PAX3 and FOXO1 junction spanning reads indicating that human PAX3-FOXO1 is expressed in fluorescent tumors (Figure 1E, Figure 1—figure supplement 2). These data demonstrate that the human fusion-oncogene is transforming in zebrafish when expressed in a variety of cellular contexts and that some, but not all, cellular identities are susceptible to transformation.
The beta actin, CMV, and ubiquitin promoter restricting PAX3-FOXO1 expression have unique latency, penetrance, and spectrum of tumor development (Figure 1F–G; Supplementary file 1). Tumor incidence curves are shown for the beta actin promoter driving PAX3-FOXO1 in a wild-type background, and for the CMV promoter driving PAX3-FOXO1 in the tp53M214K/M214K mutant background (Figure 1F–G). Rhabdomyosarcoma only develops in the tp53M214K background, predominantly with the CMV promoter as compared to the beta actin promoter. Since human PAX3-FOXO1 is active in zebrafish and produces discrete functional readouts, we next applied our model to identify PAX3-FOXO1 targets that are engaged during development to induce tumorigenesis.
PAX3-FOXO1 has a different impact on embryonic development than PAX3
The PAX3-FOXO1 fusion-oncogene functions as an aberrant transcription factor due to the PAX3 DNA-binding domain being linked to the FOXO1 transactivation domain (Linardic, 2008; Cao et al., 2010). To identify activity unique to PAX3-FOXO1 versus the normal PAX3 transcription factor, we performed a detailed analysis of the functional effects of their expression during zebrafish development. We utilized a mosaic model system, in which plasmid DNAs containing the beta actin promoter driving GFP, or GFP-tagged PAX3 or PAX3-FOXO1 were injected into developing zebrafish using the Tol2 transposon system (Figure 2A). The expression of each construct was tracked using a viral 2A linked fluorescent protein, and mosaic integration of the construct can be observed with fluorescence by 24 hr post-injection (Figure 2B). During the first three days of life, PAX3-FOXO1 injected zebrafish exhibited reduced survival as compared to PAX3 injected zebrafish (Figure 2C). Furthermore, PAX3-FOXO1-induced unique embryonic phenotypes, including cyclopia, which was present in 30% of injected embryos. To address if cyclopia was due to PAX3-FOXO1 expression, we co-injected GFP2A-PAX3FOXO1 with a morpholino that targets the expression construct. In this scenario, the morpholino binds the start codon of GFP and inhibits the ribosome’s association with the mRNA transcript, thus inhibiting the translation of the GFP-viral2A-PAX3FOXO1 mRNA. Morpholino activity was tracked by quantifying the reduced number of GFP positive embryos after injection with titrating doses of morpholino (Figure 2D). Co-injection of PAX3-FOXO1 and a morpholino that knocks-down GFP-PAX3-FOXO1 expression eliminated the cyclopia phenotype (Figure 2D). This is significantly different from the uninjected, GFP injected, and the PAX3 injected groups, suggesting that PAX3-FOXO1 is regulating distinct developmental phenotypes.
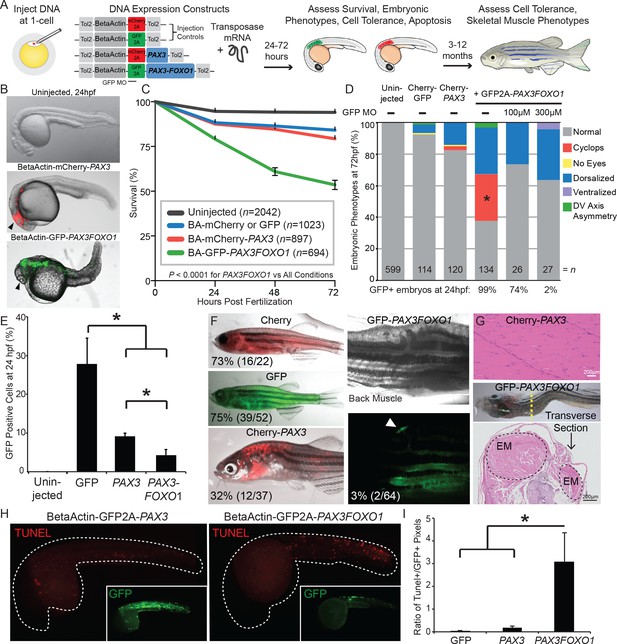
PAX3 and PAX3-FOXO1 have distinct impacts and tolerance during embryonic development and in adult zebrafish.
(A) Strategy for assessing functional differences of beta-actin-driven PAX3 and PAX3-FOXO1 in a vertebrate system. (B) Representative images at 24 hr post-fertilization of Uninjected, mCherry2A-PAX3, and GFP2A-PAX3FOXO1-injected zebrafish. (C) Survival curve of Uninjected, GFP2A or mCherry2A injected controls, mCherry2A-PAX3, and GFP2A-PAX3FOXO1. Error bars represent SE. Log rank test, p<0.0001 for PAX3FOXO1 versus all other conditions. (D) Embryonic phenotypes scored at 3 days post-injection. * indicates p<0.05, for PAX3 vs PAX3FOXO1, Fisher’s exact test. MO- morpholino. DV- Dorso-Ventral. (E) Percentage of GFP + cells from dissociated zebrafish embryos as quantified by fluorescent activated cell sorting (FACS). Error bars represent SD across three independent experiments. * indicates p<0.05, two-tailed Student’s t-test. (F) Adult zebrafish over 3 months of age robustly expressed beta-actin-driven Cherry, GFP, or Cherry2A-PAX3 and developed normally. Zebrafish injected with BetaActin-GFP2A-PAX3FOXO1 displayed developmental defects or developed tumors. Arrow denotes GFP + area. The percentage indicates zebrafish with detectable fluorescence at adulthood. (G) Hematoxylin and eosin staining showed normal histology of BetaActin-PAX3 expressing skeletal muscle (sagittal section) at 299 days of age, and abnormal histology of BetaActin-PAX3FOXO1 epaxial muscle exhibiting dramatic left-right asymmetry (transverse section, asymmetry of left-right epaxial muscle (EM) marked by dotted lines) at 307 days of age. Scale bars, 200 microns. EM- epaxial muscle. (H) Representative images from zebrafish embryos injected with GFP2A-PAX3 and GFP2A-PAX3FOXO1 that are fixed at 24 hr post-injection and then TUNEL performed (rhodamine). Embryos were counter-stained for GFP to indicate transgene expression. (I) Quantification of TUNEL-positive pixels normalized to GFP positive pixels, indicated a higher proportion of PAX3-FOXO1 cells were undergoing apoptosis. Error bars represent SD, n = 6–8 embryos per group, * indicates p<0.05, two-tailed Student’s t-test.
This discrepancy between PAX3-FOXO1 and PAX3 tolerance was evident at the level of individual cells. To demonstrate this, embryos were injected with beta-actin-driven constructs containing: GFP only, GFP-tagged PAX3, or GFP-tagged PAX3-FOXO1. Injected zebrafish embryos were allowed to develop for 24 hr, and then dissociated to single-cell suspensions. Fluorescent activated cell sorting (FACS) showed that PAX3-FOXO1 expression significantly reduced the number of GFP + cells at 24 hr post-fertilization as compared to PAX3 (Figure 2E). This reduced viability of PAX3-FOXO1 + zebrafish was consistent in injected embryos that were raised to adulthood and screened at 3 months of age for detectable fluorescence. In BetaActin-mCherry or GFP fluorescent controls, fluorescence was observed in over 70% of adult zebrafish. In PAX3-injected zebrafish, mCherry fluorescence was observed in 32% of adults. We found that only 3% of adult zebrafish had detectable GFP-PAX3FOXO1 expression with resulting tumor development or asymmetric skeletal muscle (Figure 2F–G). This was not true for BetaActin-GFP or BetaActin-GFP-PAX3, which do not generate tumors or affect normal development or survival.
One explanation for this disappearance of PAX3-FOXO1 + cells during the course of development is that PAX3-FOXO1-injected zebrafish have a significant increase in the number of cells undergoing apoptosis. At 24 hr post-fertilization, TUNEL staining was performed on embryos injected with either GFP-PAX3 or GFP-PAX3FOXO1. These same embryos were counter-stained for GFP to detect the expression levels of their respective transgenes (Figure 2H). PAX3-FOXO1-injected embryos had an increase in the number of TUNEL-positive cells undergoing apoptosis as compared to GFP controls and PAX3 in wild-type zebrafish (Figure 2I). Given these data and the propensity to generate RMS we mechanistically evaluated the contribution of tp53 mutations to early PAX3-FOXO1-induced apoptosis (Figure 1C and G; Figure 2—figure supplement 1A). We found that tp53M214K mutant zebrafish embryos are unable to invoke an appropriate response to PAX3-FOXO1 + cells, resulting in a gross reduction in survival by 3 days of age (Figure 2—figure supplement 1B). Further, injected CMV-PAX3FOXO1 + cells are more abundant in tp53M214K injected mutant zebrafish as compared to their wild-type counterparts by 28 hr post fertilization (Figure 2—figure supplement 1C–D). This significant increase in the number of GFP-PAX3FOXO1 + cells is coupled with a reduction in the ratio of TUNEL-positive cells, indicating that tp53M214K mutant zebrafish provide a susceptible environment for PAX3-FOXO1 + cellular persistence and ultimately tumor development (Figure 2—figure supplement 1E–G; Figure 2—figure supplement 2). This dichotomy of cellular tolerance and apoptosis is not true for wildtype or tp53M214K mutant zebrafish injected with GFP controls, or GFP-PAX3 (Figure 2—figure supplement 1B–G). Therefore, at both an organismal and individual cell level, ectopic expression of PAX3-FOXO1 was less tolerated than the normal PAX3 gene. Understanding the exact mechanisms for cell tolerance, and defining these discrepancies, could identify the earliest events in RMS transformation or alternatively tumor suppressive responses.
her3 is a novel developmental target of human PAX3-FOXO1.
Given that PAX3-FOXO1 is uniquely tumorigenic and has different embryonic effects than PAX3 this warranted a more thorough study of the specific developmental pathways and targets that were mediating these early outcomes. To accomplish this, we injected zebrafish embryos with DNA expression constructs in which the beta actin promoter drives either a GFP control, or GFP-tagged PAX3 and PAX3-FOXO1, and allowed the embryos to develop for 24 hr. The GFP + cell population from zebrafish embryos was then FACS sorted, total RNA isolated, and microarrays were used to determine differential gene expression signatures. After comparing differentially expressed genes versus the GFP injected control for PAX3 and PAX3-FOXO1, the most highly enriched Gene Ontology (GO) terms indicated that PAX3 and PAX3-FOXO1 act as transcription factors (Figure 3A). This was expected given their well-documented roles in the literature and suggested that the mammalian forms are active in zebrafish systems. Furthermore, DAVID analysis of the enriched genes during normal development suggested that these sorted cell populations identify different subsets of zebrafish embryonic tissues. PAX3-positive cells were most indicative of the neural plate, neuroectoderm, or ectoderm, whereas PAX3-FOXO1 was indicative of a somite, segmental plate, or optic vesicle and immature eye (Figure 3B). Even though PAX3 and PAX3-FOXO1 were injected under the control of the same promoter, cells expressing these genes represent either (1) different cell populations or (2) identical cell populations that have differential transcriptional responses as early as 24 hr post-fertilization.
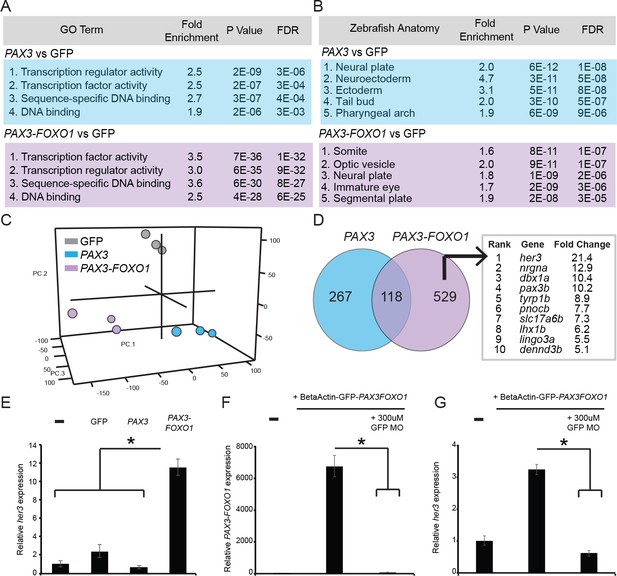
PAX3 and PAX3-FOXO1 induce distinct gene expression signatures during development, identifying a unique PAX3-FOXO1 target, her3.
Microarray analysis identified differentially expressed gene signatures in 24 hr old zebrafish FACS sorted embryonic cells that expressed either GFP2A-PAX3FOXO1 or GFP2A-PAX3. (A) Gene ontology terms associated with PAX3 or PAX3-FOXO1. (B) Embryonic tissues indicative of PAX3 or PAX3-FOXO1 gene sets. (C) 3D principal components analysis (PCA) of GFP injected controls, PAX3, and PAX3-FOXO1. (D) Intersection of up-regulated genes for PAX3 and PAX3-FOXO1 as compared to GFP controls. Uniquely up-regulated PAX3-FOXO1 target genes were included in downstream analysis only if they possessed a human ortholog. Shown are genes rank ordered based on the fold-change of their expression. (E) qRT-PCR of her3 levels from 24-hr-old zebrafish embryos that are either uninjected controls or injected with GFP, PAX3, or PAX3-FOXO1. (F) qRT-PCR for PAX3-FOXO1 mRNA levels from 24-hr-old zebrafish embryos injected with PAX3-FOXO1, or PAX3-FOXO1 in combination with a GFP morpholino (GFP MO) that inhibits transgene expression. (G) Same samples as in F, but qRT-PCR was performed for her3. In E-G the SD is derived from technical triplicates, * indicates p<0.05, two-tailed Student’s t-test.
We then performed a principal component analysis (PCA) and found that PAX3 and PAX3-FOXO1 expression changes are distinct and show statistical significance in a developmental context (Figure 3C). This does not contradict the fact that there are a large number of expression changes that are similar between these two transcription factors. Therefore, unique developmental and oncogenic targets of PAX3-FOXO1 could be ascertained by directly comparing its activity to that of PAX3. To determine what these PAX3-FOXO1 developmental targets might be, we looked at the overlap of induced genes for PAX3 or PAX3-FOXO1 versus the GFP controls. We found that PAX3 and PAX3-FOXO1 up-regulated 118 shared genes, whereas PAX3-FOXO1 uniquely induced 529 genes. These 529 genes were of interest because of their association with the fusion-oncogene and potentially tumorigenesis. Of these 529 genes, those without a human ortholog were eliminated from further analysis due to our interest in human disease. This produced a list of primarily developmental transcription factors and homeobox genes, including her3, pax3b, dbx1a, and lhx1b. The most highly induced gene unique to PAX3-FOXO1 expression was her3, with a 21-fold increase as compared to the GFP control (Figure 3D).
We validated that her3 expression was induced by the human PAX3-FOXO1 fusion-oncogene in zebrafish by injection of GFP control, PAX3, and PAX3-FOXO1. Embryos were allowed to develop and then total RNA was isolated from an embryo pool, cDNA transcribed, and a qRT-PCR performed for her3 expression levels. This analysis indicated that her3 induction was unique to PAX3-FOXO1 (Figure 3E). To verify that the up-regulation of her3 was dependent on PAX3-FOXO1, we co-injected the BetaActin-PAX3FOXO1 construct with a morpholino that inhibits its early expression by targeting the GFP2A-PAX3FOXO1 transcript. At 24 hr of age, PAX3-FOXO1 mRNA levels were measured by qRT-PCR, and co-injection of the morpholino significantly knocked down PAX3-FOXO1 expression (Figure 3F). A qRT-PCR was performed on these same samples to evaluate her3 expression levels, and the results replicated those of PAX3-FOXO1 (Figure 3G). These data suggest that her3 is a novel target of PAX3-FOXO1 in vivo, that her3 is not induced by PAX3, and that her3 expression is dependent on the PAX3-FOXO1 oncogene.
HES3, the human ortholog of her3, inhibits in vivo myogenesis and supports inappropriate persistence of PAX3-FOXO1 + cells
A hallmark of rhabdomyosarcoma is that tumors express early markers along the skeletal muscle lineage such as MYOD1, MYOG and Desmin; however, these tumors fail to terminally differentiate indicating a developmental arrest (Parham and Ellison, 2006; Saab et al., 2011). To determine if this was recapitulated in our zebrafish model, we performed a mosaic co-injection strategy with the beta actin promoter driving GFP-mCherry controls, human HES3 alone, human PAX3-FOXO1 alone, or a co-injection of PAX3-FOXO1 and HES3 (Figure 4A). Co-injection of transgenes linked to highly expressed promoters generated mosaic integration that overlaid with muscle proteins such as myosin by 24 hr post-fertilization (Figure 4B). Based on this observation, zebrafish were injected and allowed to develop for 24 hr, and then fluorescent embryos harvested and total RNA isolated and reverse transcribed for qRT-PCR analysis of muscle marker genes. This survey of temporally stereotyped muscle markers included early markers myod and myog and terminal muscle differentiation markers myl1 and myhz2. We found that PAX3-FOXO1 did not affect the mRNA levels of myod or myog; however, by 24hpf there was a significant reduction in the expression of myl1 and myhz2. Surprisingly, overexpression of HES3 alone had a similar effect of inhibiting terminal muscle differentiation, indicating that her3/HES3 contribute to maintaining a more primitive cellular state. Together, PAX3-FOXO1 and HES3 had no significant additive effect, suggesting they are functioning in a linear pathway with her3/HES3 being downstream of PAX3-FOXO1 (Figure 4C). Hence, PAX3-FOXO1 and HES3 inhibit differentiation in vivo in a vertebrate system via a shared mechanism.
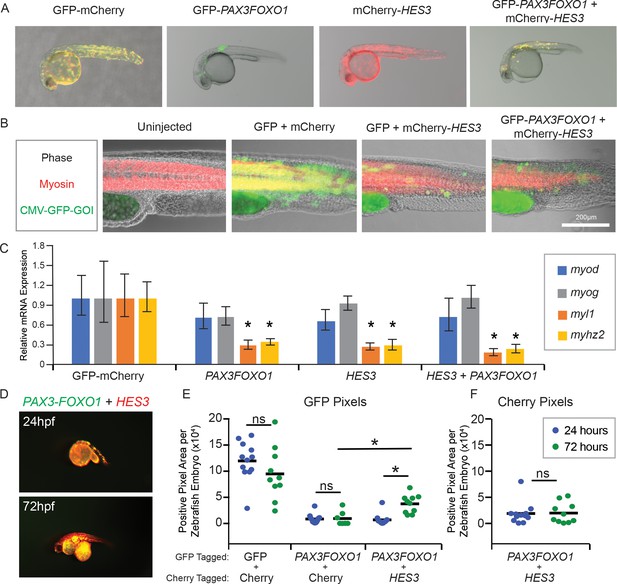
HES3 inhibits myogenic differentiation in developing zebrafish and supports persistence of PAX3-FOXO1-positive cells.
(A) Zebrafish embryos were injected at the single-cell stage with the beta actin promoter driving GFP-Cherry, GFP-PAX3FOXO1, mCherry-HES3, or combined mCherry-HES3 and GFP-PAX3FOXO1. Shown are representative embryos at 24 hr post fertilization with indicated transgene expression. (B) Representative overlays of zebrafish embryo musculature that were fixed at 24 hr and immunofluorescence performed for myosin (red) and injected genes of interest (GOI; green). (C) Pools of n = 5 embryos were harvested at 24 hr and markers of myogenesis assessed by qRT-PCR, including myod, myog, myl1, and myhz2. SD is derived from technical triplicates. * indicates significant differences between treatment group and the GFP-mCherry control at a threshold of p<0.05, two-tailed Student’s t-test. (D) Representative overlay of images from co-injections of mCherry-HES3 and GFP-PAX3FOXO1 from the same embryo at 24 and 72 hr post-fertilization. Images were taken with the same exposure settings and objective. (E) Quantification of the number of positive pixels for each embryo imaged at 24 and 72 hr post-fertilization. GFP-positive pixels are plotted after the same settings are applied for imaging and analysis. Each marker represents a single zebrafish embryo at 24 or 72 hr post fertilization, n = 6–12 embryos per group. Black bar is the mean, and * indicates p<0.05, two-tailed Student’s t-test. ns- not significant. (F) Same analysis as in E but for mCherry positive pixels.
The majority of the PAX3-FOXO1 and HES3 co-injected cells overlaid indicating that this mosaic system is a powerful model to study the interactions of two genes within the same cell (Figure 4A and D). Given this capacity, we next determined if HES3 modified the behavior of GFP-PAX3FOXO1 + cells during embryogenesis. Zebrafish embryos were co-injected at the single-cell stage with two independent plasmids that were dually integrated into the genome using the Tol2-transposon-based system. The conditions included the beta actin promoter driving: (1) mCherry and GFP, (2) GFP and mCherry-HES3, (3) mCherry and GFP-PAX3FOXO1, and (4) GFP-PAX3FOXO1 and mCherry-HES3. Both GFP and mCherry images were taken using the same settings at both 24 hr post-fertilization and 72 hr post-fertilization to evaluate the capacity of the cells to survive (Figure 4D). The number of GFP and mCherry positive pixels were calculated independently and plotted for both timepoints (Figure 4E–F). Co-injection of HES3 allowed PAX3-FOXO1 cells to survive and/or proliferate, with a greater number of GFP-positive pixels and thus PAX3-FOXO1 positive cells persisting by 72hpf (Figure 4E). This phenomenon was unique to the co-injection of HES3 + PAX3FOXO1, as the number of pixels that were positive between 24hpf and 72hpf for mCherry + GFP-PAX3FOXO1 was not significantly different. Additionally, the number of mCherry positive pixels from 24hpf to 72hpf was insignificantly different for mCherry-HES3 + GFP-PAX3FOXO1 indicating that this observation was not globally applicable, but unique to the persistence of PAX3-FOXO1 (Figure 4F). These results suggest that HES3 promotes a more tolerant cellular environment allowing for survival with expression of the highly toxic PAX3-FOXO1.
One hypothesis is that the co-expression of HES3 allows for PAX3-FOXO1 expressing cells to circumvent apoptosis. By implementing our CMV injected mosaic zebrafish models, we found that HES3 overexpression in zebrafish allows for PAX3-FOXO1+ cells to either inappropriately persist or divide, resulting in an increased number of observable GFP-PAX3FOXO1 + pixels by 24 hr post-fertilization (Figure 4—figure supplement 1A–C). We performed TUNEL assays on these same zebrafish embryos and found that HES3 alone does not induce apoptosis. Further, there is no significant difference in the normalized ratio of TUNEL cells for PAX3-FOXO1-injected embryos as compared to PAX3-FOXO1 + HES3 (Figure 4—figure supplement 1D–E). These data indicate that PAX3-FOXO1 + HES3 cooperation facilitates PAX3-FOXO1 + cells to persist during embryogenesis independently of apoptosis inhibition at this timepoint (Figure 4—figure supplement 2).
HES3 inhibits myogenesis in mammalian myoblasts
Impaired skeletal muscle differentiation is a signature feature of human RMS tumors. To evaluate how HES3 is contributing to oncogenesis in fusion-positive RMS, we determined its capacity to inhibit differentiation in mammalian cell culture (Figure 5A). Stable mouse C2C12 myoblast cell lines were generated by transfection and selection of CMV-HES3 or CMV-Empty control, and the resulting cell lines were evaluated for overexpression of HES3 by qRT-PCR (Figure 5B). Myoblasts were viable with HES3 overexpression and were next tested to determine if there was a temporal or functional difference in their myogenic capacity. Cells were seeded onto porcine gelatin coated plates and collected at one and 6 days post plating after being exposed to low-serum concentrations and supplemented with insulin to promote fusion. The expression levels of selected genes were evaluated to represent the spectrum of myogenic differentiation, including MyoD, MyoG, Myl1, and Myh1 (Figure 5A). By day 6 of differentiation, MyoD, MyoG, Myl1, and Myh1 exhibited significant inhibition of expression in C2C12-HES3 overexpressing cells as compared to the C2C12-Empty controls. Moreover, Myl1 and Myh1, which represent terminal muscle differentiation, were the most affected by HES3 overexpression at fusion day 6, with a 2.9–3.2 fold decrease in expression levels, as compared to a 1.8–1.9 fold decrease observed for MyoD and MyoG (Figure 5B).
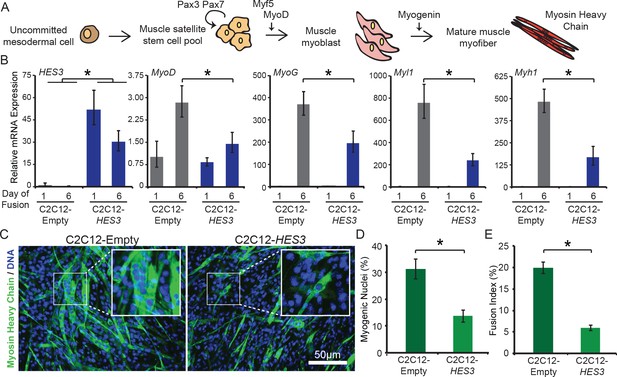
HES3 overexpression inhibits myogenic differentiation of mouse muscle myoblasts.
(A) Schematic of muscle development. Specifically noted are key skeletal muscle transcription factors and proteins assessed in B. Muscle satellite cells that self-renew express Pax3/Pax7. During development, they are activated to further differentiate by expression of MyoD, the master muscle regulator, after which they become myoblasts, an intermediate cell type. Myoblasts proliferate and fuse together to form multi-nucleated and contractile myofibers that express structural proteins, such as myosin heavy chain. (B) C2C12-Empty and C2C12-HES3 overexpressing cells were seeded in growth media and after 24 hr exposed to differentiation media. At this point Day 1 of fusion was collected. Following 5 additional days in fusion media the final timepoint was collected. qRT-PCR was performed for HES3, and the following muscle marker genes: MyoD, MyoG, Myl1, Myh1. SD is derived from technical triplicates. * indicates p<0.05, two-tailed Student’s t-test. (C) C2C12-Empty and C2C12-HES3 cells were differentiated for five days, fixed, and immunofluorescence performed for Myosin Heavy Chain (MyHC) protein. Cells were counterstained with DAPI to detect DNA. Shown are representative images of the fusion capacity for each condition. (D) Quantification of the differentiation capacity of HES3 overexpressing mouse myoblasts. Plotted are the number of myogenic nuclei (# of nuclei within a MyHC + cell divided by the total nuclei) with n = 3 technical replicates per group ± SD. Experiment was repeated with biological replicates. * indicates p<0.05, two-tailed Student’s t-test. (E) Same data as in D represented as the fusion capacity which focuses on multi-nucleation. The fusion capacity was calculated by including nuclei with n > 3 nuclei per MyHC + cell divided by the total number of nuclei. Plotted is the mean of n = 3 technical replicates per group ± SD. Experiment was repeated with biological replicates. * indicates p<0.05, two-tailed Student’s t-test.
A decrease in the mRNA expression of markers of muscle identity translated to a reduction in the functional capacity to fuse into multi-nucleated myofibers. C2C12-HES3 overexpressing cells and C2C12-Empty controls were differentiated in low-serum conditions, and immunofluorescence performed to determine protein expression and localization of myosin heavy chain (Figure 5C). Both C2C12-Empty and C2C12-HES3 cells exhibited the capacity to fuse into multi-nucleated myotubes that express myosin heavy chain upon terminal differentiation. However, there was a stark difference in the success of the differentiation. After analyzing the percentage of myogenic nuclei, or the nuclei within a myofiber, there was 2.3X more myogenic nuclei in C2C12-Empty as compared to the C2C12-HES3 overexpressing cells (Figure 5D). Furthermore, the fusion index (myogenic nuclei with > 3 nuclei present per myofiber) indicated a significant reduction in the fusion capacity of HES3 overexpressing cells (Figure 5E). HES3 overexpression also modified the kinetics of C2C12 differentiation, with myosin protein being undetectable at day 4 of fusion in C2C12-HES3 overexpressing cells and detectable in C2C12 controls (Figure 5—figure supplement 1A–C). These contrasting fusion capacities cannot be fully attributed to a dominant negative effect on MyoD and inhibition of MyoD expression during fusion initiation. In fact, there were comparable levels of MyoD protein at day 3 of C2C12 fusion. By day 9 there is a trend towards decreased MyoD in HES3 overexpressing C2C12 myotubes (Figure 5—figure supplement 2A–C). Hence, HES3 is inhibiting the differentiation process and promoting a more primitive cellular state in mammalian systems.
HES3 overexpression increases pro-tumorigenic features in cell culture systems
To determine if HES3 overexpression confers properties indicative of tumorigenic capacity in cell culture systems, we analyzed HES3’s impact on C2C12 mouse myoblasts and human rhabdomyosarcoma cells. We focused on the human cell line, Rh30, which is derived from a bone marrow metastasis of ARMS and contains the PAX3-FOXO1 fusion (Hinson et al., 2013). Stable Rh30 cell lines were generated by transfection and selection of either the CMV-Empty or CMV-HES3 expression construct. A qRT-PCR verified HES3 overexpression and that PAX3-FOXO1 was present (Figure 6A–B). This overexpression strategy was pursued because of the low levels of baseline HES3 in Rh30. We examined a panel of rhabdomyosarcoma cell lines and have observed low HES3 levels by RNAseq, and no detectable endogenous HES3 protein expression with commercially available antibodies (data not shown). To determine if HES3 modified the proliferation kinetics of Rh30, cells were plated at a low density and timepoints evaluated from days 1 to 6. We found that in this context HES3 accelerated the cellular accumulation of Rh30 (Figure 6C). Additionally, overexpression of HES3 modified the expression of a panel of genes implicated in metastasis and more aggressive disease. Notably, there was > 300 fold down-regulation of Missing in Metastasis (MTSS1), and an up-regulation of matrix metallopeptidases 3 and 9 (MMP3 and MMP9) (Figure 6D).
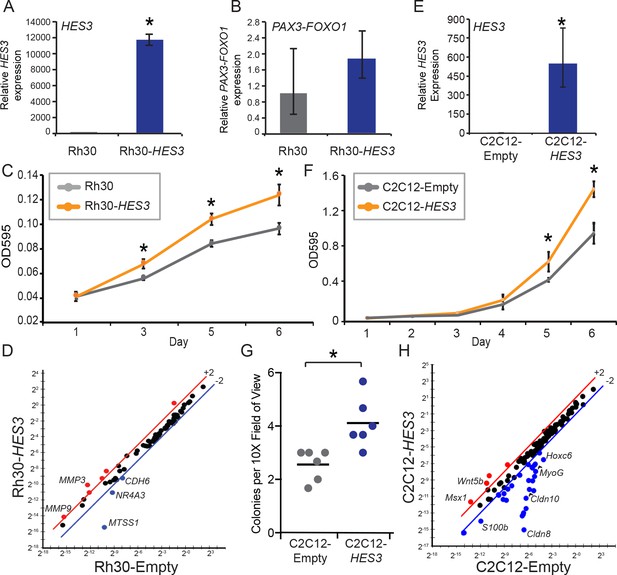
HES3 overexpression promotes pro-tumorigenic features in PAX3-FOXO1 RMS patient cells and mouse myoblasts.
(A) Rh30 cells stably express CMV-HES3 as evaluated by qRT-PCR. SD is derived from technical triplicates. (B) PAX3-FOXO1 levels were assessed by qRT-PCR on the same samples in A. SD is derived from technical triplicates. (C) Cells were seeded at a low density and timepoints taken on Days 1, 3, 5, 6 to assess cellular accumulation. At each timepoint, cells were fixed and stained with crystal violet. Plotted is the absorbance from each timepoint with n = 4 technical replicates ± SE. Each experiment was repeated two times with biological duplicates. (D) qPCR array of metastasis associated genes for Rh30-HES3 versus Rh30-Empty. Red indicates > 2-fold change above the mean, blue indicates > 2-fold change below the mean. (E) C2C12 cells stably expressed CMV-Empty or CMV-HES3, and qRT-PCR confirmed HES3 overexpression. SD is derived from technical triplicates. (F) C2C12 cells were seeded at a low density and timepoints taken on Days 1–6 to assess cellular accumulation. At each timepoint, cells were fixed and stained with crystal violet. Plotted is the absorbance from each timepoint with n = 4 technical replicates ± SD. Each experiment was repeated two times with biological duplicates. (G) Soft-agar colony formation assay was performed over 30 days for C2C12-Empty versus C2C12-HES3. Each data point is the mean of three images per well, across six technical replicates. Black bar indicates the mean. (H) qPCR array of rhabdomyosarcoma associated genes for C2C12-HES3 versus C2C12-Empty. Red indicates > 2-fold change above the mean, blue indicates > 2-fold change below the mean. In all panels * indicates p<0.05, two-tailed Student’s t-test.
Complementary results were obtained when C2C12 cells were transfected with either an empty control vector or CMV-HES3. Stable C2C12-HES3 cells overexpressed HES3 as compared to the C2C12-Empty control (Figure 6E). HES3 was then evaluated for its ability to modulate cellular accumulation. HES3 significantly accelerated the growth kinetics of C2C12 cells when plated at sub-confluent levels over the course of 6 days (Figure 6F). These same cells were plated in suspension in soft agar and cultured for one month to determine the colony formation capacity. HES3 overexpression significantly increased the number of colonies that formed (Figure 6G). This is complemented by data indicating that HES3 alters the expression of genes that are PAX3-FOXO1 direct targets or whose expression is modified by PAX3-FOXO1; including, MyoG, Igf2, and Hoxc6 (Figure 6H) (Khan et al., 1999; Lagha et al., 2010). Potentially, HES3 is acting as a transcriptional activator and repressor, consistent with its role in normal development.
HES3 is overexpressed in patient tumors, predicts reduced survival, and identifies potential therapeutic targets
We evaluated the clinical significance of HES3 by determining HES3 expression levels in human RMS tumors. We analyzed RNA-Seq and HuEx array data from three independent RMS tumor cohorts, together representing the largest collection of over 200 sequenced RMS tumors (Chen et al., 2013; Shern et al., 2014). We found that HES3 had higher expression levels in fusion-positive RMS as compared to fusion-negative RMS (Figure 7A–C). Significantly, in RMS patients, the overexpression of HES3 is associated with reduced overall survival in PAX3/7-FOXO1 fusion-positive RMS patients, but not in fusion-negative RMS patients (Figure 7D). RMS tumors from Shern et al. (2014) were evaluated for HES3 expression levels, and then differentially expressed genes expounded between HES3 high and low tumors to identify molecular targets with potential translational benefit. A subset of clinically relevant kinases and molecular targets was identified as being concordantly overexpressed with HES3; namely, FGFR4, ALK, and PARP1 (Figure 7E). Taken together, these data are consistent with our results showing that her3/HES3 is a PAX3-FOXO1 target, and that HES3 contributes to a more aggressive phenotype in rhabdomyosarcoma.
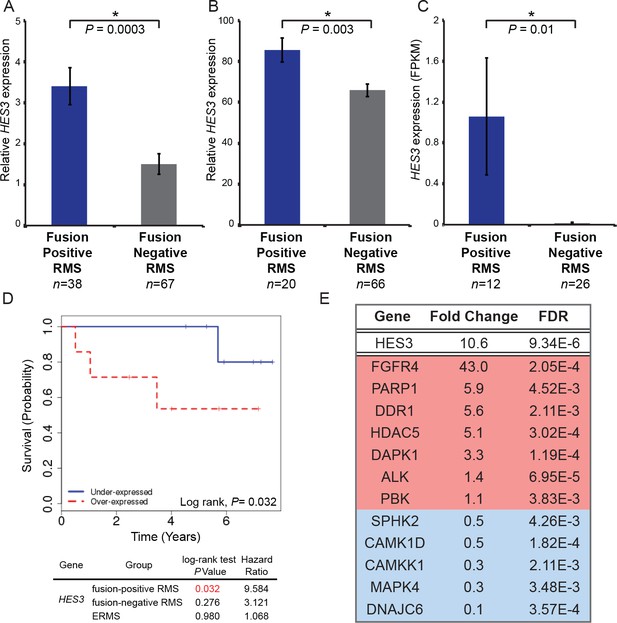
HES3 is overexpressed in fusion-positive RMS patient tumors, predicts reduced overall survival, and identifies potential therapeutic targets.
HES3 expression levels determined in fusion-positive ARMS versus fusion-negative ARMS/ERMS. (A) RNAseq data from Shern et al. (2014) . (B) HuEx data from Triche T, Skapek S; GEO: Accession Number GSE114621. (C) RNAseq data from Chen et al. (2013). p-Values were calculated based on two-tailed Student’s t-test (D) Survival curve of fusion-positive ARMS in the context of HES3 expression status (n = 7 per group). HES3 expression is derived from the HuEx data presented in B. (E) RMS patient tumors from Shern et al. (2014) were stratified based on high (FPKM > 1, n = 25) or low (FPKM < 1, n = 80) HES3 expression and these subsets evaluated for the most differentially expressed genes (FDR < 0.005). Listed are a subset of kinases or other druggable targets that are up-regulated (red) or down-regulated (blue) in the context of HES3 overexpression. Genomic datasets from analysis of human subjects are dbGaP controlled-access data, available to users submitting an approved data access request through the dbGaP Authorized Access System (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login.
Discussion
Despite the identification of the primary oncogenic driver PAX3/7-FOXO1 over 20 years ago, there have been few changes to the primary treatment and prognosis of the disease (Barr et al., 1993; Galili et al., 1993; Shapiro et al., 1993; Davis et al., 1994). Developing complementary ARMS animal models will aid in the understanding of disease biology and identify new therapeutic avenues. Zebrafish ARMS models provide a relevant vertebrate developmental context with experimental advantages such as lineage tracing of the ARMS cell of origin, identifying PAX3-FOXO1 transcriptional targets, and providing a platform for a high-throughput drug screens. Here, we describe a genetic zebrafish model of human disease, in which the human fusion-oncogene, PAX3-FOXO1, is mosaically integrated into the zebrafish genome. We found that the human fusion is oncogenic in zebrafish under the control of the beta actin, CMV, and ubiquitin promoters, resulting in PNETs, RMS, and histologically undifferentiated sarcoma. Surprisingly, PAX3-FOXO1 transformation capacity is not limited to myogenic cells, reflecting the lineage specific control of PAX3 which is relevant in the developing brain and muscle. Specific lineages that have been postulated as possible RMS tissues of origin, such as endothelial, neural crest and differentiated muscle (represented here by the fli1, mitfa, and unc503 promoters) did not develop tumors, highlighting the requirement for high-expressing ubiquitous promoters in this model. Whereas in patient genomic data, PAX7-FOXO1 undergoes copy number gain to increase its expression levels, this is not the case for PAX3-FOXO1. Likely, PAX3-FOXO1 must increase its expression level for transformation via transcriptional upregulation (Barr et al., 1996). To achieve these higher levels of transcription and transformation, we opted to utilize high-level promoters. This threshold was likely not reached with the fli1, mitfa, and unc503 endogenous promoters, or, the lineages and/or temporal kinetic of transgene expression are not relevant to the disease.
In our zebrafish model, the relevant PAX3-FOXO1 RMS cell type is supportive of CMV-mediated gene expression and requires a tp53 mutation. These observations are substantiated by the mouse Pax3-Foxo1 RMS model, in which deletion of Tp53 or Cdkn2a is required for tumor development (Keller et al., 2004). In our zebrafish ARMS model, tp53 mutations allow for PAX3-FOXO1 + cells to survive during embryogenesis by inhibiting apoptosis resulting in later tumor formation (Figure 2—figure supplements 1–2). This may represent a unique necessity in animal models as both fusion-positive RMS vertebrate models of the disease require that tp53 is mutant or lost. In zebrafish, beta-actin-driven PAX3-FOXO1 is sufficient for tumorigenesis, providing a cleaner background for its functional assessment and greater conservation with the genetics of the human disease. One possible explanation is that zebrafish do not have p14 ARF orthologs, making it an unique model to understand the function of the fusion alone. Therefore, we pursued embryonic analyses of PAX3-FOXO1 function using both the beta actin and CMV promoters due to their tumorigenic effects and unique genetic requirements. We found by performing analogous studies with BetaActin-PAX3FOXO1 and CMV-PAX3FOXO1 that these expression constructs behaved comparably in an embryonic environment (Figure 2, Figure 2—figure supplements 1–2, Figure 4, Figure 4—figure supplements 1–2). Focusing on robust and conserved early oncogenic mechanisms that are consistent across multiple promoter models limited the possibility of artifactual observations.
By studying PAX3-FOXO1 signaling in a complex developmental context, we identified a novel fusion-positive RMS biomarker, her3/HES3, that predicts more aggressive disease. her3 is a transcription factor that contains a DNA-binding domain, an orange domain, and a tetrapeptide motif at the C terminus (Hans et al., 2004). In zebrafish and mouse models, her3 is expressed in the neuroepithelial stem cells of the neural plate and inhibits their premature differentiation (Hatakeyama et al., 2004). The human ortholog of her3, HES3, is a member of the basic helix-loop-helix (bHLH) transcription factor family, which includes MYOD1 and MYOG, that are known to be transcriptionally targeted by PAX3-FOXO1 (14, 33). HES3 acts as both a transcriptional repressor and activator, by indirectly or directly influencing transcription depending on its sub-cellular localization. PAX3-FOXO1 regulation of HES3 is likely indirect. Chip-seq data from rhabdomyosarcoma cell culture indicates that PAX3-FOXO1 does not bind the HES3 promoter; however, these results may differ in a dynamic developmental context (Cao et al., 2010). Copy number analysis of fusion-positive patient RMS tumors finds no amplification of HES3, indicating up-regulation is at the transcriptional level. Our data suggest that PAX3-FOXO1 and HES3 function linearly, with HES3 being downstream of PAX3-FOXO1. The exact mechanisms of regulation will be addressed in future studies.
HES3 contributes to more aggressive disease in patients when overexpressed in the context of the PAX3-FOXO1 fusion, likely by a multi-variate platform of: (1) inhibiting terminal muscle differentiation, (2) modifying cellular plasticity to make cells that acquire the PAX3-FOXO1 fusion more tolerant of its expression, and (3) inducing additional transcriptional changes. PAX3-FOXO1 induction of HES3 expression in a mesodermal lineage is ectopic, and HES3 plays no known role in normal somitogenesis or myogenesis. We have found that mis-expression of HES3 during embryonic development inhibits muscle differentiation. We note that HES3 allows for PAX3-FOXO1 cells to inappropriately persist during embryogenesis independently of inhibiting apoptosis. In human fusion-positive RMS, which typically lack TP53 mutations (Shern et al., 2014), HES3 overexpression could represent an alternative mechanism to facilitate PAX3-FOXO1 + cell persistence and tumor initiation events. This role for HES3 promoting PAX3-FOXO1 + embryonic persistence is suggestive of HES3’s capacity in normal development to alter cellular plasticity. PAX3-FOXO1 in combination with HES3 could be challenging the epigenetic identity of mutated cells. Finally, we provide evidence that HES3 may induce a more aggressive and/or metastatic program in PAX3-FOXO1 + patient RMS, including the down-regulation of MTSS1 and up-regulation of MMP3 and MMP9 (34, 35). Previous studies indicate that MTSS1 is a target of the PAX3-FOXO1 fusion (Ebauer et al., 2007); therefore, the modification of MTSS1 expression, and additional genes integral in cell invasion could occur in part through PAX3-FOXO1 induction of HES3. Most significantly, stratifying patient tumor data between HES3 high and low expressing RMS elucidated translational molecular targets with immediate potential applications in the clinic such as co-expression of FGFR4, ALK, and PARP1.
Overall, this new PAX3-FOXO1 zebrafish model of rhabdomyosarcoma identifies her3/HES3 as a mediator of tumorigenesis. Further application of this model determined mechanisms of PAX3FOXO1/HES3 cooperation that result in more aggressive disease. Given the strengths of the zebrafish system, we can now apply it for drug screening and repurposing efforts. We envision the utility of our zebrafish PAX3-FOXO1 RMS model being applied for transplant experiments, where generated tumors can be propagated and grafted into adult zebrafish and in developing zebrafish embryos for further study. Zebrafish embryos containing PAX3-FOXO1 + tumor cells can be exposed to single or combinations of small molecules that effectively couple efficacy for tumor elimination with toxicity data in a vertebrate whole animal model. Additionally, zebrafish cancer models are a complementary approach to ongoing sequencing efforts of patient tumors. The functional significance of cooperating genetic mutations can be determined quickly and robustly. In our study, we find that human PAX3-FOXO1 and HES3 are active in zebrafish, and that biomarkers identified in this system are relevant in the human disease. We anticipate that additional application of this new PAX3-FOXO1 driven tumor model will elucidate novel tumorigenic mechanisms and avenues for treatment.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | ||||
|---|---|---|---|---|---|---|---|---|
| Gene (Homo sapiens) | HES3 | NA | NM_001024598 | |||||
| Gene (Homo sapiens) | PAX3-FOXO1 | PMID:8275086; PMID:8221646 | NM_181457; NM_002015.3 | exons 1–7 of NM_181457 fused to exons 2–3 of NM_002015.3 | ||||
| Gene (Mus musculus) | Pax3 | NA | NM_008781 | |||||
| Gene (Danio rerio) | her3 | NA | NM_131080 | |||||
| Strain, strain background (Danio rerio AB) | AB Wildtype | Zebrafish International Resource Center (ZIRC) | ZIRC:ZL1 | https://zebrafish.org/fish/lineAll.php | ||||
| Strain, strain background (Danio rerio AB/TL) | AB\TL Wildtype | NA | Cross of AB and TL | |||||
| Strain, strain background (Danio rerio TL) | TL Wildtype | ZIRC | ZIRC:ZL86 | https://zebrafish.org/fish/lineAll.php | ||||
| Strain, strain background (Danio rerio WIK) | WIK Wildtype | ZIRC | ZIRC:ZL84 | https://zebrafish.org/fish/lineAll.php | ||||
| Strain, strain background (Danio rerio tp53M214K mutant) | tp53M214K mutant | PMID:15630097; available from ZIRC | ZIRC:ZL1057 | https://zebrafish.org/fish/lineAll.php | ||||
| Genetic reagent | Tol2 transposase mRNA | PMID:16959904 | Injected at 50 ng/uL | |||||
| Cell line (Mus musculus) | C2C12 | ATCC | ATCC:CRL-1772 | Maintained in DMEM + 10% FBS+1X Antimycotic-Antibiotic | ||||
| Cell line (Homo sapiens) | Rh30 | ATCC | ATCC:CRL-2061 | Maintained in RPMI-1640 + 10% FBS+1X Antimycotic-Antibiotic | ||||
| Transfected construct (Homo sapiens) | CMV-HES3 (MYC and FLAG tagged) | Origene | Origene:RC224630 | |||||
| Transfected construct (NA) | CMV-Empty | this paper | HES3 excised using EcoR1 and Mlu1 and re-ligated | |||||
| Antibody | MF20 (mouse monoclonal) | Developmental Studies Hybridoma Bank | DSHB:MF20c | 1:40 in cells, 1:100 in zebrafish embryo whole mounts, 1:1000 Western blot | ||||
| Antibody | alpha-Tubulin DM1A (mouse monoclonal) | Cell Signaling | Cell Signaling:3873 | 1:1000 dilution | ||||
| Antibody | MyoD 5.8A (mouse monoclonal) | Thermo | Thermo:MA5-12902 | 1:1000 dilution | ||||
| Antibody | MYC 71D10 (rabbit monoclonal) | Cell Signaling | Cell Signaling:2278 | 1:1000 dilution | ||||
| Antibody | GFP (rabbit polyclonal) | MBL International Corporation | MBL:598 | 1:1000 dilution | ||||
| Antibody | GFP-488 (rabbit polyclonal) | Thermo | Thermo:A-21311 | 1:500 dilution | ||||
| Antibody | Alexa 488 or 594 secondaries | Thermo | 1:500 dilution | |||||
| Antibody | HRP conjugate secondaries | BioRad | 1:20000 dilution | |||||
| Other | DAPI stain (ProLong Gold Antifade mounting media with DAPI) | Thermo | Thermo:P36931 | |||||
| Recombinant DNA reagent | p5E beta actin | PMID:17937395 | ||||||
| Recombinant DNA reagent | p5E cmv | PMID:17937395 | ||||||
| Recombinant DNA reagent | p5E mcs | PMID:17937395 | ||||||
| Recombinant DNA reagent | p5E ubi | PMID:21138979; available from Addgene | Addgene:27320 | |||||
| Recombinant DNA reagent | p5E unc503 | PMID:23444339; available from Addgene | Addgene:64020 | |||||
| Recombinant DNA reagent | p5E fli1a | PMID:17948311; available from Addgene | Addgene:31160 | |||||
| Recombinant DNA reagent | p5E mitfa | James Lister; available from Addgene | Addgene:81234 | |||||
| Recombinant DNA reagent | pmE beta globin splice acceptor | PMID:15239961 | ||||||
| Recombinant DNA reagent | pmE GFP2A | PMID:17941043 | ||||||
| Recombinant DNA reagent | pmE mCherry2A | PMID:17941043 | ||||||
| Recombinant DNA reagent | p3E PAX3-FOXO1 | this paper | attb2r/attb3 sites added with primers in Supplementary file 3 by high-fidelity PCR | |||||
| Recombinant DNA reagent | p3E Pax3 | this paper | attb2r/attb3 sites added with primers in Supplementary file 3 by high-fidelity PCR | |||||
| Recombinant DNA reagent | p3E HES3 | this paper | attb2r/attb3 sites added with primers in Supplementary file 3 by high-fidelity PCR | |||||
| Recombinant DNA reagent | p3E SV40 late poly A | PMID:17937395 | ||||||
| Recombinant DNA reagent | p3E 2A-mCherry | PMID:23462469; available from Addgene | Addgene:26031 | |||||
| Recombinant DNA reagent | pDONRP2R-P3 (3′ donor vector; attP2R-P3 flanking chlor/ccdB cassette) | Invitrogen | Invitrogen:pDONR P2R-P3 | Used to generate p3E's from this paper | ||||
| Recombinant DNA reagent | pDestTol2pA2 destination vector | PMID:17937395 | ||||||
| Recombinant DNA reagent | BetaActin-GFP2A-pA | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-mCherry2A-pA | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-GFP2A- 2AmCherry | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-mCherry2A-Pax3 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-GFP2A-Pax3 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-mCherry2A-HES3 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | BetaActin-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | CMV-GFP2A-pA | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | CMV-mCherry2A-pA | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | CMV-GFP2A-Pax3 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | CMV-mCherry2A-HES3 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | CMV-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | ubi-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | mitfa-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | fli1-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | unc503-GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Recombinant DNA reagent | SpliceAcceptor- GFP2A-PAX3FOXO1 | this paper | Generated by Gateway Cloning | |||||
| Sequence-based reagent | GFP Morpholino | Gene Tools | Gene Tools: GFP Morpholino | (5' ACAGCTCCTCGCCC TTGCTCACCAT 3') | ||||
| Commercial assay or kit | ApopTag Red In Situ Apoptosis Detection Kit | Millipore | Millipore:S7165 | |||||
| Commercial assay or kit | Affymetrix Zebrafish Gene 1.1 ST Array strip | Affymetrix | Affymetrix:901802 | |||||
| Commercial assay or kit (Mus musculus) | M384 Rhabdomy osarcoma 384 well panel | BioRad | BioRad:M384 Rhabdomyosarcoma | |||||
| Commercial assay or kit (Homo sapiens) | H384 Tumor Metastasis (SAB Target List) 384 well panel | BioRad | BioRad:H384 Tumor Metastasis (SAB Target List) | |||||
| Chemical compound, drug | Geneticin (G418) | Thermo | Thermo:10131027 | Select at 1 mg/mL | ||||
| Software, algorithm | ImageJ | http://imageJ.nih.gov/ij | ||||||
| Software, algorithm | GraphPad Prism 7.0 c | https://www.graphpad.com | ||||||
| Software, algorithm | Rv3.3.1 | https://www.R-project.org | ||||||
Zebrafish Husbandry
Request a detailed protocolDanio rerio were maintained in an Aquaneering aquatics facility according to industry standards. Vertebrate animal work is accredited by AALAC and overseen by the UT Southwestern IACUC committee. AB, WIK, TL, and AB/TL were the wild-type lines used and were obtained from the Zebrafish International Resource Center (https://zebrafish.org). The p53 mutant line, tp53M214K, was a kind gift from Tom Look (Berghmans et al., 2005).
Plasmids and cloning
Request a detailed protocolMouse Pax3 coding sequence and human PAX3-FOXO1 coding sequence was a gift from Steve Skapek. The HES3 coding sequence with a MYC-DDK C-terminal tag was obtained from ORIGENE (RC224630). Pax3, PAX3-FOXO1, and HES3 were cloned into the Gateway expression system (Thermo) by adding 5’ and 3’ ATT sites (attb2r/attb3) with primers and high-fidelity PCR (Thermo; Supplementary file 3). Purified PCR products were cloned into a 3’entry clone using the described protocol (Kendall and Amatruda, 2016). The tol2 kit beta actin promoter, cmv promoter, multiple cloning site, hsp70l promoter, 3’ SV40 late poly A signal construct, and pDestTol2pA2 destination vector were used for construct generation and expression in zebrafish (Kwan et al., 2007). The ubi promoter was a kind gift from Len Zon (Addgene #27320) (Mosimann et al., 2011), the unc503 promoter from Peter Currie (Addgene #64020) (Berger et al., 2013), the fli1 promoter and 3’ entry 2A-mCherry from Nathan Lawson (Addgene #31160 and #26031) (Villefranc et al., 2007; Villefranc et al., 2013), and the mitfa promoter from James Lister (Addgene #81234). Middle entry beta globin intron and splice acceptor was from Koichi Kawakami (Kawakami et al., 2004). The plasmids containing a GFP or mCherry viral 2A sequence were a gift from Steven Leach (Provost et al., 2007), and were sub-cloned into a middle entry Gateway expression system. Tol2 mRNA was synthesized from pCS2FA-transposase which was from Koichi Kawakami (Urasaki et al., 2006). The constructs generated and utilized in the described studies include: BetaActin-GFP2A-pA, BetaActin-mCherry2A-pA, BetaActin-GFP2A-2AmCherry, BetaActin-mCherry2A-Pax3, BetaActin-GFP2A-Pax3, BetaActin-mCherry2A-HES3, BetaActin-GFP2A-PAX3FOXO1, CMV-GFP2A-pA, CMV-mCherry2a-pA, CMV-GFP2A-Pax3, CMV-mCherry2A-HES3, CMV-GFP2A-PAX3FOXO1, ubi-GFP2A-PAX3FOXO1, mitfa-GFP2A-PAX3FOXO1, fli1-GFP2A-PAX3FOXO1, unc503-GFP2A-PAX3FOXO1, SpliceAcceptor-GFP2A-PAX3FOXO1.
Zebrafish embryo injections
Request a detailed protocolZebrafish were injected at the single-cell stage with equimolar ratios of the described DNA constructs. Injection mixes contained 50 ng/µL Tol2 transposase mRNA, 25 or 50 ng/µL of BetaActin-GFP2A-PAX3FOXO1 or CMV-GFP2A-PAX3FOXO1, and equivalent molar amounts of comparative plasmid DNA, 0.1% phenol red, and 0.3X Danieau’s buffer. In knock-down experiments, 300 µM of GFP morpholino (5’ ACAGCTCCTCGCCCTTGCTCACCAT 3’; Gene Tools), was added to the injection mixes in combination with the plasmid DNA.
Zebrafish embryo survival and embryonic cellular persistence
Request a detailed protocolFor survival analysis of embryos, the total number of injected fish was counted, and then the resulting dead or alive embryos subsequently determined at 24, 48, and 72 hpf. Survival curves were plotted using GraphPad Prism 7.0 c (La Jolla, CA). For cellular persistence analysis during embryogenesis, embryos were injected at the single-cell stage with equimolar amounts of plasmid DNA and Tol2 transposase mRNA. At 24 hr post fertilization zebrafish embryos were dechorionated, and then individually imaged at 24 and 72 hr for GFP and mCherry expression using the exact same settings. In ImageJ, the positive pixel threshold was determined and then applied to quantify the number of GFP or mCherry-positive pixels for each embryo.
Zebrafish embryo FACS sorting
Request a detailed protocolFor FACS sorting of GFP-positive zebrafish cells, zebrafish embryos were injected with purified DNA constructs and transposon mRNA at the single-cell stage and allowed to develop for 24 hr, after which they were deyolked and dissociated to single cells (Manoli and Driever, 2012). Using a MoFlo Cell Sorter (Beckman Coulter Life Sciences) live cells were gated, and then GFP + zebrafish cells were sorted out and collected in 1X Phosphate Buffered Saline (Thermo) on ice. Total RNA was immediately isolated using the RNeasy Microkit (Qiagen) and its integrity confirmed by Nanodrop on a spectrophotometer and a Bioanalyzer RNA Nanochip (Agilent). Total RNA was utilized for the Zebrafish Affymetrix Gene 1.1 Microarray strips which was run in-house.
Zebrafish tumor collection, processing for RNA/DNA and histology, and tumor incidence
Request a detailed protocolZebrafish with tumors were humanely euthanized and screened under a Nikon SMZ25 fluorescent stereomicroscope to detect the fluorescent protein indicative of transgene expression. Fresh GFP + tumor tissue was resected and snap frozen in liquid nitrogen. Frozen tissue was subjected to DNA isolation with the DNeasy Kit (Qiagen) or total RNA isolation using the RNeasy Microkit (Qiagen). The remaining tumor specimen was placed in histology cassettes and fixed in 4% paraformaldehyde/1XPBS for 48 hr at 4°C (Fisher). They were then de-calcified in 0.5M EDTA for 5 days and mounted in paraffin blocks for microtome sectioning. Hematoxylin and eosin staining was performed on de-paraffinized slides. For tumor incidence curves, zebrafish were injected and then only those surviving past 30 days of age were included in the analysis. All zebrafish were screened under the fluorescent microscope to determine if they were GFP positive or negative. Zebrafish without GFP fluorescence were considered to be negative for transgene-dependent tumor formation. Zebrafish that were GFP positive were collected as described earlier, and the presence of malignancies confirmed by hematoxylin and eosin staining and visual review by a pathologist.
Detection of apoptosis in whole-mount zebrafish embryos
Request a detailed protocolInjected zebrafish embryos were fixed at 24 hr post fertilization in 4% paraformaldehyde/1XPBS for 24 hr at 4°C or for 2 hr at room temperature in scintillation vials. TUNEL staining was performed using the ApopTag Red In Situ Apoptosis Detection Kit (Millipore). A GFP counter-stain was performed with an anti-GFP antibody at 1:1000 (MBL International Corporation) and an Alexa Fluor 488 goat anti-rabbit IgG secondary (H + L) at 1:500 (Thermo), or an anti-GFP polyclonal antibody directly conjugated to Alexa Fluor 488 at 1:500 (Thermo). Images for rhodamine/GFP were taken using the same settings across embryos on a Nikon SMZ25 fluorescent stereomicroscope, and number of positive pixels in each embryo determined and analyzed in ImageJ.
Microarray
Request a detailed protocolGene expression signatures of injected zebrafish embryos were compared for GFP controls, Pax3, or PAX3-FOXO1 using the Affymetrix Zebrafish Gene 1.1 ST Array strip (Cat #901802). Each sample was run with technical triplicates. Microarray analyses were conducted using the R v3.3.1 environment (R core Team 2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (https://www.R-project.org). Expression profiles were extracted and normalized using Robust Microarray Average (Irizarry et al., 2003). The intersection of statistically significant up-regulated genes for Pax3 and PAX3-FOXO1 conditions as compared to an injected GFP control was determined. Significance was assessed using a Welsh Two Sample t-tests. Genes unique to PAX3-FOXO1 expression were eliminated if they did not contain a human ortholog (based on NBCI HomoloGene build 68 database), and genes were then rank ordered and prioritized based on fold change. Genes with statistically significant expression changes were analyzed using DAVID (https://david.ncifcrf.gov/) (Huang et al., 2009a; Huang et al., 2009b) to identify enriched Gene Ontology (GO) terms (http://www.geneontology.org/).
RNA sequencing
Request a detailed protocolApproximately 2 µg of total RNA was utilized for poly-A RNA enrichment and subsequent library preparation. Sequencing was performed on the Illumina NextSeq 500 Sequencing System with 2 × 75 bp paired end reads. For RNA-seq data, adapter removal and quality-filtering was conducted by Cutadapt (Martin, 2011). Alignment to the reference zebrafish genome, build GRCz10, was performed by Bowtie2 (Langmead and Salzberg, 2012), an ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences. TopHat2 (Kim et al., 2013) was used for alignment. De novo assembly of reads into transcripts (including mRNA, lincRNA, and many other RNA species) and differential expression analysis were performed by Cufflinks and Cuffdiff (Trapnell et al., 2012). To detect the human PAX3-FOXO1 fusion, we realigned all reads to the known junction sequence within PAX3-FOXO1 fusion, which is shown in Figure 1—figure supplement 2. To reduce false positives, we required that (1) junction-spanning reads should contain no mismatch with known PAX3-FOXO1 junction sequence and (2) junction-spanning reads with less than 6 bp matches on either gene was discarded, as suggested by an earlier study (Li et al., 2011).
Immunofluorescence
Request a detailed protocolIn cells, the MF20 primary antibody was utilized for detection of myosin heavy chain at a dilution of 1:40 (Developmental Studies Hybridoma Bank; DSHB) in combination with an Alexa Fluor 488 goat anti-mouse IgG (H + L) secondary antibody at a dilution of 1:500 (Thermo). DNA was detected using ProLong Gold Antifade mounting media with DAPI (Thermo). MF20 staining in cells was performed as in Kendall et al., 2012. In zebrafish embryo whole mounts, the MF20 antibody was used for the detection of myosin at a concentration of 1:100 in combination with an anti-GFP polyclonal antibody directly conjugated to Alexa Fluor 488 at 1:500 (Thermo). The secondary antibody used was Alexa Fluor 594 goat anti-mouse IgG (H + L) at 1:500 (Thermo). Images were taken on a Keyence BZ-X700 fluorescent microscope.
RNA extraction, cDNA synthesis, and qRT-PCR
Request a detailed protocolTotal RNA from zebrafish embryos, tumor tissues, and cells was isolated using the RNeasy Mini or Microkit (QIAGEN). cDNA was reverse transcribed from 200 ng-2µg of total RNA with the RT2 HT First Strand Synthesis kit (QIAGEN). qRT-PCR was performed on an ABI 7900HT using the SYBRGreen Master Mix (BioRad) and a 10 µL total volume in 384 well plates. See Supplementary file 3 for primer sequences. Delta-delta Ct was calculated, and the calibrator plotted on the far left for every graph. Error bars indicate standard deviation and a student’s t-test was performed on normalized Ct replicates to determine significance. The following BioRad qPCR arrays were utilized, M384 Rhabdomyosarcoma for mouse C2C12 cells and H384 Tumor Metastasis (SAB Target List) for Rh30 human RMS cells. Input cDNA was synthesized using the RT2 HT First Strand Synthesis kit (QIAGEN) including a synthetic mRNA as an internal quality control (BioRad). qRT-PCR arrays were run on the BioRad CFX384. Plotted is the fold change above and below the mean gene expression levels from all genes on the array.
Protein extraction and western blots
Request a detailed protocolCells were harvested and lysed in radioimmunoprecipitation assay buffer (RIPA) with cOmplete Mini Protease Inhibitor Cocktail inhibitors (Sigma), and protein levels quantified using Qubit 3.0 Fluorometer (Thermo). Twenty micrograms of protein was denatured and loaded on a 4–20% gradient gel (BioRad), and then transferred to PVDF membranes at 4°C. Membranes were blocked in Casein + 0.1% Tween-20 (Thermo), and incubated overnight at 4°C with the following antibodies with agitation: MYOD monoclonal antibody 5.8A at 1:1000 (Thermo), MYC 71D10 monoclonal antibody at 1:1000 (Cell Signaling), alpha-Tubulin DM1A monoclonal antibody at 1:1000 (Cell Signaling), and MF20 at 1:1000 (Developmental Studies Hybridoma Bank). Goat anti-mouse IgG (H + L)-HRP conjugate and goat anti-rabbit IgG (H + L)-HRP conjugate secondaries were utilized at 1:20,000 (BioRad). Signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Fisher). Membranes were imaged on the BioRad GelDoc XR + and quantification was performed in ImageJ.
Cell culture and transfection
Request a detailed protocolC2C12 and Rh30 were a kind gift from Steve Skapek. All cell lines were mycoplasma tested with the MycoAlert Mycoplasma Detection Kit (Lonza) within 6 months of their use and were negative. C2C12 was maintained in DMEM (Gibco) with 10% Fetal Bovine Serum (FBS; Sigma) and 1X Antibiotic-Antimycotic (Gibco) at 37°C in 5% CO2. Rh30 were maintained in RPMI-1640 (ATCC) with 10% FBS (Sigma) and 1X Antibiotic-Antimycotic (Gibco) at 37°C in 5% CO2. Differentiation media for C2C12s included DMEM (Gibco) with 2% Horse Serum (Sigma) and 10 µg/mL of insulin (Fisher). Cells were transfected with Fugene HD Transfection reagent (Promega) of 3 µL:1 µg of DNA. Cells were selected for 30 days as sub-populations in 1 mg/mL of G418 (Thermo) prior to experiments and were maintained in 1 mg/mL of G418.
Myogenic differentiation in cell culture
Request a detailed protocolC2C12 cells were differentiated by plating 50,000 or 150,000 cells per well on 0.01% porcine/PBS-coated plates (24 wells or six welsl) in growth media + G418. If plating density varied from this it is indicated in the figure. For immunofluorescence C2C12 cells were plated on porcine coated glass coverslips. After 24 hours, cells were washed in 1XPBS, and differentiation media + G418 added. Cells were then fused for 6 days in differentiation media, with fresh media being added every other day. For fusion timepoints, cells were collected and pelleted after incubation in TrypLE Express Enzyme (Fisher), and frozen at −80°C for later total RNA isolation. For immunofluorescence, cells were fixed directly in the well with a 1:1 ratio of 4% paraformaldehyde:media at 37°C for 15 min. Subsequent washing and staining steps are detailed in Kendall et al., 2012.
Cellular proliferation assays
Request a detailed protocolCells were seeded at 5,000 cells per well for C2C12 and Rh30 in a 24-well plate with four replicates per timepoint. Cells were maintained in GM supplemented with 1 mg/mL of G418 (Gibco) during the course of the experiment. At each timepoint, cells were fixed with 4% PFA for 15 min at room temperature, and stained with 0.0025% crystal violet (Alfa Aesar) in 20% methanol (Sigma) for 15 min. They were then rinsed and crystals were solubilized with 10% Acetic Acid (VWR). Absorbance was read on a plate reader at 595 nm.
Soft agar colony formation assay
Request a detailed protocolColony formation assay strategy was adapted from Borowicz et al. (2014). A bottom layer of 1.2% noble agar (Difco) in GM was plated with 1 mg/mL of G418 and allowed to solidify, followed by a top layer of 0.6% noble agar in GM with 1 mg/mL of G418 with 5000 cells per well in a six-well plate. Each well was maintained in 100 µL of GM + G418 that was changed three times per week. On day 30, wells were imaged for analysis, with three images being taken at 10X per well and then averaged. Six technical replicates were performed per sub-clonal population. Three biological replicates representing independent sub-clonal populations were included per experiment.
Statistics
Statistical analysis for survival and tumor incidence curves, and Fisher’s exact test, was performed using GraphPad Prism 7.0 c (La Jolla, CA). All other calculations were performed using two-tailed student’s t test in Microsoft Excel Version 15.38. Sample sizes are provided in the figures or figure legends.
Data availability
-
Exon level expression analysis from rhabdomyosarcoma patient tumors.Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE11462).
-
Data files for SJRHB RNA-SeqPublicly available at the European Genome-phenome Archive (accession no. EGAD00001000878).
-
Genomic sequencing of Pediatric Rhabdomyosarcomaphs000720.v2.p1; Genomic datasets from analysis of human subjects are dbGaP controlled-access data, available to users submitting an approved data access request through the dbGaP Authorized Access System (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login).
-
Genomic sequencing of Pediatric Rhabdomyosarcomaphs000720.v2.p1; Genomic datasets from analysis of human subjects are dbGaP controlled-access data, available to users submitting an approved data access request through the dbGaP Authorized Access System (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login).
References
-
In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcomaHuman Molecular Genetics 5:15–21.https://doi.org/10.1093/hmg/5.1.15
-
The soft agar colony formation assayJournal of Visualized Experiments 92:e51998.https://doi.org/10.3791/51998
-
Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcomaCancer Research 54:2869–2872.
-
Matrix metalloproteinases and tumor metastasisCancer and Metastasis Reviews 25:9–34.https://doi.org/10.1007/s10555-006-7886-9
-
Muscling in: Uncovering the origins of rhabdomyosarcomaNature Medicine 16:171–173.https://doi.org/10.1038/nm0210-171
-
Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancerInternational Journal of Oncology 31:461–466.https://doi.org/10.3892/ijo.31.2.461
-
Zebrafish as a model for the study of solid malignanciesMethods in Molecular Biology 1451:121–142.https://doi.org/10.1007/978-1-4939-3771-4_9
-
Dantrolene enhances antisense-mediated exon skipping in human and mouse models of duchenne muscular dystrophyScience Translational Medicine 4:164ra160.https://doi.org/10.1126/scitranslmed.3005054
-
The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructsDevelopmental Dynamics 236:3088–3099.https://doi.org/10.1002/dvdy.21343
-
Effects of RAS on the genesis of embryonal rhabdomyosarcomaGenes & Development 21:1382–1395.https://doi.org/10.1101/gad.1545007
-
Fast gapped-read alignment with Bowtie 2Nature Methods 9:357–359.https://doi.org/10.1038/nmeth.1923
-
A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesisDisease Models & Mechanisms 5:95–106.https://doi.org/10.1242/dmm.007401
-
Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndromeJournal of the National Cancer Institute 43:1365–1373.
-
PAX3-FOXO1 fusion gene in rhabdomyosarcomaCancer Letters 270:10–18.https://doi.org/10.1016/j.canlet.2008.03.035
-
Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolationCold Spring Harbor Protocols 2012:pdb.prot069633.https://doi.org/10.1101/pdb.prot069633
-
RAS signaling dysregulation in human embryonal RhabdomyosarcomaGenes, Chromosomes and Cancer 48:975–982.https://doi.org/10.1002/gcc.20702
-
Rhabdomyosarcomas in adults and children: an updateArchives of Pathology & Laboratory Medicine 130:1454–1465.https://doi.org/10.1043/1543-2165(2006)130[1454:RIAACA]2.0.CO;2
-
Myogenesis and Rhabdomyosarcoma the Jekyll and Hyde of skeletal muscleCurrent Topics in Developmental Biology 94:197–234.https://doi.org/10.1016/B978-0-12-380916-2.00007-3
-
Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar RhabdomyosarcomaCancer Research 53:5108–5112.
-
Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reactionCancer Research 49:6324–6327.
-
Gateway compatible vectors for analysis of gene function in the zebrafishDevelopmental Dynamics 236:3077–3087.https://doi.org/10.1002/dvdy.21354
Article and author information
Author details
Funding
Cancer Prevention and Research Institute of Texas (RP120685-P3)
- James F Amatruda
American Association for Cancer Research (QuadW Foundation-AACR Fellowship for Clinical/Translational Sarcoma Research)
- Genevieve C Kendall
Alex's Lemonade Stand Foundation for Childhood Cancer (Young Investigator Grant)
- Genevieve C Kendall
Hartwell Foundation (Postdoctoral Fellowship)
- Genevieve C Kendall
Ligue National Contre le Cancer
- Olivier Delattre
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This project is funded by a grant from Cancer Prevention and Research Institute of Texas (RP120685-P3) to JFA and the Ligue National Contre le Cancer (équipe labellisée). G Kendall is funded by a CPRIT postdoctoral fellowship through the UTSW Cancer Intervention and Prevention Discoveries training program, a QuadW-AACR Postdoctoral Fellowship for Clinical/Translational Sarcoma Research, a Young Investigator Grant from Alex’s Lemonade Stand, and a Hartwell Foundation Postdoctoral Fellowship. S Watson was a recipient of a Ph.D. fellowship from Institut Curie. We thank the UTSW Molecular Pathology core and the UTSW Flow Cytometry core for exceptional services and for their expertise. We thank Tim Triche and Javed Khan for sharing RMS genomic data, and members of the Amatruda lab.
Ethics
Human subjects: This study analyzed RNASeq and HuEx data from deidentified patient rhabdomyosarcoma samples. Samples were deidentified and histologic diagnosis and clinical information were compiled. All studies involving human subjects were conducted under a protocol approved by the UT Southwestern Institutional Review Board (approval # STU 102011-034).
Animal experimentation: UT Southwestern is a registered research facility with the US Department of Agriculture and is committed to comply with the Guide for the Care and Use of Laboratory Animals and all applicable federal, state and local regulations. These pertain to the purchase, transportation, housing and research use of animals. In addition, UT Southwestern is an AAALAC accredited institution that has developed institutional standards for the humane care and use of animals, which are maintained through published policies. Zebrafish research described in this study has been approved and conducted under the oversight of the UT Southwestern Institutional Animal Care and Use Committee (approval # 2016-101805). UT Southwestern uses the "Guide for the Care and Use of Laboratory Animals" when establishing animal research standards.
Copyright
© 2018, Kendall et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 4,131
- views
-
- 475
- downloads
-
- 53
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 53
- citations for umbrella DOI https://doi.org/10.7554/eLife.33800






