Agonist-mediated switching of ion selectivity in TPC2 differentially promotes lysosomal function
Figures
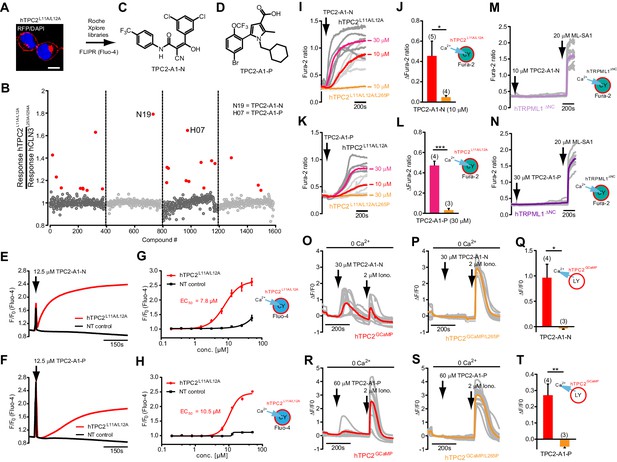
Identification of novel small molecule agonists of TPC2.
(A) The chemical screening was performed using cells stably expressing human TPC2 re-routed to the plasma membrane (hTPC2L11A/L12A) and loaded with the fluorescent Ca2+ indicator, Fluo-4 (CY = cytosol). For counter-screening, a stable cell line expressing cell surface human CLN3 (hCLN3L253A/I254A) was used. (B) A library comprising 80.000 compounds was screened. Shown are the primary screen results of four plates (with 384 compounds, each) where the response to a given compound in cells expressing hTPC2L11A/L12A is expressed relative to that in cells expressing hCLN3L253A/I254A. Primary hits are shown as red spots. Subsequent confirmatory and follow-up assays led to the identification of the two hit compounds, N19 (TPC2-A1-N) and H07 (TPC2-A1-P). (C–D) Figures of TPC2-A1-N and TPC2-A1-P. (E–F) Representative FLIPR-generated Ca2+ signals (Fluo-4) in TPC2L11A/L12A–expressing cells (red lines) or non-transfected (NT) control cells (black) after addition of TPC2-A1-N (E) or TPC2-A1-P (F). (G–H) Concentration-effect relationships for Ca2+ increases (Fluo-4) in response to different concentrations of TPC2-A1-N (G) and TPC2-A1-P (H). Each concentration was tested in duplicates in two to four independent experiments, each. (I) Representative Ca2+ signals recorded from HeLa cells loaded with the ratiometric Ca2+ indicator, Fura-2 and stimulated with the indicated concentration of TPC2-A1-N. Cells were transiently transfected with plasma-membrane-targeted human TPC2 (hTPC2L11A/L12A) or a pore dead mutant (TPC2L11A/L12A/L265P). Colored lines represent the mean response from a population of cells. Traces in grey represent responses of single cells. (J) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM) with the number of independent transfections in parentheses. An unpaired t-test was applied. *p<0.05. (K–L) Similar to I and J except that cells were stimulated with TPC2-A1-P. ***p<0.001. Traces in grey represent single cells responding to TPC2-A1-P. (M–N) Representative Ca2+ signals recorded from HeLa cells transiently transfected with human TRPML1ΔNC. Cells were sequentially stimulated with TPC2-A1-N (M) or TPC2-A1-P (N) and the TRPML agonist ML-SA1. (O–T) Ca2+ signals recorded from HeLa cells transiently transfected with GCaMP6(s) fused to human TPC2 (hTPC2GCaMP) or a pore-dead mutant (hTPC2GCaMP/L265P). Cells were sequentially stimulated with TPC2-A1-N (O and P) or TPC2-A1-P (R and S) and the Ca2+ ionophore, ionomycin in the absence of extracellular Ca2+. Statistical analysis of the change in fluorescence intensity (mean ± SEM; Q and T). An unpaired t-test was applied. *p<0.05 and **p<0.01. CY = cytosol, LY = lysosome.
-
Figure 1—source data 1
Identification of TPC2 hit compounds.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-data1-v3.xlsx
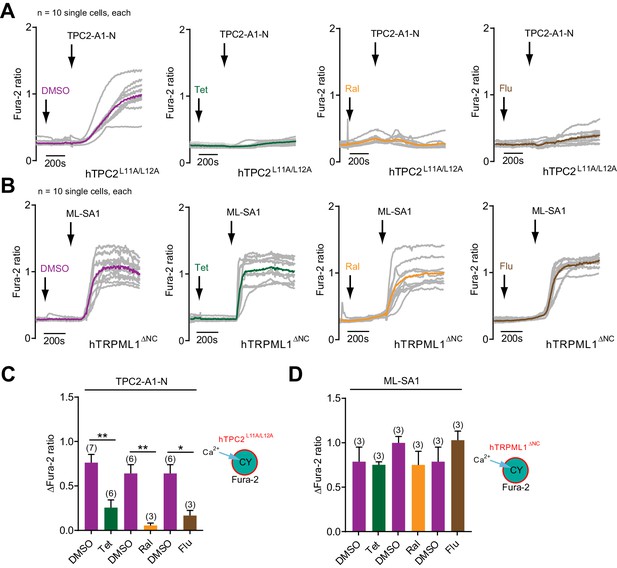
Effect of different TPC2 channel blockers on TPC2-A1-N and ML-SA1 mediated changes in intracellular calcium.
(A–B) Representative Ca2+ signals recorded from HeLa cells loaded with the ratiometric Ca2+ indicator, Fura-2 and sequentially stimulated with the indicated channel blockers, tetrandrine (Tet, 10 µM) and raloxifene (Ral, 10 µM), fluphenazine (Flu, 10 µM) or vehicle (DMSO), followed by TPC2-A1-N (10 µM) or ML-SA1 (3.2 µM). Cells were transiently transfected with human TPC2 (hTPC2L11A/L12A, A) or human TRPML1 (hTRPML1ΔNC, B) targeted to the plasma membrane. Coloured lines represent the mean response from a population of cells. (C–D) Statistical analysis of the change in fluorescence ratio (mean ± SEM) of experiments as shown in A-B. An unpaired t-test was applied. *p<0.05 and **p<0.01.
-
Figure 1—figure supplement 1—source data 1
Effect of different TPC2 channel blockers on TPC2-A1-N and ML-SA1 mediated changes in intracellular calcium.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp1-data1-v3.xlsx
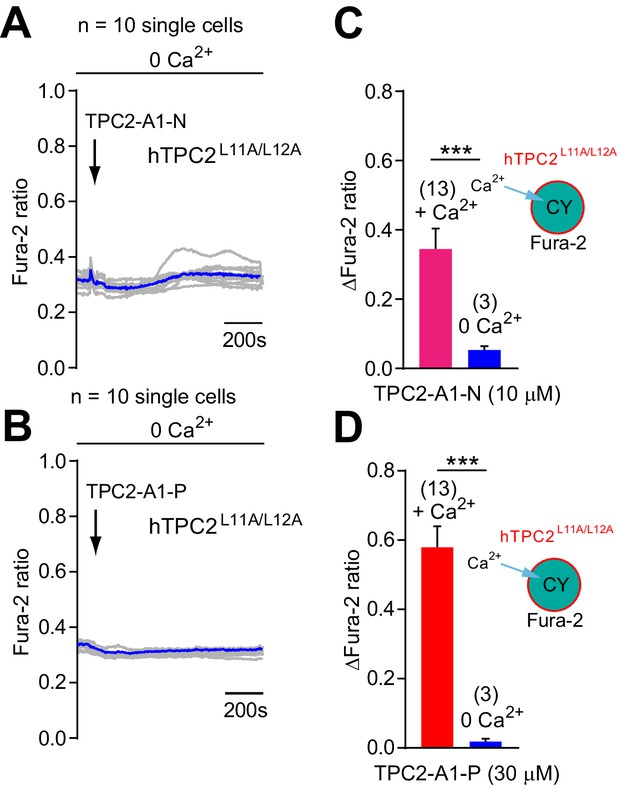
TPC2-A1-N and TPC2-A1-P mediated changes in intracellular calcium in the absence of extracellular calcium.
(A–B) Representative Ca2+ signals recorded from HeLa cells loaded with the ratiometric Ca2+ indicator, Fura-2 and stimulated with TPC2-A1-N (10 µM, A) or TPC2-A1-P (30 µM, B) in nominally Ca2+-free medium. Cells were transiently transfected with plasma membrane targeted human TPC2 (hTPC2L11A/L12A). (C–D) Statistical analysis of the change in fluorescence ratio (mean ± SEM) of experiments as shown in A-B. An unpaired t-test was applied. **p<0.01 and ***p<0.001.
-
Figure 1—figure supplement 2—source data 1
TPC2-A1-N and TPC2-A1-P mediated changes in intracellular calcium in the absence of extracellular calcium.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp2-data1-v3.xlsx
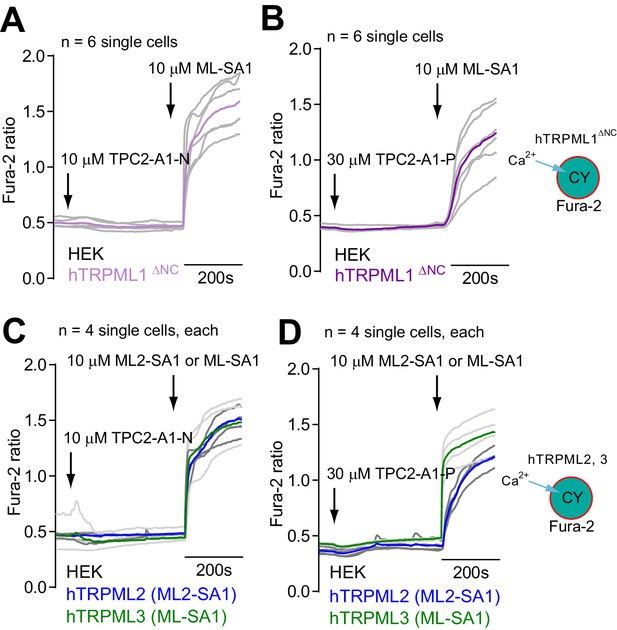
Effects of TPC2-A1-N and TPC2-A1-P on TRPML1, 2, and 3.
(A–B) Representative Ca2+ signals recorded from HEK293 cells loaded with the ratiometric Ca2+ indicator, Fura-2 and sequentially stimulated with TPC2-A1-N (10 µM, (A) or TPC2-A1-P (30 µM, (B) and the TRPML agonist ML-SA1 (10 µM). Cells were transiently transfected with human TRPML1ΔNC. (C–D) Experiment as in A-B, but HEK293 cells transiently transfected with human TRPML2, or TRPML3. Cells were sequentially stimulated with TPC2-A1-N (10 µM, (C) or TPC2-A1-P (30 µM, (D) and the TRPML agonist ML-SA1 (10 µM) or the TRPML2 selective agonist ML2-SA1 (10 µM).
-
Figure 1—figure supplement 3—source data 1
Effects of TPC2-A1-N and TPC2-A1-P on TRPML1, 2, and 3.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp3-data1-v3.xlsx
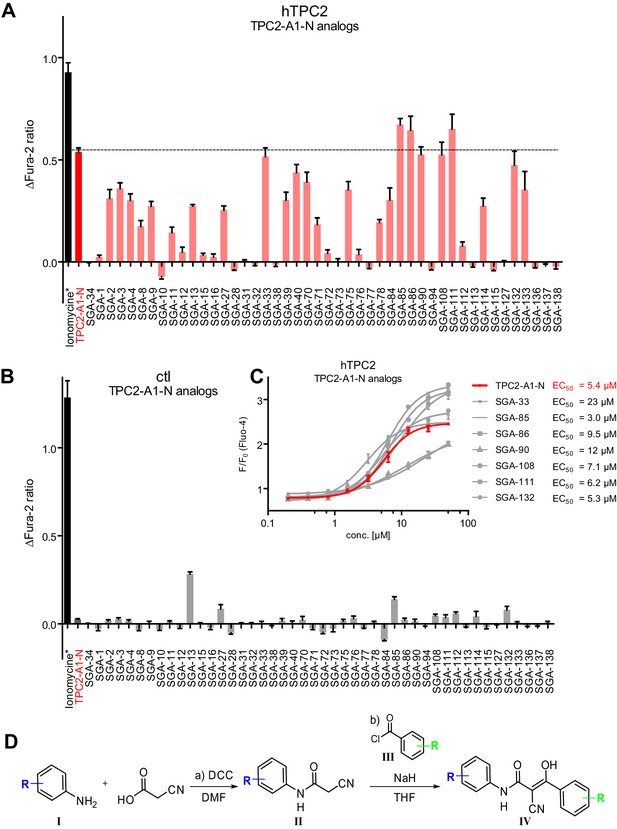
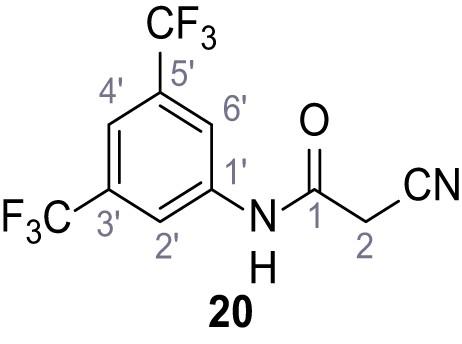
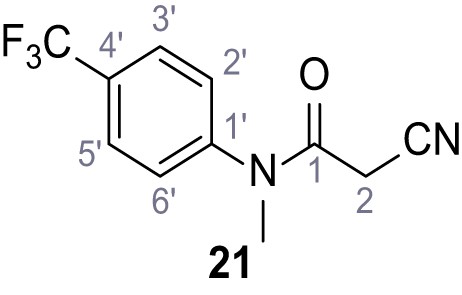
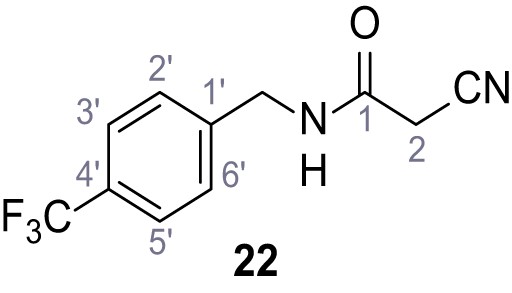
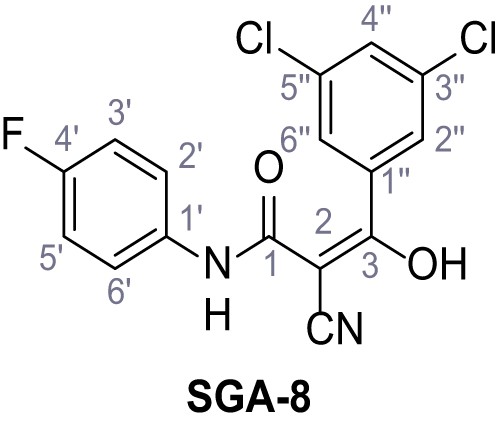
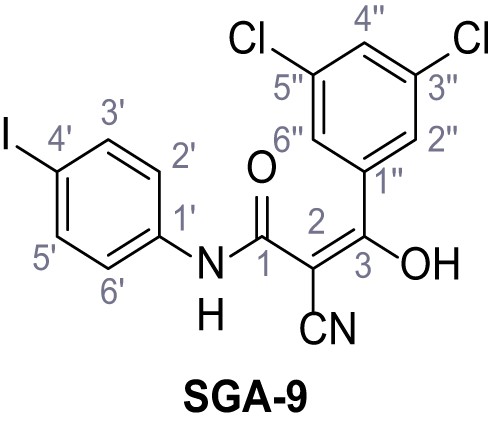
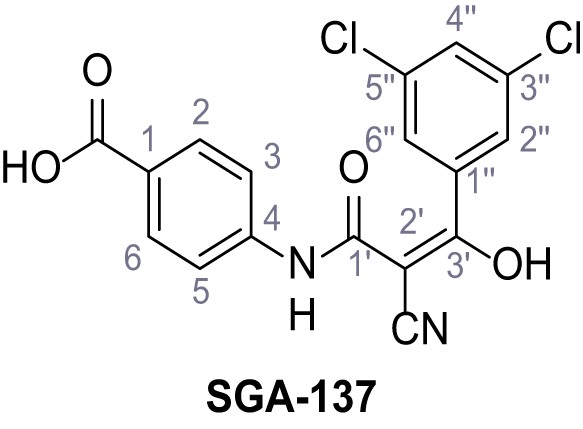
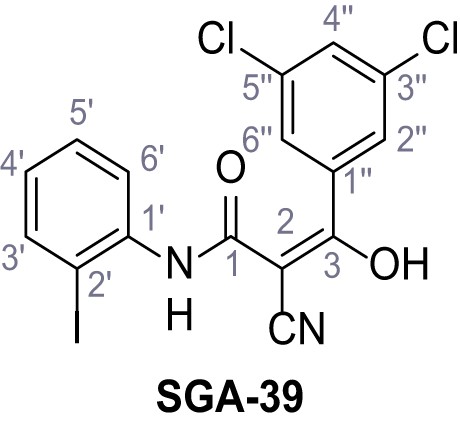
Synthesis and functional analysis of TPC2-A1-N analogs.
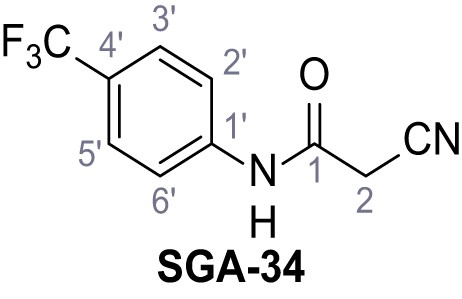
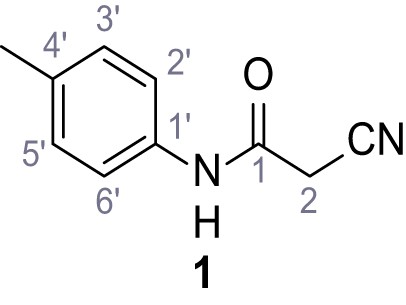
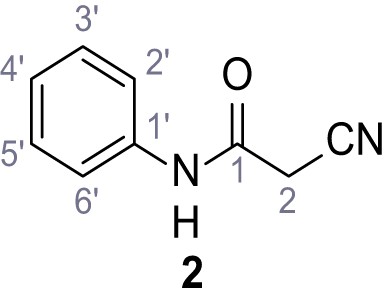
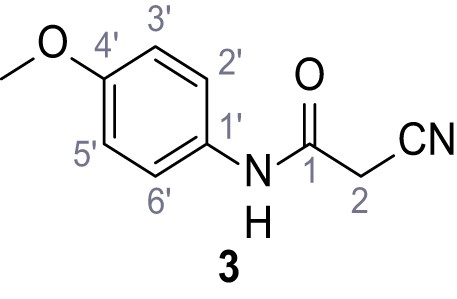
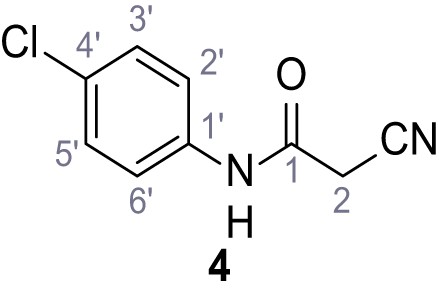
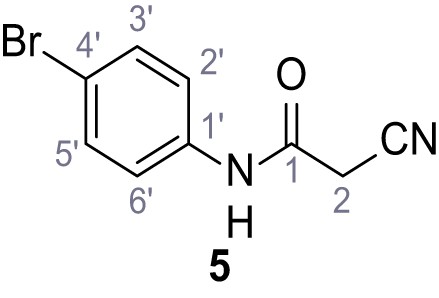
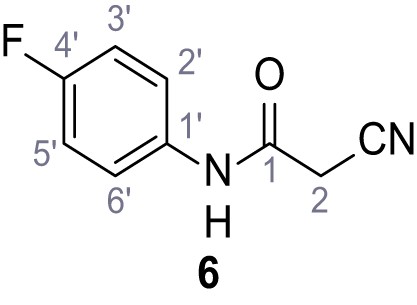
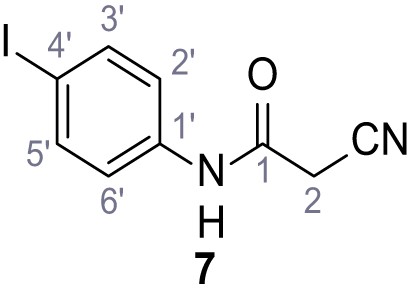
(A–B) Fura-2 based Ca2+ imaging results showing the effect of TPC2-A1-N and its analogs (10 µM) on HEK293 cells transiently transfected with hTPC2L11/12A-YFP (A, red) and control cells (B, grey). Red arrow indicates effect on control cells and green arrow no effect on these. Mean values normalized to basal (400 s after compound application)± SEM of at least three independent experiments with 3–10 cells each are shown. (C) Fluo-4 based concentration-effect relationships for Ca2+ increases in response to different concentrations of the strongest analogs (grey) compared to TPC2-A1-N (red). Each concentration was tested in duplicates in two to four independent experiments, each. (D) General scheme for the synthesis of TPC2-A1-N and its analogs (IV), derived from different aniline (I) and benzoic acid chloride (III) building blocks. Standard amide coupling conditions using different anilines (I) yield amides of type II). Following acylation with different acid chlorides (III) results in TPC2-A1-N and analogs (IV); a) substituted aniline (I), 1.0 eq.), 2-cyanoacetic acid (1.0 eq.), DCC (1.0 eq.) in DMF, 0°C – rt, 1 hr; b) amide (II), 1.0 eq.), appropriate acid chloride (III), 1.1 eq.), NaH (2.3 eq.) in THF, 0°C, 1.25 hr.
-
Figure 1—figure supplement 4—source data 1
Functional analysis of TPC2-A1-N analogs.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp4-data1-v3.xlsx
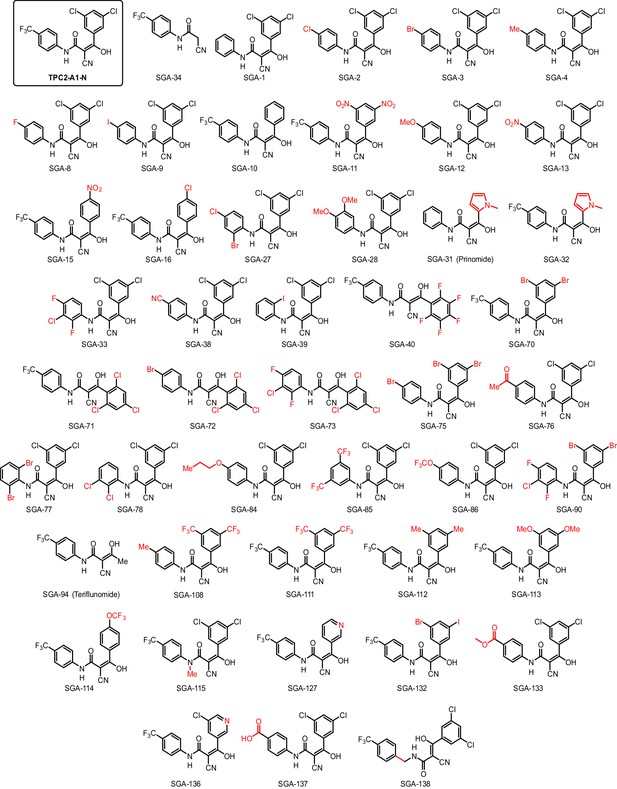
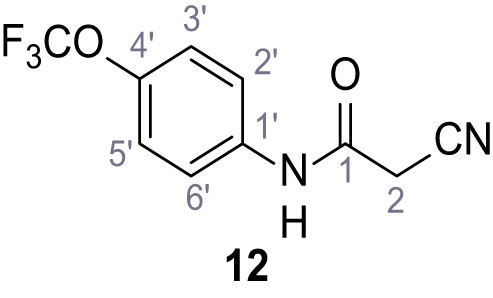
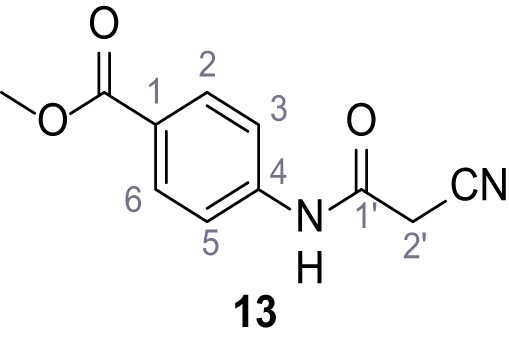
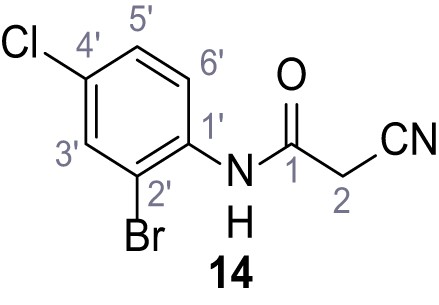
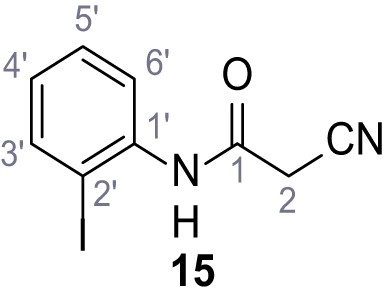
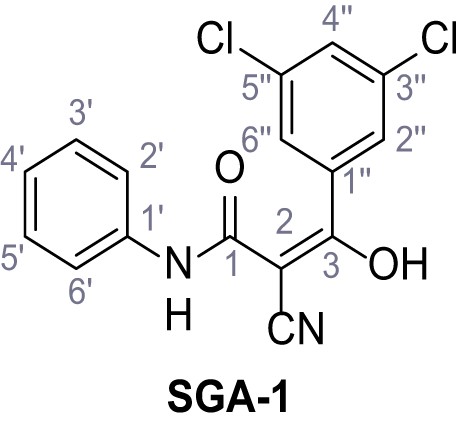
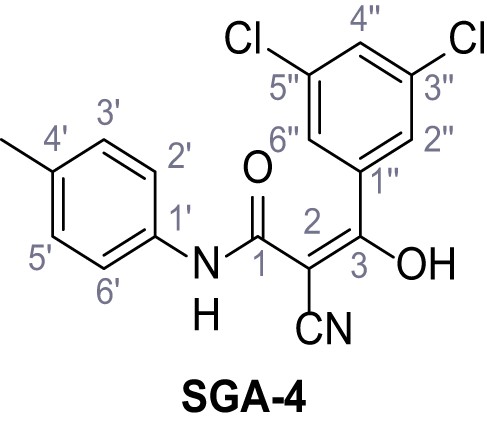
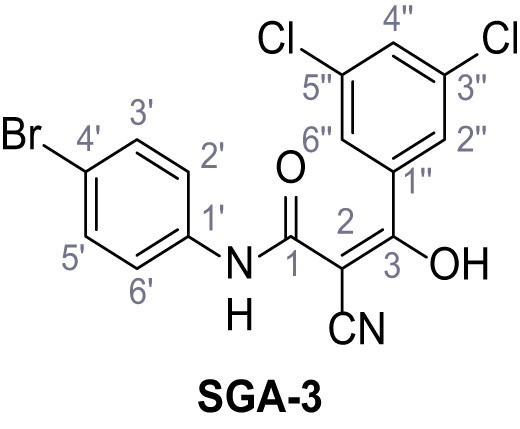
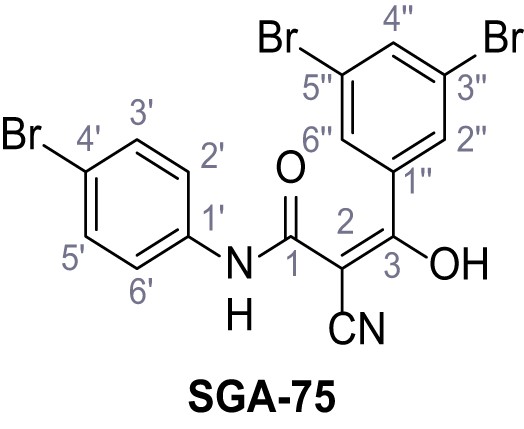
Structures of TPC2-A1-N and analogs.
In total 46 analogs were synthesized. Differences in single structures are marked in red.

Synthesis and functional analysis of TPC2-A1-P analogs.
(A–B) Fura-2 based Ca2+ imaging results showing the effect of TPC2-A1-P and its analogs (10 µM) on HEK293 cells transiently transfected with hTPC2L11/12A-YFP (A, red) and control cells (B, grey). Mean values normalized to basal (400 s after compound application)± SEM of at least three independent experiments with 3–10 cells each are shown. (C) Structures of TPC2-A1-P and analogs. In total 19 analogs were synthesized. Differences in single structures are marked in red. (D) General scheme for the synthesis of TPC2-A1-P and its analogs (IV and V), derived from different α-halogenated acetophenone (I), β-ketoester (II) and primary amine building blocks. Halogenated acetophenones (I) and β-ketoesters (II) give intermediates (III), which directly undergo a Paal-Knorr pyrrole synthesis using different primary amines to yield esters of type IV). Alkaline deprotection yields the free carboxylic acids (V); a) α-halogenated acetophenone (I), 1.0 eq.), β-ketoester (II), 1.1 eq.), NaH (1.5 eq.), KI (1.0 eq.) in THF 0°C – rt, 2 hr; then appropriate primary amine (2.0 eq.) in acetic acid, 80°C, 18 hr; b) appropriate ester (IV), 1.0 eq.), LiOH (10 eq.) in dioxane/H2O (5:1), mw, 1–18 hr.
-
Figure 1—figure supplement 6—source data 1
Functional analysis of TPC2-A1-P analogs.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp6-data1-v3.xlsx
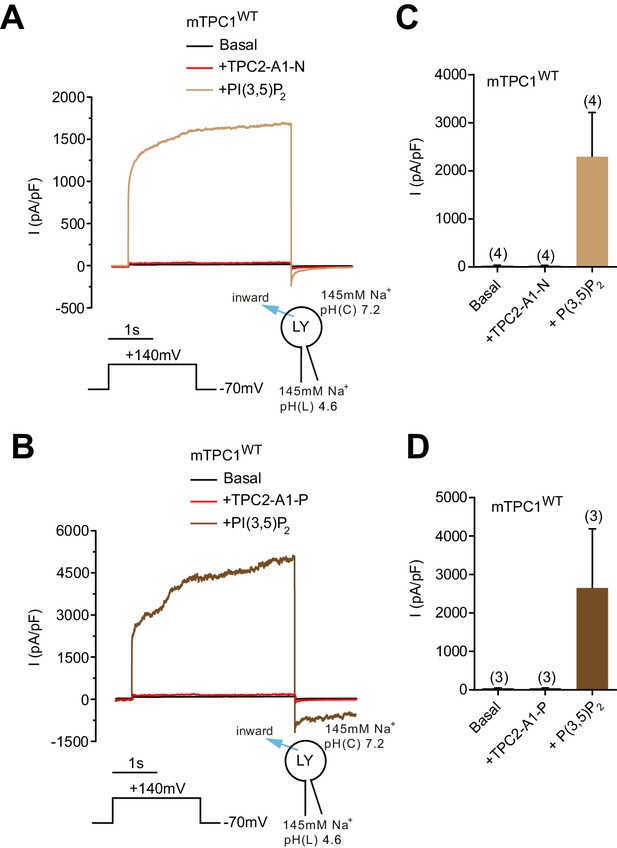
Effect of TPC2-A1-N and TPC2-A1-P on TPC1.
(A–B) Representative currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing mouse TPC1 (MmTPC1-YFP) evoked by TPC2-A1-N (10 µM, A) or TPC2-A1-P (10 µM, B), followed by PI(3,5)P2 (1 µM). Recordings were carried out using standard bath and pipette solutions and applying a one-step protocol +140 mV over 2 s at a holding potential of −70 mV. (C–D) Statistical analysis of current densities (mean ± SEM, n = 3, each) recorded at +140 mV for endo-lysosomes expressing mTPC1 activated with TPC2-A1-N (C) or TPC2-A1-P (D), followed by PI(3,5)P2 application.
-
Figure 1—figure supplement 7—source data 1
Effects of TPC2-A1-N and TPC2-A1-P on TPC1 in endo-lysosomal patch-clamp experiments.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-figsupp7-data1-v3.xlsx
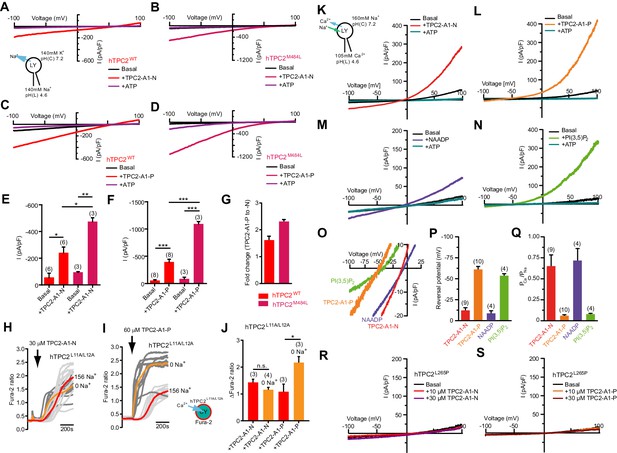
TPC2 agonists alter Ca2+/Na+ permeability of TPC2.
(A–B) Representative TPC2-A1-N-evoked currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing human TPC2 (hTPC2) (A) or a gain-of-function variant, (hTPC2M484L) (B). Currents were obtained before and after addition of 10 µM TPC2-A1-N in the absence or presence of 1 mM ATP. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV. (C–D) Similar to A and B except that TPC2-A1-P (10 µM) was used. (E–G) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV for endo-lysosomes expressing TPC2 or TPC2M484L for TPC2-A1-N (E) and TPC2-A1-P (F), and the fold-change in current evoked by TPC2-A1-P versus TPC2-A1-N in the two variants (G). Red bars are WT and maroon bars are mutant. An unpaired t-test was applied. *p<0.05, **p<0.01, and ***p<0.001 (n > 10 endo-lysosomes per condition). (H–I) Ca2+ signals evoked by TPC2-A1-N (H) or TPC2-A1-P (I) in Fura-2-loaded HEK293 cells stably expressing hTPC2L11A/L12A. Recordings (mean responses from three to four independent experiments) were obtained in standard or Na+-free extracellular solution. (J) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM). An unpaired t-test was applied. *p<0.05. (K–N) Agonist-evoked cation currents from enlarged endo-lysosomes isolated from HEK293 cells stably expressing human TPC2 under bi-ionic conditions: 160 mM Na+ in the cytosol (bath) and 105 mM Ca2+ in the lumen (pipette). Representative current-voltage curves before and after stimulation with either 10 μM TPC2-A1-N (K), 10 μM TPC2-A1-P (L), 50 nM NAADP (4 recordings out of 8 attempts) (M), or 1 μM PI(3,5)P2 (N). ATP (1 mM) was added at the end of each experiment. Recordings were carried out using ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of 0 mV. (O) Expanded views of K-N, showing the reversal potentials (Erev) of currents evoked by the indicated agonist. (P–Q) Statistical analyses of Erev (P) and the calculated relative cationic permeability ratios (PCa/PNa) (Q). Shown are mean values ± SEM. (R–S) Representative TPC2-A1-N- (R) or TPC2-A1-P- (S) evoked currents from endo-lysosomes isolated from HEK293 cells transiently expressing hTPC2L265P under bi-ionic conditions.
-
Figure 2—source data 1
TPC2 agonists alter Ca/Na permeability of TPC2.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig2-data1-v3.xlsx
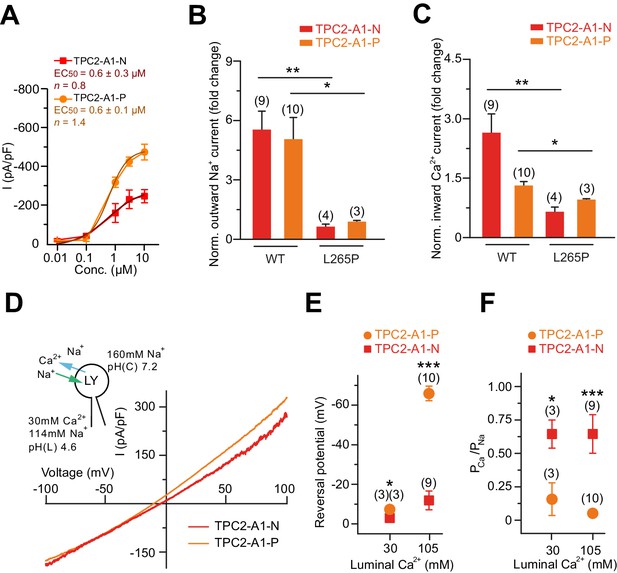
Dose response curves for TPC2-A1-N and TPC2-A1-P in endo-lysosomal patch-clamp experiments and endo-lysosomal patch-clamp analysis of compound effects under different conditions.
(A) Dose response curves of compounds using conditions as shown in Figure 2A and C. EC50 for TPC2-A1-P is 0.6 μM (Hill slope n = 1.4), EC50 for TPC2-A1-N is 0.6 μM (Hill slope n = 0.8) (n > 3). (B) Outward Na+ currents from Figure 2K,L,R and S normalized to basal currents (at +100 mV). (C) Inward Ca2+ currents from Figure 2K,L,R and S normalized to basal currents (at −100 mV). (D) Agonist-evoked cation currents from enlarged endo-lysosomes isolated from HEK293 cells stably expressing human TPC2 using the following conditions: luminal solution containing 114 mM Na+ and 30 mM Ca2+, pH 4.6; bath solution containing 160 mM Na+, pH 7.2 (n = 3, each). (E–F) Statistical analyses of Erev (E) and permeability ratio (PCa/PNa) (F) using either bi-ionic conditions as shown in Figure 2P–Q or conditions as used in D. In B-D, B/C and in E/F unpaired t-tests were applied. *p<0.05, **p<0.01 and ***p<0.001.
-
Figure 2—figure supplement 1—source data 1
Dose response curves for TPC2-A1-N and TPC2-A1-P in endo-lysosomal patch-clamp experiments and endo-lysosomal patch-clamp analysis of compound effects under different conditions.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig2-figsupp1-data1-v3.xlsx
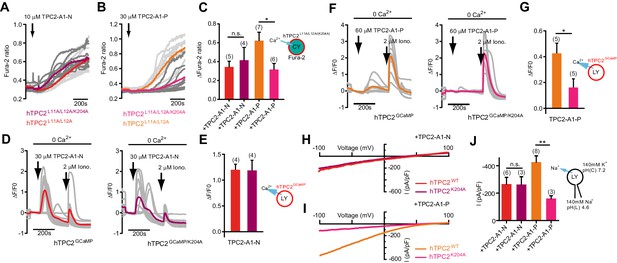
TPC2 agonists activate TPC2 through distinct sites.
(A–B) Representative Ca2+ signals recorded from Fura-2-loaded HeLa cells (n = 10, each) stimulated with the indicated concentration of TPC2-A1-N or -P. Cells were transiently transfected with plasma membrane targeted human TPC2 (hTPC2L11A/L12A) or the PI(3,5)P2-insensitive mutant (TPC2L11A/L12A/K204A). (C) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM) with the number of independent transfections in parentheses. An unpaired t-test was applied. *p<0.05. (D–F) Representative Ca2+ signals recorded from HeLa cells (n = 10, each) transiently transfected with GCaMP6(s) fused to wild-type TPC2 (TPC2GCaMP) or the PI(3,5)P2-insensitive mutant (TPC2GCaMP/K204A). Cells were sequentially stimulated with TPC2-A1-N or -P and the Ca2+ ionophore ionomycin in the absence of extracellular Ca2+. (E–G) Statistical analysis of the change in fluorescence intensity (mean ± SEM) of experiments as shown in D and F. An unpaired t-test was applied. *p<0.05. (H–I) Representative TPC2-A1-N or -P-evoked currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing human TPC2 (hTPC2) or the PI(3,5)P2-insensitive mutant, (hTPC2K204A). Currents were obtained before and after addition of 10 µM TPC2-A1-N or -P. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV. (J) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV for endo-lysosomes expressing TPC2 or TPC2K204A using TPC2-A1-N or TPC2-A1-P to activate, respectively. *p<0.05 and **p<0.01, using one-way ANOVA followed by Tukey’s post hoc test.
-
Figure 3—source data 1
TPC2 agonists activate TPC2 through distinct sites.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig3-data1-v3.xlsx
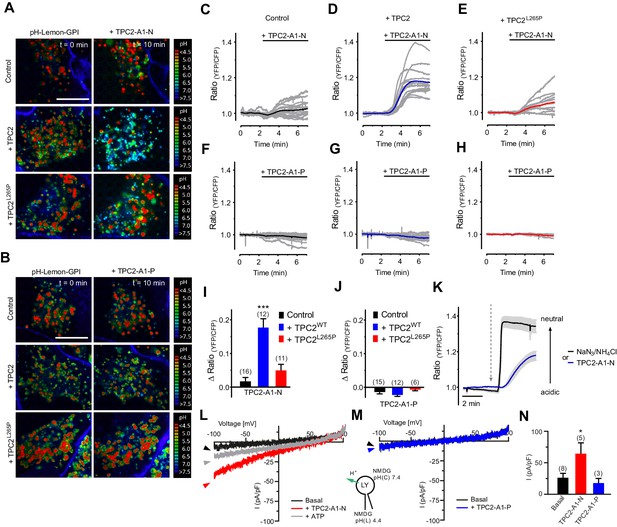
TPC2 agonists differentially affect lysosomal pH and proton conductance.
(A) Representative pseudo colored ratio images (mTurquoise2/EYFP) of vesicle targeted pH-Lemon-GPI in control HeLa cells (upper panel, n = 4/141 cells), HeLa cells positive for wild-type TPC2-mCherry (middle panel, c = 3/70 cells) and HeLa cells positive for TPC2L265P-mCherry (lower panel, n = 3/74 cells) in the absence (t = 0 min; left images) and upon treatment for 10 min with 10 µM TPC2-A1-N (t = 10 min; right images). Scale bar = 5 µm. (B) Representative pseudo colored ratio images (mTurquoise2/EYFP) of vesicle targeted pH-Lemon-GPI in control HeLa cells (upper panel, n = 3/121 cells), HeLa cells positive for wild-type TPC2-mCherry (middle panel, n = 3/59 cells) and HeLa cells positive for TPC2L265P-mCherry (lower panel, n = 3/71 cells) in the absence (t = 0 min; left images) and upon treatment for 10 min with 10 µM TPC2-A1-P (t = 10 min; right images). Scale bar = 5 µm. (C–E) Normalized single cell responses (grey curves) and the respective average response (colored curves) of pH-Lemon-GPI upon treatment with 10 µM TPC2-A1-N in a region of high vesicle density. Shown are ratio curves of pH-Lemon-GPI expressed in control HeLa cells (n = 16 cells; C), in HeLa cells co-expressing wild-type TPC2-mCherry (n = 12 cells; D), and HeLa cells co-expressing TPC2L265P-mCherry (n = 11 cells; E). (F–H) Experiments as in C-E for TPC2-A1-P. Shown are ratio curves of pH-Lemon-GPI expressed in control HeLa cells (n = 15 cells; F), in HeLa cells co-expressing wild-type TPC2-mCherry (n = 12 cells; G), and HeLa cells co-expressing TPC2L265P-mCherry (n = 6 cells, (H). (I–J) Columns represent delta ratio values of pH-Lemon-GPI at min six from curves as shown in panels C-E and F-H, respectively. ***p<0.001 using unpaired student’s t-test. (K) Average ratio signals over time of pH-Lemon-GPI in HeLa cells expressing wild-type TPC2-mCherry in response to either 0.5% NaN3 and 50 mM NH4Cl (black average curve ± SEM, n = 12 cells) or 10 µM TPC2-A1-N (blue average curve ± SEM, n = 12 cells). Cells were treated with compounds as indicated. (L–M) Representative agonist-evoked inward H+ currents from enlarged endo-lysosomes using hTPC2-expressing HEK293 cells. Both bath and pipette solutions contained 1 mM HCl, 5 mM HEPES, 5 mM MES, 150 mM NMDG, pH as indicated, adjusted with MSA. Ramp protocol (−100 mV to +100 mV in 500 ms, holding voltage = 0 mV) was used. Currents were obtained before and after addition of 10 μM TPC2-A1-N (L) or TPC2-A1-P (M). ATP (1 mM) was added at the end of the experiment to block TPC2 (L). (N) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV (inward H+ conductance; H+ from luminal to cytosolic site) as shown in L and M. *p<0.05, using one-way ANOVA followed by Bonferroni’s post hoc test.
-
Figure 4—source data 1
Effect of TPC2-A1-N on vesicular pH.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig4-data1-v3.xlsx
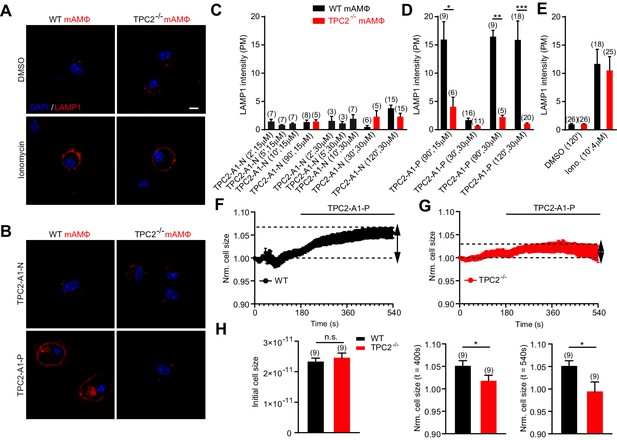
TPC2 agonists differentially affect lysosomal exocytosis.
(A–B) Representative images of LAMP1 translocation assay using murine wild-type and TPC2 KO alveolar macrophages. Shown are results obtained after 120 min treatment with either DMSO, TPC2-A1-P (30 μM) or TPC2-A1-N (30 μM), or 10 min treatment with ionomycin (4 μM). (C–E) Statistical analysis of experiments as shown in A and B at different compound concentrations and incubation times as indicated. Shown are mean values ± SEM. **p<0.01, and ***p<0.001, using two-way ANOVA followed by Bonferroni’s post hoc test. (F–G) Electrophysiological measurements of membrane capacitance via the whole-cell patch-clamp technique as an estimate of cell size (in picofarad [pF]) were used to record fusion of vesicles of primary alveolar macrophages isolated from WT (black, n = 9, F) or TPC2 KO (red, n = 9, G) mice. The normalized, averaged cell size was plotted versus time of the experiment. TPC2-A1-P (30 µM) was applied at 180 s until the end of the experiment indicated by the black bar. Data are shown as mean ± SEM. (H) Bar graphs show mean cell sizes at different time points. The initial cell size was measured immediately after whole cell break in pF and was used for normalization (left panel). Statistical analysis at 400 s (middle panel) and at 540 s (right panel). *p<0.05, unpaired student’s t-test.
-
Figure 5—source data 1
TPC2 agonists differentially affect lysosomal exocytosis.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig5-data1-v3.xlsx
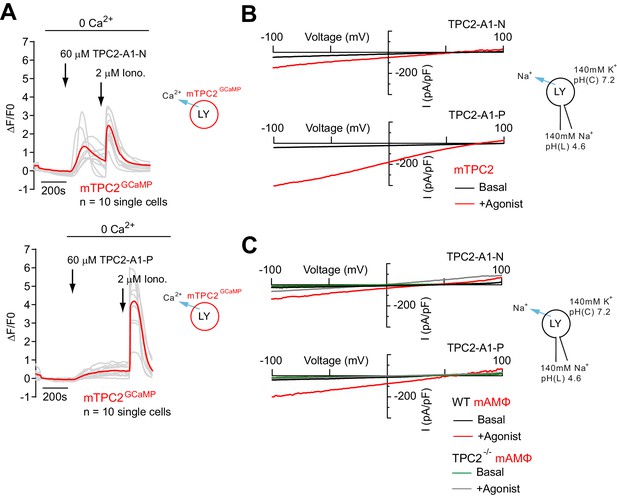
Effect of TPC2-A1-N and TPC2-A1-P on mouse TPC2 in overexpressing and endogenously expressing cells.
(A) Representative Ca2+ signals recorded from HeLa cells transiently transfected with GCaMP6(s) fused to wild-type murine TPC2 (mTPC2GCaMP). Cells were stimulated with TPC2-A1-N or -P and the Ca2+ ionophore ionomycin in the absence of extracellular Ca2+. (B) Representative TPC2-A1-N- or TPC2-A1-P-evoked currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing murine TPC2 (mTPC2). Currents were obtained before and after addition of 10 µM TPC2-A1-N or -P. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV. (C) Representative TPC2-A1-N- or TPC2-A1-P-evoked currents from enlarged endo-lysosomes isolated from murine wild-type and TPC2 KO alveolar macrophages. Currents were obtained before and after addition of 10 µM TPC2-A1-N or -P. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV.
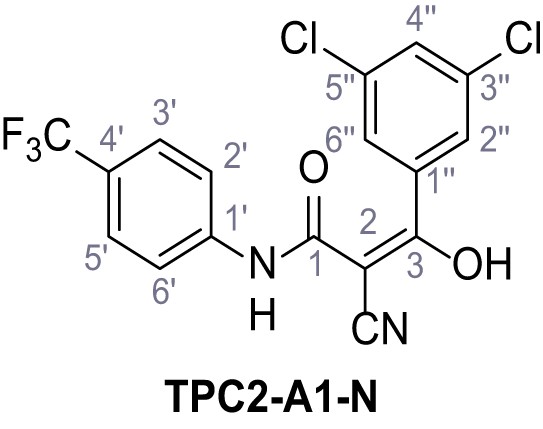
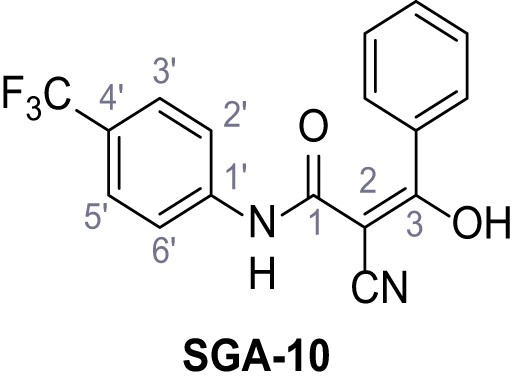
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – TPC2-A1-N.
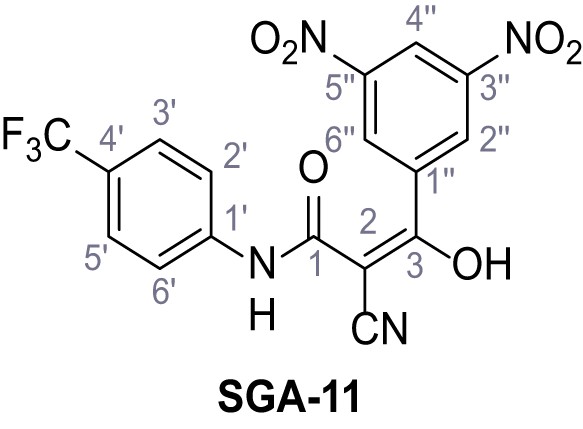
2-Cyano-3-(3,5-dinitrophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-11.
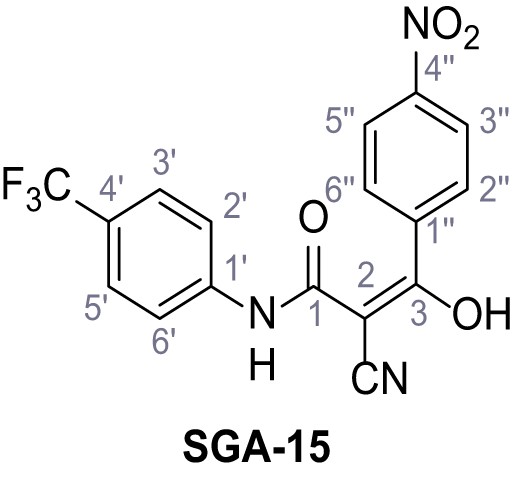
2-Cyano-3-(4-nitrophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-15.
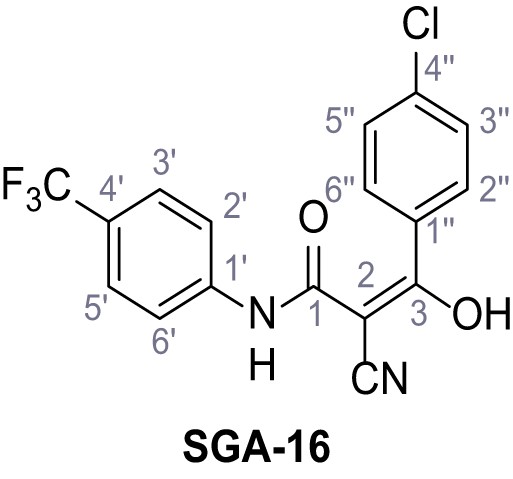
3-(4-Chlorophenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-16.
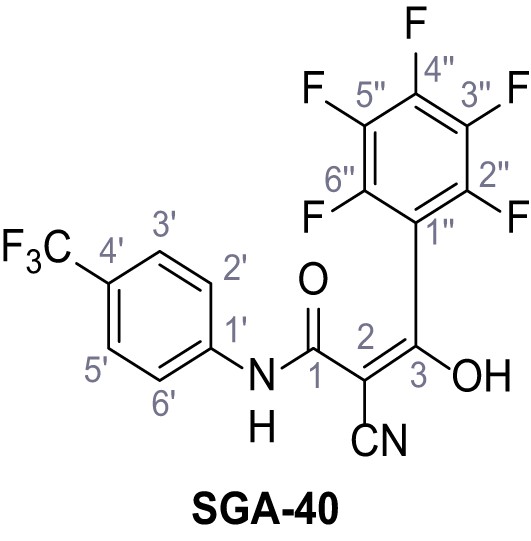
2-Cyano-3-hydroxy-3-(2,3,4,5,6-pentafluorophenyl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-40.
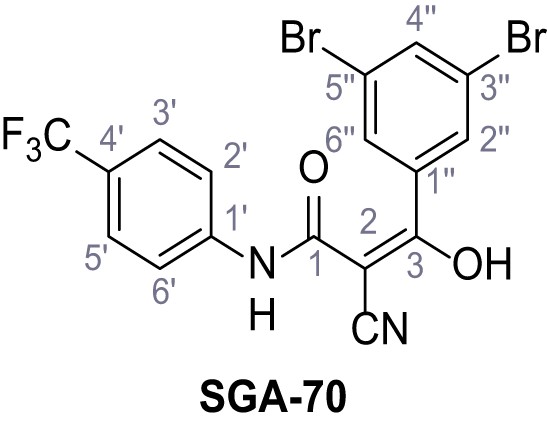
2-Cyano-3-(3,5-dibromophenyl)-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-70.
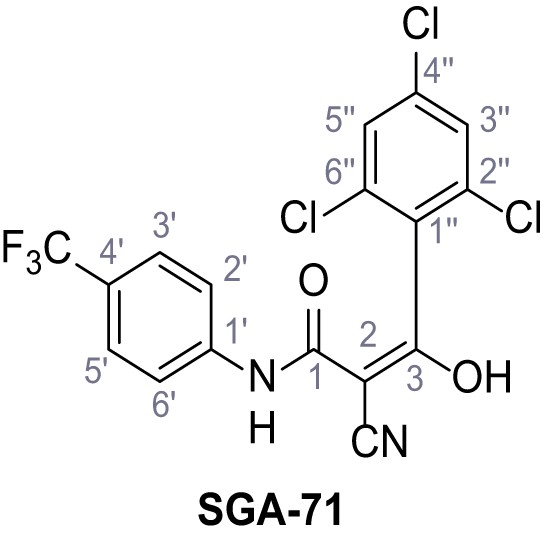
2-Cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-71.
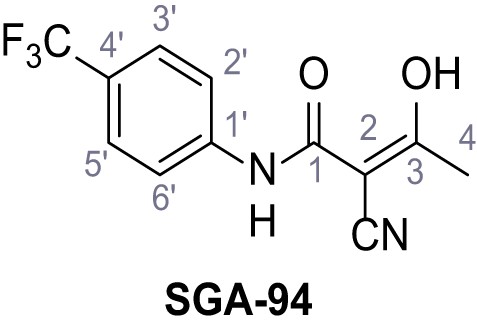
2-Cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)but-2-enamide (Teriflunomide) – SGA-94.
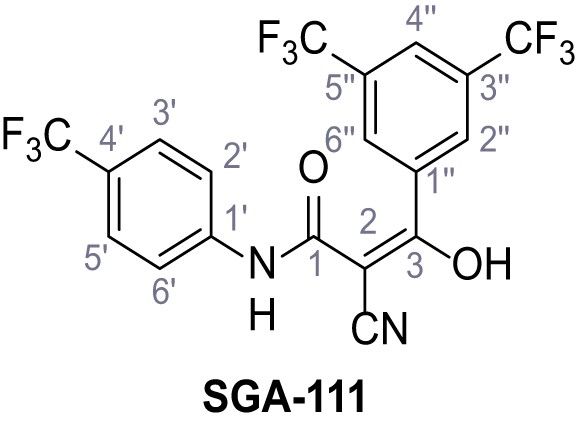
3-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-111.
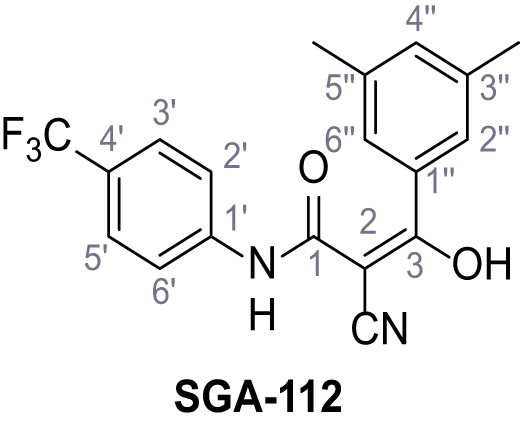
2-Cyano-3-(3,5-dimethylphenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-112.
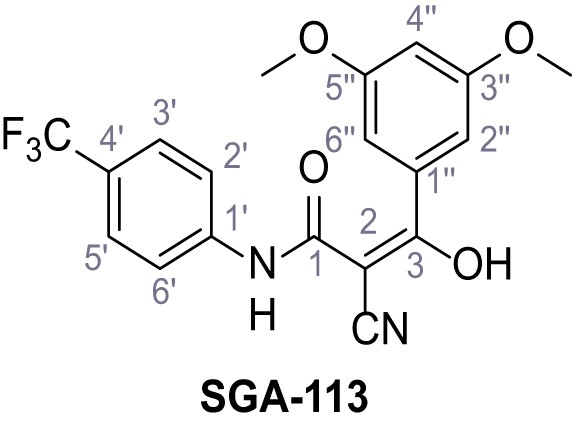
2-Cyano-3-(3,5-dimethoxyphenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-113.
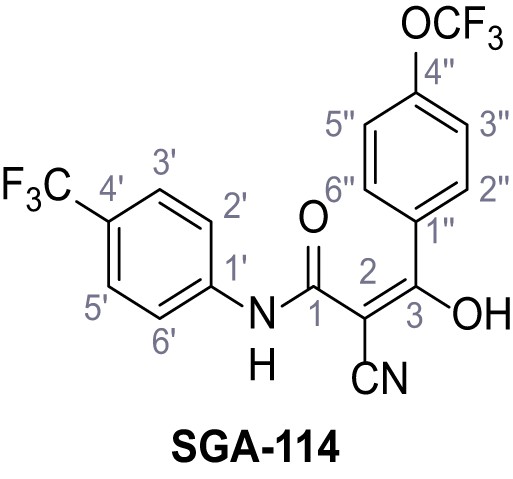
2-Cyano-3-hydroxy-3-(4-(trifluoromethoxy)phenyl)-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-114.
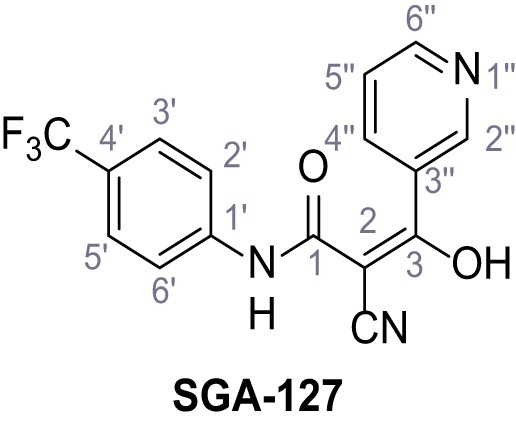
2-Cyano-3-hydroxy-3-(pyridin-3-yl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-127.
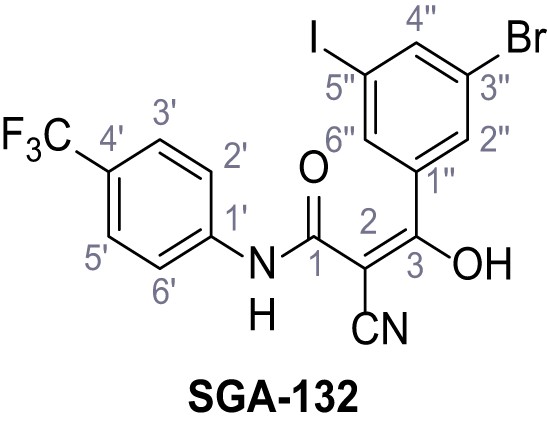
3-(3-Bromo-5-iodophenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-132.
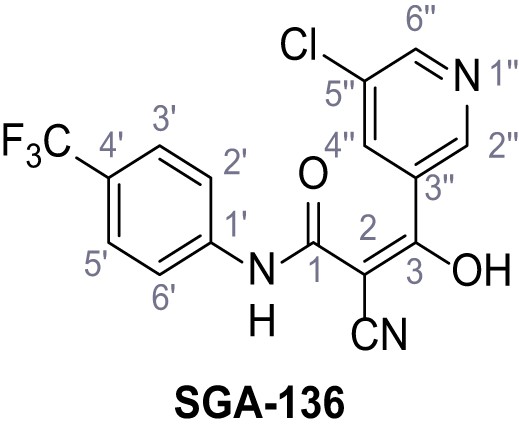
3-(5-Chloropyridin-3-yl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-136.
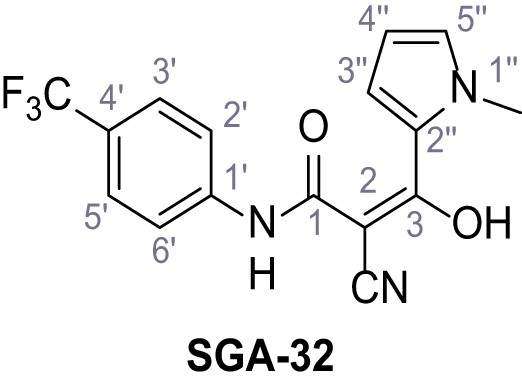
2-Cyano-3-(1-methyl-1H-pyrrol-2-yl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-32.
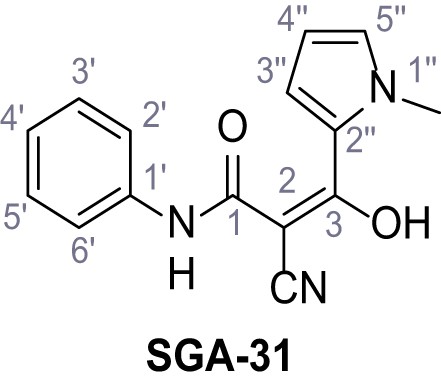
2-Cyano-3-(1-methyl-1H-pyrrol-2-yl)−3-hydroxy-N-phenylacrylamide (Prinomide) – SGA-31.
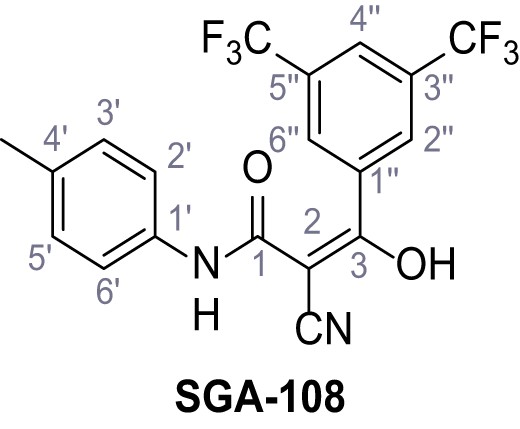
3-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-hydroxy-N-(p-tolyl)acrylamide – SGA-108.
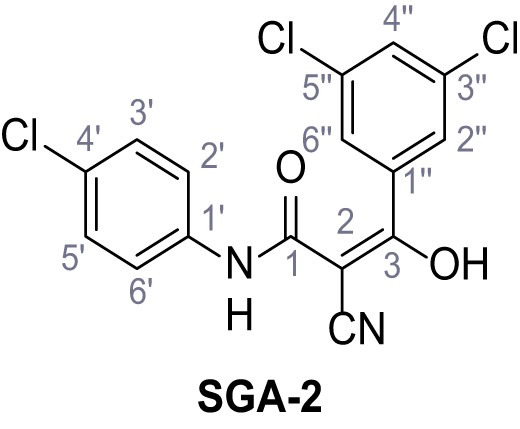
N-(4-Chlorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-2.
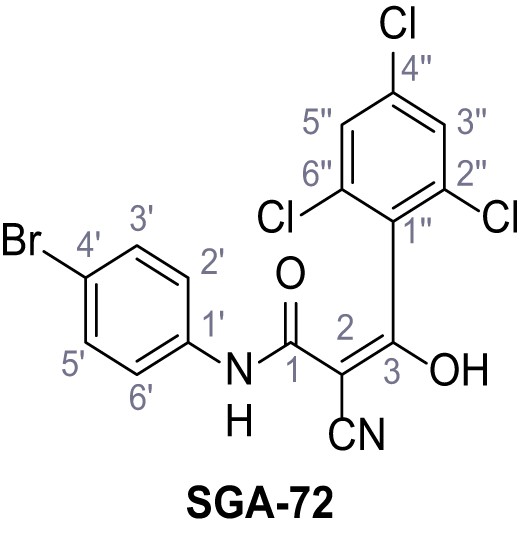
N-(4-Bromophenyl)−2-cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)acrylamide – SGA-72.
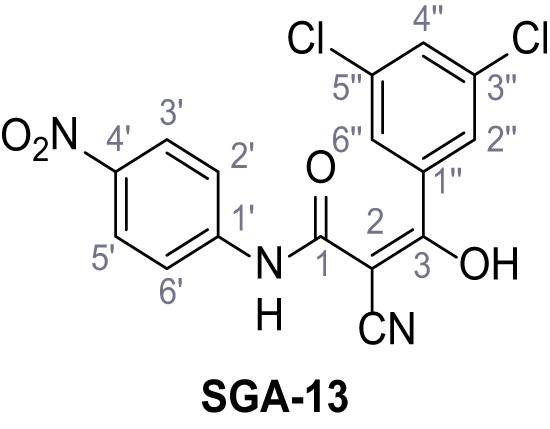
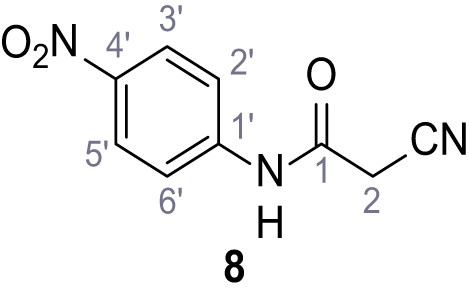
2-Cyano-3-(3,5-dichlorophenyl)-N-(4-nitrophenyl)−3-hydroxyacrylamide – SGA-13.
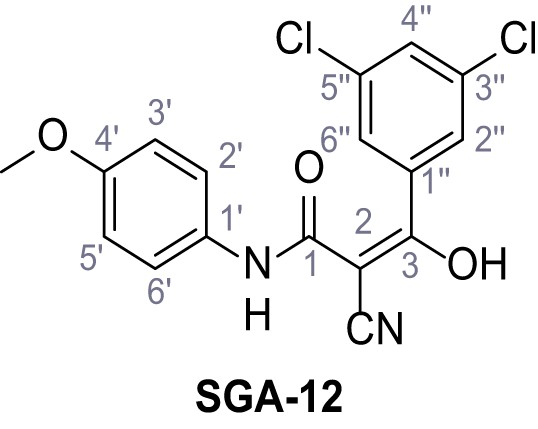
2-Cyano-3-(3,5-dichlorophenyl)-N-(4-methoxyphenyl)−3-hydroxyacrylamide – SGA-12.
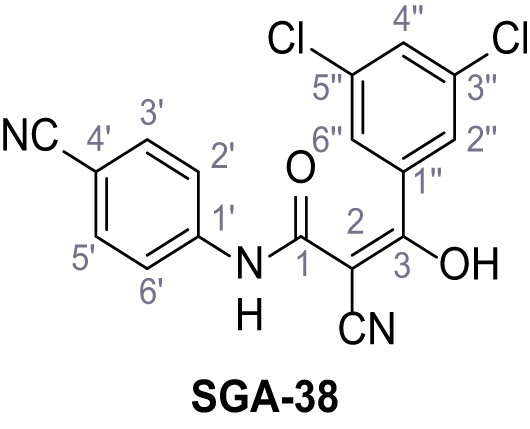
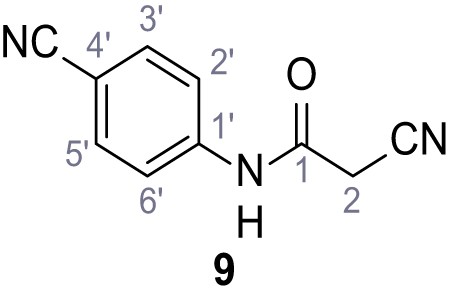
2-Cyano-N-(4-cyanophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-38.
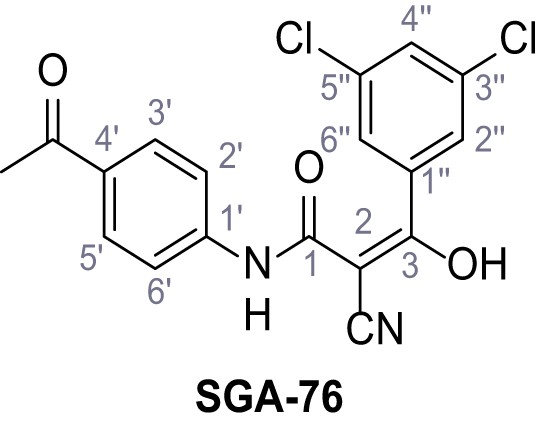
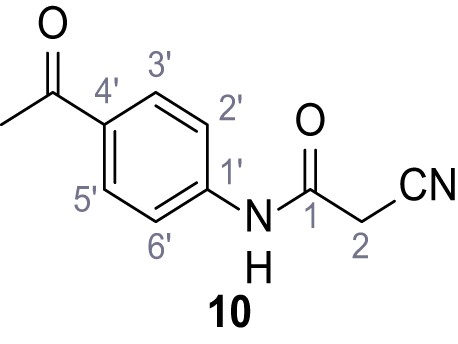
N-(4-Acetylphenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-76.
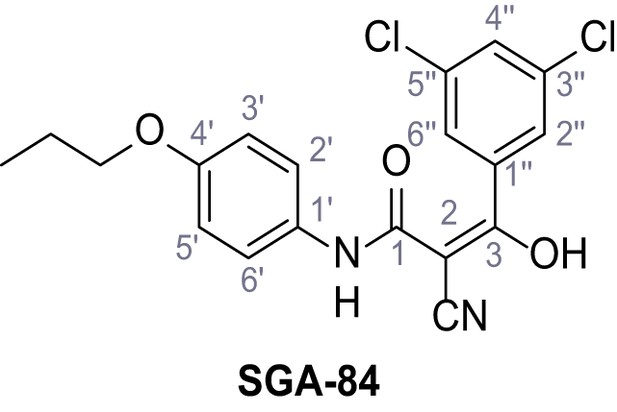
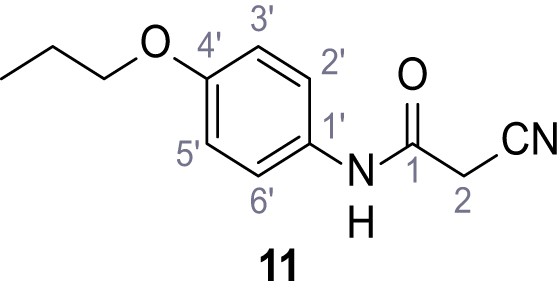
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-propoxyphenyl)acrylamide – SGA-84.
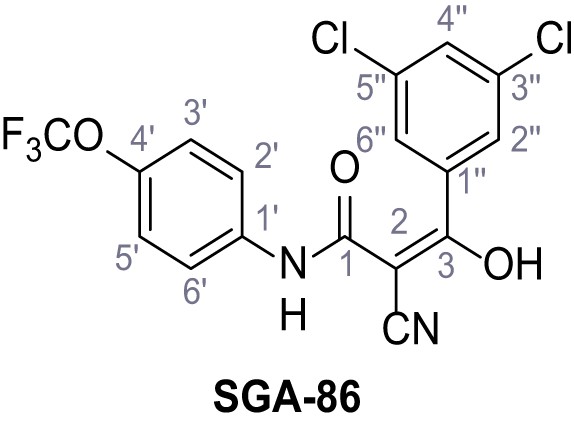
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethoxy)phenyl)acrylamide – SGA-86.
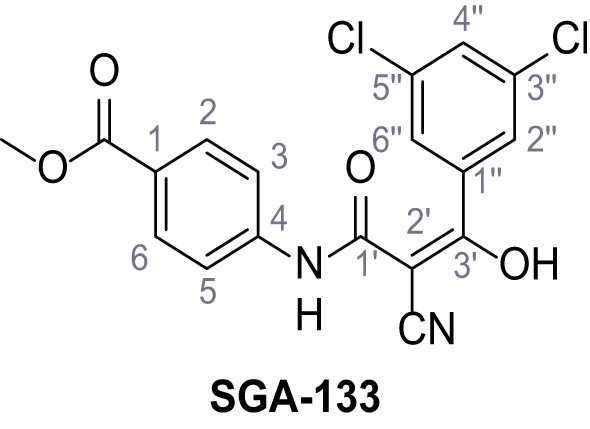
Methyl 4-(2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamido)benzoate – SGA-133.
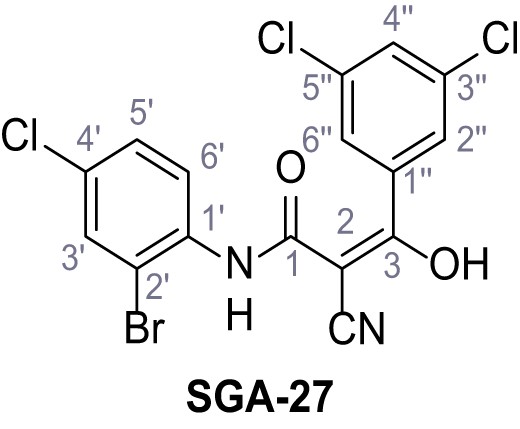
N-(2-Bromo-4-chlorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-27.
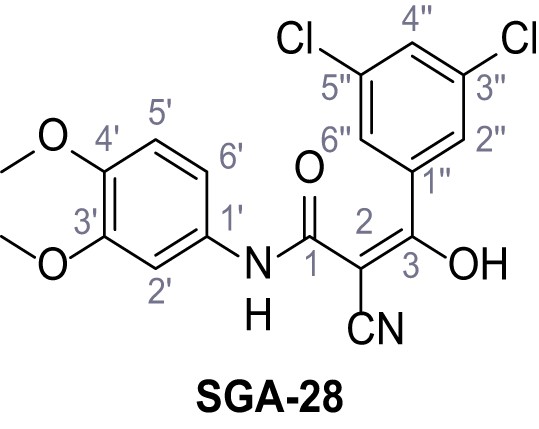
2-Cyano-3-(3,5-dichlorophenyl)-N-(3,4-dimethoxyphenyl)−3-hydroxyacrylamide – SGA-28.
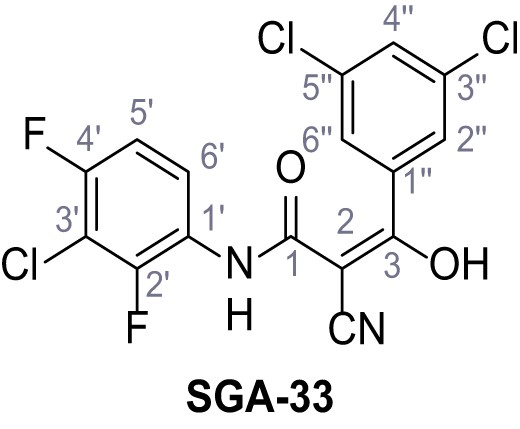
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-33.
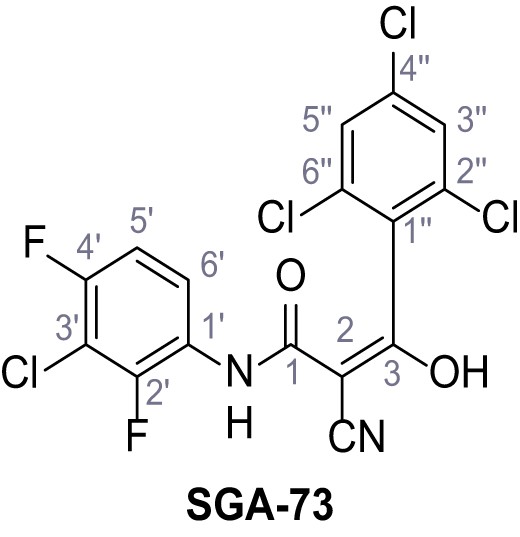
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)acrylamide – SGA-73.
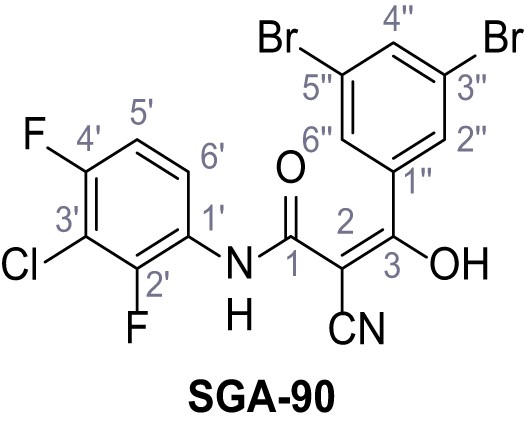
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-(3,5-dibromophenyl)−3-hydroxyacrylamide – SGA-90.
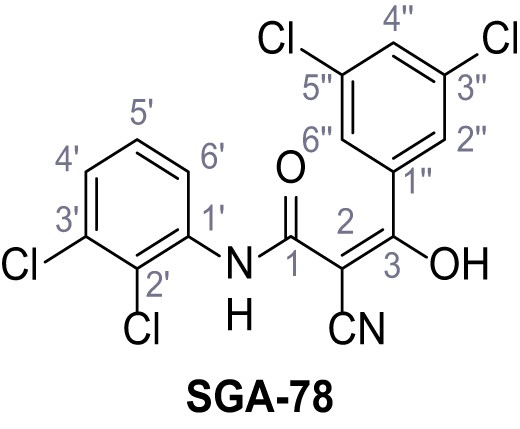
2-Cyano-N-(2,3-dichlorophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-78.
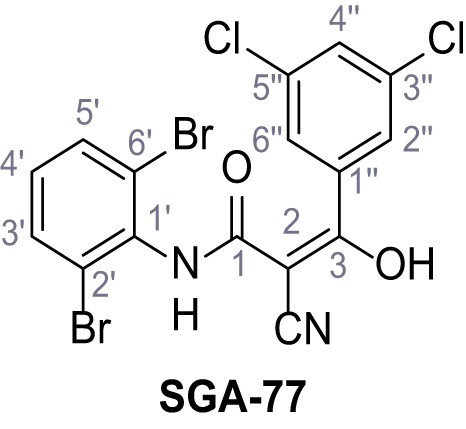
2-Cyano-N-(2,6-dibromophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-77.
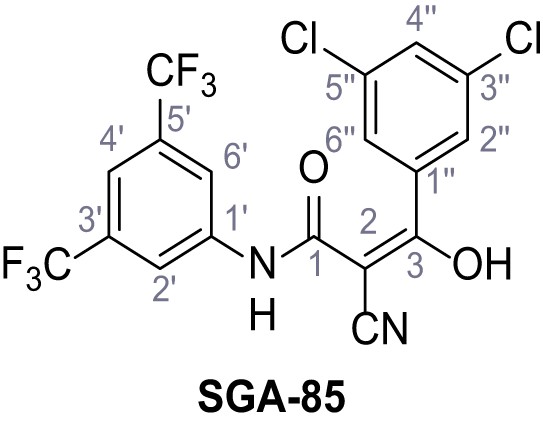
N-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-85.
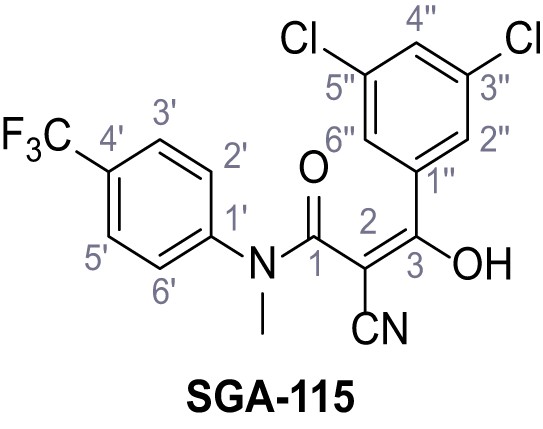
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-methyl-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-115.
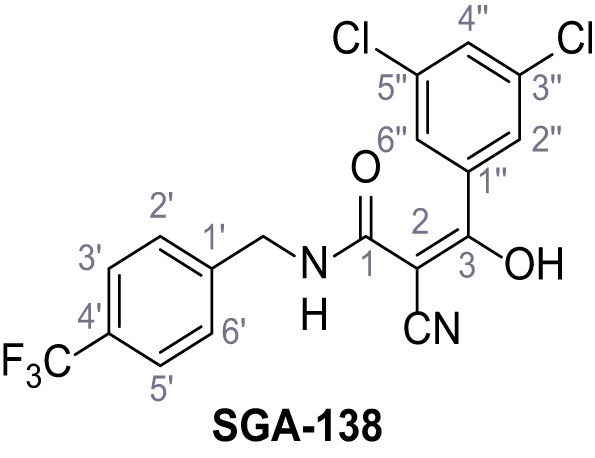
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethyl)benzyl)acrylamide – SGA-138.
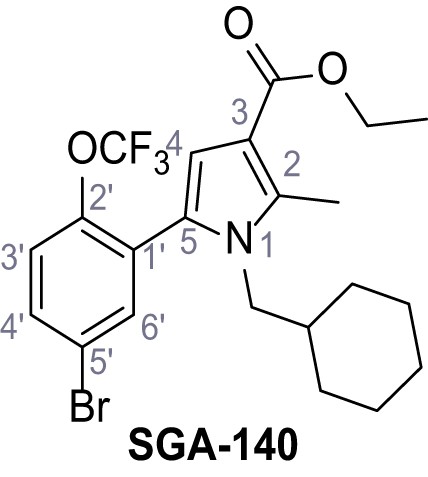
Ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylate.
– SGA-140.
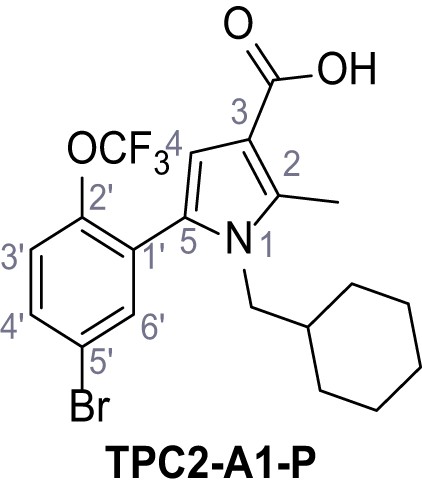
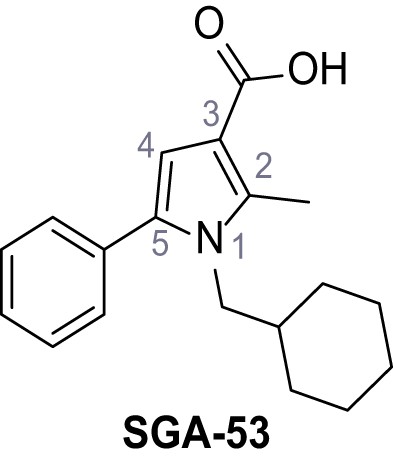
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylic acid – TPC2-A1-P.
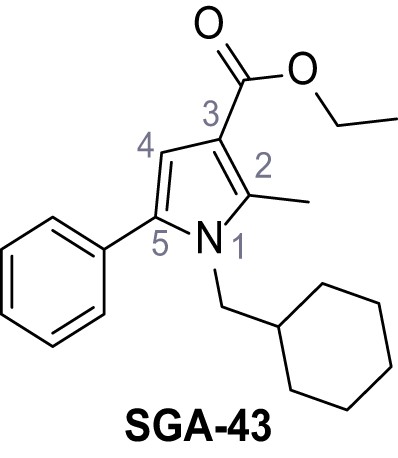
Ethyl 1-(cyclohexylmethyl)−2-methyl-5-phenyl-1H-pyrrole-3-carboxylate – SGA-43.
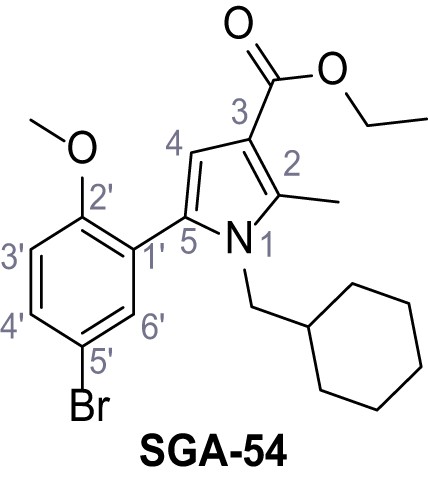
Ethyl 5-(5-bromo-2-methoxyphenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-54.
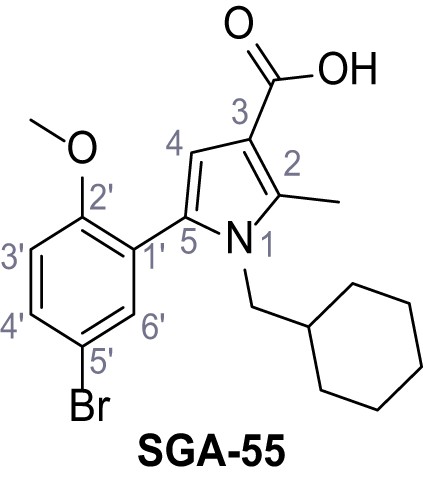
5-(5-Bromo-2-methoxyphenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-55.
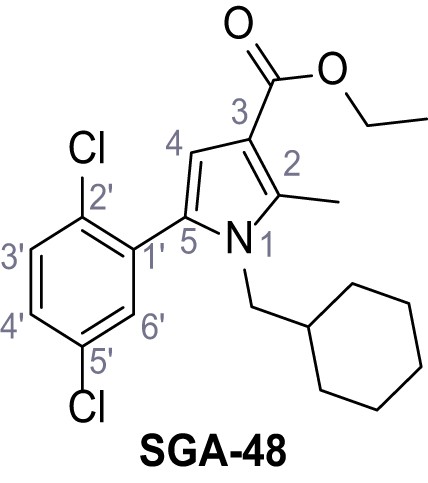
Ethyl 1-(cyclohexylmethyl)−5-(2,5-dichlorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-48.
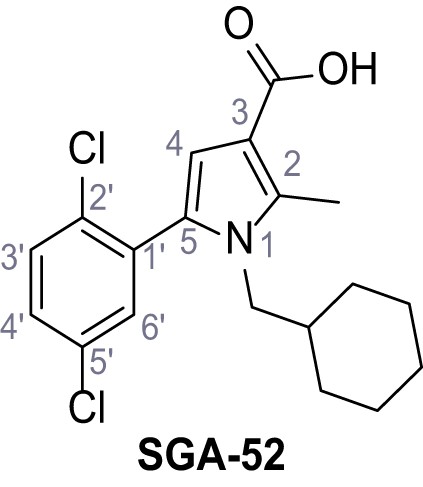
1-(Cyclohexylmethyl)−5-(2,5-dichlorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-52.
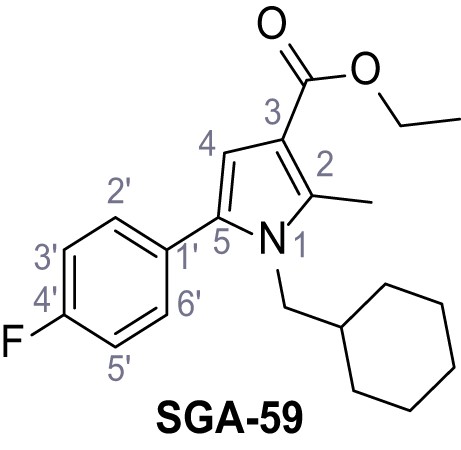
Ethyl 1-(cyclohexylmethyl)−5-(4-fluorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-59.
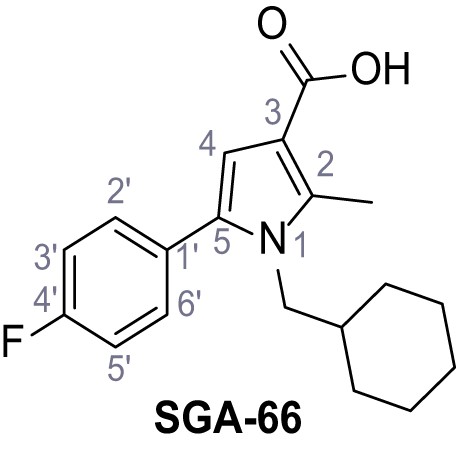
1-(Cyclohexylmethyl)−5-(4-fluorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-66.
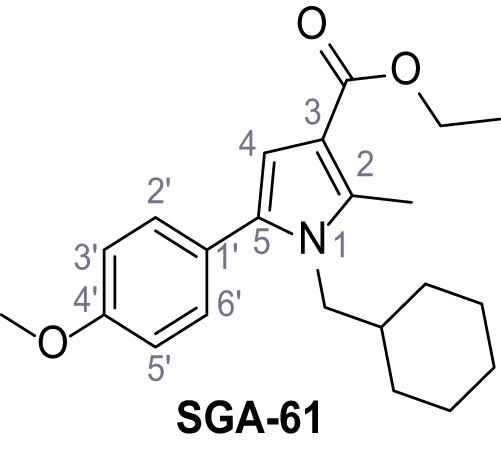
Ethyl 1-(cyclohexylmethyl)−5-(4-methoxyphenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-61.
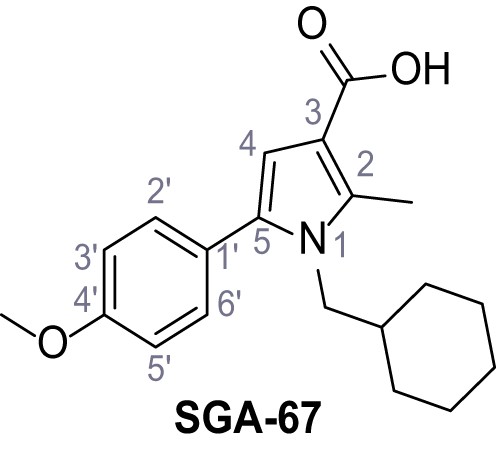
1-(Cyclohexylmethyl)−5-(4-methoxyphenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-67.
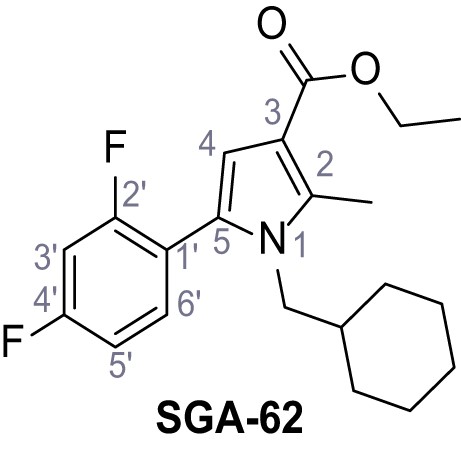
Ethyl 1-(cyclohexylmethyl)−5-(2,4-difluorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-62.
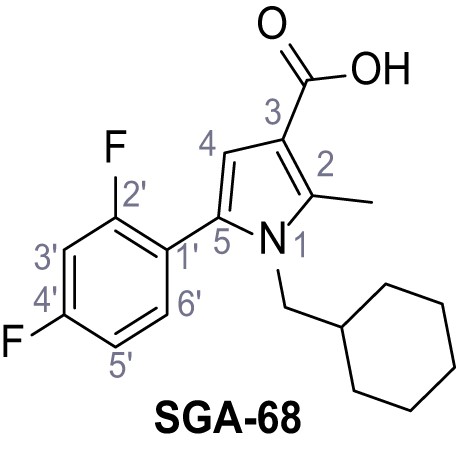
1-(Cyclohexylmethyl)−5-(2,4-difluorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid - SGA-68.
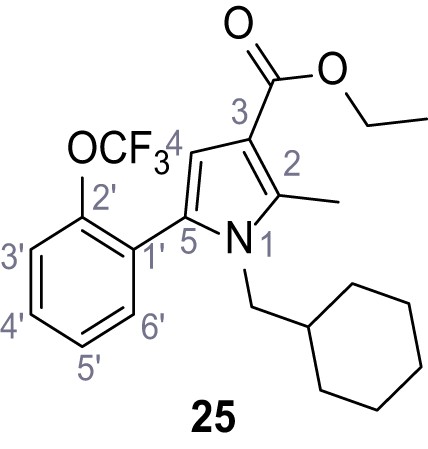
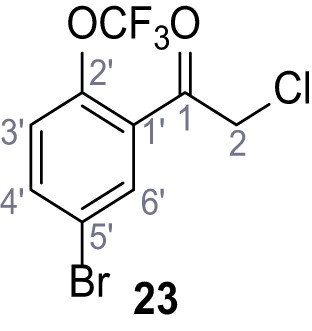
Ethyl 1-(cyclohexylmethyl)−2-methyl-5-(2-(trifluoromethoxy)phenyl)−1H-pyrrole-3-carboxylate (25).
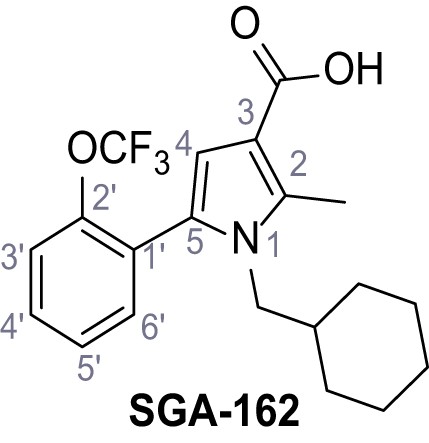
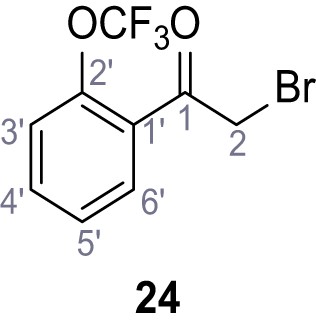
1-(Cyclohexylmethyl)−2-methyl-5-(2-(trifluoromethoxy)phenyl)−1H-pyrrole-3-carboxylic acid – SGA-162.
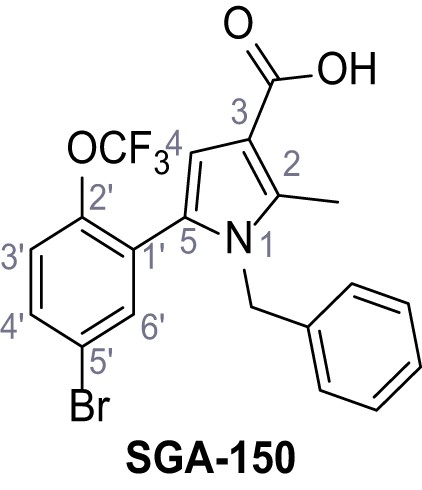
1-Benzyl-5-(5-bromo-2-(trifluoromethoxy)phenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-150.
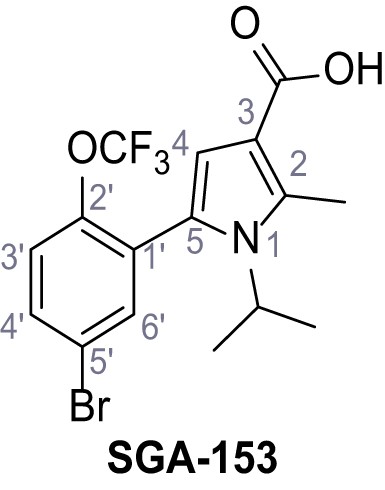
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-isopropyl-2-methyl-1H-pyrrole-3-carboxylic acid – SGA-153.
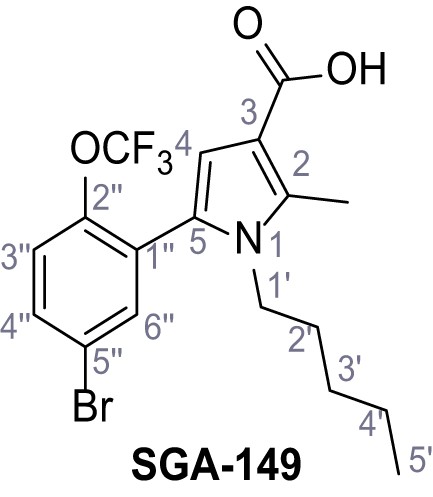
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−2-methyl-1-pentyl-1H-pyrrole-3-carboxylic acid – SGA-149.
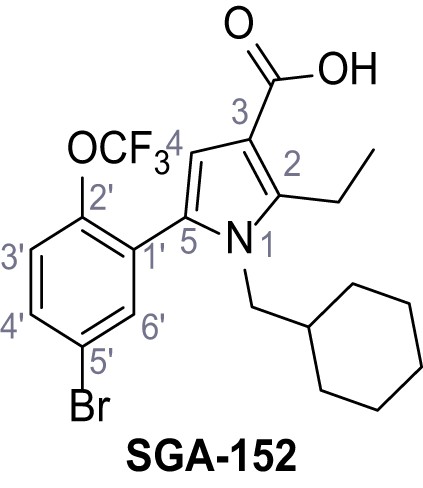
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-ethyl-1H-pyrrole-3-carboxylic acid – SGA-152.
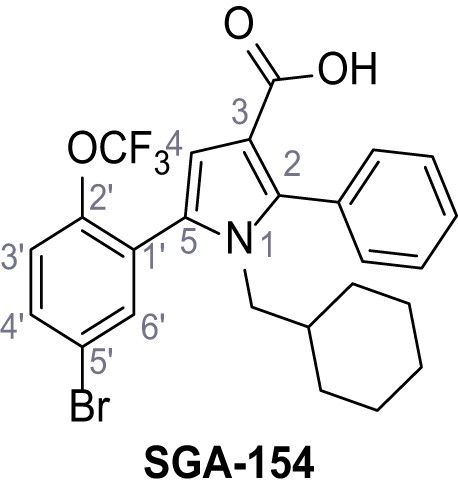
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-phenyl-1H-pyrrole-3-carboxylic acid – SGA-154.
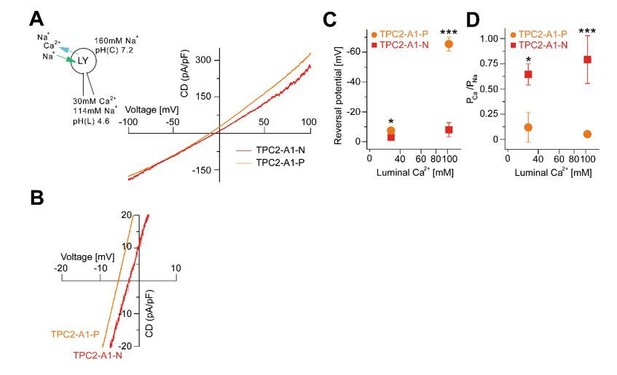
Agonist-evoked TPC2 cation currents from enlarged endo-lysosomes under different bi-ionic conditions.
(A) Agonist-evoked cation currents from enlarged endo-lysosomes isolated from HEK293 cells stably expressing human TPC2 using the following conditions: luminal solution containing 114 mM Na+ and 30 mM Ca2+, pH 4.6; bath solution containing 160 mM Na+, pH 7.2 (n = 3, each). (B) Expanded view of A. (C-D) Statistical analyses of Erev (C) and permeability ratio (PCa/PNa) (D) using either bi-ionic conditions as shown in Figure 2K-S or conditions as used in A and B.
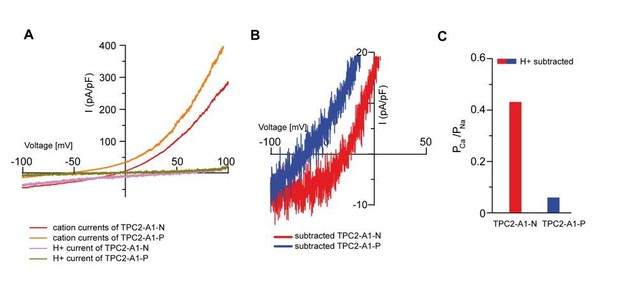
Proton permeablity does not substantially change estimates of relative Ca/Na permeabilities.
(A) I-Vs from Figure 2K/L (Na+/Ca2+ bi-ionic condition) and Figure 4L/M (NMDG+/H+ condition). (B) Expanded views of I-Vs after subtracting the average H+ current from the average Na+/Ca2+ currents from A. (C) Relative cationic permeability ratios (PCa/PNa) calculated from data in B.
Videos
TPC2-A1-N effectively increases vesicular pH in HeLa cells.
Dynamic pseudo-colored ratio (mTurquoise2/EYFP) video of HeLa cells stably expressing pH-Lemon-GPI and co-expressing TPC2 WT upon addition of TPC2-A1-N at the time-point indicated. Pseudo-colored ratio scale with estimated pH values is shown on the right. Scale bar represents 10 µm.
Vesicle movement is impaired upon treatment with TPC2-A1-N, but not with TPC-A1-P.
HeLa cells stably expressing pH-Lemon-GPI and co-expressing either TPC2 WT (upper left panel and lower left panel) or TPC2L265P (upper right panel and lower right panel) were analyzed. Shown are the mTurquoise2 fluorescence signals of pH-Lemon over time upon treatment with TPC2-A1-N (upper panels) and TPC2-A1-P (lower panels) at the time-points indicated. Time-lapse stacks were acquired using wide-field microscopy. Scale bar represents 10 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | HEK 293 | DSMZ | ACC 305 | |
| Cell line (Homo-sapiens) | HeLa S3 | ATCC | CCL-2.2 | |
| Cell line (Homo-sapiens) | HeLa cells stably expressing pH-Lemon-GPI | This paper | Generated by selective antibiotic G418 disulfate salt from Sigma Aldrich (Cat#A1720). After 4 weeks of cultivation in800 µg/mL, positive cells were selected by FACS analysis. | |
| Cell line (Homo-sapiens) | HEK 293 stably expressing TPC2L11A/L12A -RFP | This paper | Generated by selective antibiotic neomycin from Invitrogen (Cat#10486–025) following the guideline V790-20/V795-20 (Invitrogen) | |
| Cell line (Homo-sapiens) | HEK 293 stably expressing CLN3L253A/I254A -RFP | This paper | Generated by selective antibiotic neomycin from Invitrogen (Cat#10486–025) following the guideline V790-20/V795-20 (Invitrogen) | |
| Recombinant DNA reagent | HsTRPML1-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTRPML2-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTRPML3-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTPC2-YFP | (Chao et al., 2017) PMID:28923947 | ||
| Recombinant DNA reagent | HsTPC2M484L-YFP | (Chao et al., 2017) PMID:28923947 | ||
| Recombinant DNA reagent | HsTPC2L11/12A-YFP | This paper | Generated by site-directed mutagenesis from WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2L11/12A/L265P-YFP | This paper | Generated by site-directed mutagenesis of WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2L11/12A/K204A-YFP | This paper | Generated by site-directed mutagenesis of WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2-GCaMP6s | This paper | Generated by subcloning (Chao et al., 2017) WT TPC2 construct into Addgene vector #40753 | |
| Recombinant DNA reagent | HsTPC2L265P-GCaMP6s | This paper | Generated by site-directed mutagenesis of WT GCaMP6s plasmid | |
| Recombinant DNA reagent | HsTPC2K204A-GCaMP6s | This paper | Generated by site-directed mutagenesis of WT GCaMP6s plasmid | |
| Recombinant DNA reagent | MmTPC2-GCaMP6s | This paper | Generated by subcloning GCaMP6s from Addgene vector #40753 into WT MmTPC2 vector | |
| Recombinant DNA reagent | HsTPC2L11/12A-GFP | (Brailoiu et al., 2010) PMID:20880839 | ||
| Recombinant DNA reagent | HsTPC2L11/12A-RFP | (Brailoiu et al., 2010) PMID:20880839 | ||
| Recombinant DNA reagent | HsTPC2L11/12A/L265P-GFP | This paper | Generated by site-directed mutagenesis of HsTPC2L11/12A-GFP (plasmid) | |
| Recombinant DNA reagent | TRPML1ΔNC-GFP | (Yamaguchi et al., 2011) PMID:21540176 | ||
| Recombinant DNA reagent | HsTPC2L11/12A/K204A-GFP | (She et al., 2019) PMID:30860481 | ||
| Recombinant DNA reagent | MmTPC1-YFP | (Zong et al., 2009) PMID:19557428 | Generated by subcloning into TOPO pcDNA3.1-YFP, Addgene vector (#13033) | |
| Biological sample | Macrophages from TPC2 KO mouse | (Grimm et al., 2014) PMID:25144390 | ||
| Antibody | LAMP1 antibody | Santa Cruz Biotechnology | sc19992 | 1:200 |
| Transfection reagent | PolyJet | SignaGen Laboratories | SL100688 | |
| Transfection reagent | TurboFect | Thermo Fisher | R0531 | |
| Transfection reagent | Lipofectamine 2000 | Thermo Fisher | 11668 | |
| Commercial assay or kit | Fluo-4 AM, cell permeant | Thermo Fisher | F14202 | |
| Commercial assay or kit | Fura-2 AM, cell permeant | Thermo Fisher | F1221 | |
| Chemical compound, drug | NAADP | Tocris | 3905 | |
| Chemical compound, drug | PI(3,5)P2 | Echelon Biosciences | P-3508 | |
| Chemical compound, drug | Ionomycin | Sigma Aldrich and Cayman Chemical | I-0634 and 11932 | |
| Chemical compound, drug | Tetrandrine | Sigma Aldrich and Santa Cruz Biotechnology | T2695 and sc201492A | |
| Chemical compound, drug | Raloxifene | Cayman Chemical | 10011620 | |
| Chemical compound, drug | Fluphenazine | Sigma Aldrich | F4765 | |
| Chemical compound, drug | ML-SA1 | Merck | 648493 | |
| Chemical compound, drug | ATP | Sigma Aldrich | A9187 | |
| Chemical compound, drug | Sodiumazide (NaN3) | Sigma Aldrich | 09718 | |
| Chemical compound, drug | Ammoniumchloride (NH4Cl) | Sigma Aldrich | S2002 | |
| Software, algorithm | Origin8 | OriginLab | ||
| Software, algorithm | GraphPad Prism | GraphPad Software Inc | ||
| Other | Glass Bottom Dish 35 mm | ibidi | 81218 | |
| Other | Perfusion Chamber PC30 | Next Generation Fluorescence Imaging | PC30 (www.ngfi.eu) | Perfusion chamber used with gravity based perfusion system (NGFI, Graz, Austria) |
| Other | µ-Slide 8 Well | ibidi | 80826 |