Agonist-mediated switching of ion selectivity in TPC2 differentially promotes lysosomal function
Abstract
Ion selectivity is a defining feature of a given ion channel and is considered immutable. Here we show that ion selectivity of the lysosomal ion channel TPC2, which is hotly debated (Calcraft et al., 2009; Guo et al., 2017; Jha et al., 2014; Ruas et al., 2015; Wang et al., 2012), depends on the activating ligand. A high-throughput screen identified two structurally distinct TPC2 agonists. One of these evoked robust Ca2+-signals and non-selective cation currents, the other weaker Ca2+-signals and Na+-selective currents. These properties were mirrored by the Ca2+-mobilizing messenger, NAADP and the phosphoinositide, PI(3,5)P2, respectively. Agonist action was differentially inhibited by mutation of a single TPC2 residue and coupled to opposing changes in lysosomal pH and exocytosis. Our findings resolve conflicting reports on the permeability and gating properties of TPC2 and they establish a new paradigm whereby a single ion channel mediates distinct, functionally-relevant ionic signatures on demand.
Introduction
When ion channels open in response to a given stimulus, they allow the flow of a specific set of ions. The textbook view is that ion selectivity for a given ion channel is a hallmark feature that cannot be changed. But evidence has accrued suggesting that this may not always be the case. P2X receptors have long been proposed to change their ion selectivity in real time upon prolonged activation (Khakh and Lester, 1999) albeit controversially (Li et al., 2015). And in TRPV channels, the selectivity filter is thought to form a second gate thereby coupling channel opening with changes in ion selectivity (Cao et al., 2013). Other channels such as Orai channels (McNally et al., 2012) and the mitochondrial uniporter MCU1 (Kamer et al., 2018) appear to alter their ion selectivity depending on protein partners.
Two-pore channels (TPC1-3) are ancient members of the voltage-gated ion channel superfamily (Patel, 2015). TPCs are expressed throughout the endo-lysosomal system where they regulate the trafficking of various cargoes (Grimm et al., 2014; Ruas et al., 2010; Sakurai et al., 2015). Loss- and gain-of function of TPCs is implicated in a number of diseases including NAFLD (non-alcoholic fatty liver disease) and Parkinson’s (Patel, 2015; Patel and Kilpatrick, 2018). Despite their pathophysiological importance, both the ion selectivity and activating ligand(s) of TPCs are equivocal. Initial studies characterized TPCs as non-selective Ca2+-permeable channels activated by NAADP (nicotinic acid adenine dinucleotide phosphate) (Brailoiu et al., 2009; Brailoiu et al., 2010; Calcraft et al., 2009; Grimm et al., 2014; Pitt et al., 2010; Ruas et al., 2015; Schieder et al., 2010). But other studies indicate that TPCs are highly-selective Na+ channels that similar to endo-lysosomal TRP mucolipins (TRPMLs), are activated directly by PI(3,5)P2 (phosphatidylinositol 3,5-bisphosphate), and/or by voltage (Boccaccio et al., 2014; Cang et al., 2014; Guo et al., 2017; She et al., 2018; Wang et al., 2012). Co-regulation of TPCs by these disparate stimuli has been noted in some instances (Jha et al., 2014; Ogunbayo et al., 2018; Pitt et al., 2014; Rybalchenko et al., 2012).
Functional characterization of TPCs is challenging due to their intracellular location. The anionic nature of NAADP and PI(3,5)P2 prevents sufficient plasma membrane permeability, making them less suitable for experimental use in intact cells. On the other hand, studies of isolated lysosomes may result in loss of crucial accessory factors such as NAADP-binding proteins (Lin-Moshier et al., 2012). To circumvent these issues, we screened for membrane-permeable small molecule activators of TPCs. Here, we report two agonists that surprisingly switch the ion selectivity of TPC2 with different consequences for lysosomal function and integrity.
Results
Identification of novel small molecule agonists of TPC2
Mutation of the N-terminal endo-lysosomal targeting motif in human TPC2 redirects it to the plasma membrane where TPC2 reportedly mediates robust Ca2+ entry upon activation with NAADP (Brailoiu et al., 2010). We took advantage of this property to screen a cell line stably expressing TPC2L11A/L12A with a library of 80000 natural and synthetic small molecules using a FLIPR-based Ca2+ assay (Figure 1A and B). Two structurally independent hits were identified, termed TPC2-A1-N and TPC2-A1-P (Figure 1C and D), which reproducibly evoked Ca2+ signals. The signals evoked by the compounds showed different kinetics whereby the TPC2-A1-N response reached its plateau faster than TPC2-A1-P (Figure 1E and F). The structures and activities of these hits were confirmed by independent chemical syntheses and subsequent retesting. Full concentration-effect relationships for the plateau response indicated EC50 values of 7.8 μM and 10.5 μM, for TPC2-A1-N and TPC2-A1-P, respectively (Figure 1G and H). We thus identified novel small molecules that activate TPC2 and confirm its Ca2+-permeability in intact cells.
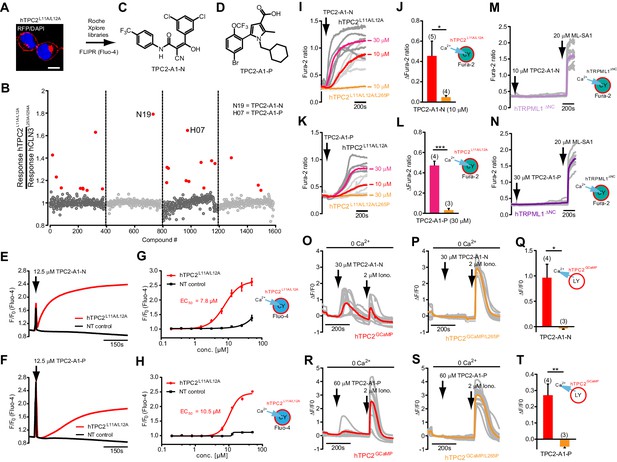
Identification of novel small molecule agonists of TPC2.
(A) The chemical screening was performed using cells stably expressing human TPC2 re-routed to the plasma membrane (hTPC2L11A/L12A) and loaded with the fluorescent Ca2+ indicator, Fluo-4 (CY = cytosol). For counter-screening, a stable cell line expressing cell surface human CLN3 (hCLN3L253A/I254A) was used. (B) A library comprising 80.000 compounds was screened. Shown are the primary screen results of four plates (with 384 compounds, each) where the response to a given compound in cells expressing hTPC2L11A/L12A is expressed relative to that in cells expressing hCLN3L253A/I254A. Primary hits are shown as red spots. Subsequent confirmatory and follow-up assays led to the identification of the two hit compounds, N19 (TPC2-A1-N) and H07 (TPC2-A1-P). (C–D) Figures of TPC2-A1-N and TPC2-A1-P. (E–F) Representative FLIPR-generated Ca2+ signals (Fluo-4) in TPC2L11A/L12A–expressing cells (red lines) or non-transfected (NT) control cells (black) after addition of TPC2-A1-N (E) or TPC2-A1-P (F). (G–H) Concentration-effect relationships for Ca2+ increases (Fluo-4) in response to different concentrations of TPC2-A1-N (G) and TPC2-A1-P (H). Each concentration was tested in duplicates in two to four independent experiments, each. (I) Representative Ca2+ signals recorded from HeLa cells loaded with the ratiometric Ca2+ indicator, Fura-2 and stimulated with the indicated concentration of TPC2-A1-N. Cells were transiently transfected with plasma-membrane-targeted human TPC2 (hTPC2L11A/L12A) or a pore dead mutant (TPC2L11A/L12A/L265P). Colored lines represent the mean response from a population of cells. Traces in grey represent responses of single cells. (J) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM) with the number of independent transfections in parentheses. An unpaired t-test was applied. *p<0.05. (K–L) Similar to I and J except that cells were stimulated with TPC2-A1-P. ***p<0.001. Traces in grey represent single cells responding to TPC2-A1-P. (M–N) Representative Ca2+ signals recorded from HeLa cells transiently transfected with human TRPML1ΔNC. Cells were sequentially stimulated with TPC2-A1-N (M) or TPC2-A1-P (N) and the TRPML agonist ML-SA1. (O–T) Ca2+ signals recorded from HeLa cells transiently transfected with GCaMP6(s) fused to human TPC2 (hTPC2GCaMP) or a pore-dead mutant (hTPC2GCaMP/L265P). Cells were sequentially stimulated with TPC2-A1-N (O and P) or TPC2-A1-P (R and S) and the Ca2+ ionophore, ionomycin in the absence of extracellular Ca2+. Statistical analysis of the change in fluorescence intensity (mean ± SEM; Q and T). An unpaired t-test was applied. *p<0.05 and **p<0.01. CY = cytosol, LY = lysosome.
-
Figure 1—source data 1
Identification of TPC2 hit compounds.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig1-data1-v3.xlsx
TPC2 agonists differentially evoke cytosolic Ca2+ signals in live cells
To validate the hits further, we performed ratiometric Ca2+ imaging of single cells transiently transfected with TPC2L11A/L12A or a ‘pore-dead’ version (TPC2L11A/L12A/L265P). As shown in Figure 1I and J, TPC2-A1-N evoked Ca2+ signals in cells expressing TPC2L11A/L12A but not TPC2L11A/L12A/L265P. These data confirm that TPC2-A1-N evoked Ca2+ influx through the TPC2 pore. In accord, the responses to TPC2-A1-N were selectively blocked by the recently identified TPC2 blockers tetrandrine (Tet), raloxifene (Ral), and fluphenazine (Flu) (Penny et al., 2019; Sakurai et al., 2015; Figure 1—figure supplement 1) and by removal of extracellular Ca2+ (Figure 1—figure supplement 2). TPC2-A1-P also induced Ca2+ signals in cells expressing TPC2 in the presence (Figure 1K and L) but not absence (Figure 1—figure supplement 2) of extracellular Ca2+. However, the responses were smaller and delayed compared to TPC2-A1-N (Figure 1I and K), consistent with the results obtained in cells stably expressing TPC2L11A/L12A (Figure 1E and F). TPC2-A1-P failed to induce Ca2+ signals in cells expressing ‘pore-dead’ TPC2L11A/L12A/L265P (Figure 1K and L). Both TPC2-A1-N and TPC2-A1-P also failed to evoke Ca2+ signals in cells expressing human TRPML1 re-routed to the plasma membrane (TRPML1ΔNC) (Grimm et al., 2010; Yamaguchi et al., 2011; Figure 1M–N and Figure 1—figure supplement 1A–B). Similar negative results were obtained with the agonists in cells expressing TRPML2 or TRPML3 (Figure 1—figure supplement 3C and D), indicating a selective, pore-dependent action of both agonists on TPC2.
The consensus logP (octanol/water; SwissADME) values for TPC2-A1-N (4.56) and TPC2-A1-P (5.35) predict that they are cell permeable. To determine whether TPC2-A1-N and TPC2-A1-P were able to release Ca2+ from lysosomes, we expressed TPC2 fused to the genetically encoded Ca2+ indicator, GCaMP6(s) both with (TPC2GCaMP) and without (TPC2GCaMP/L265P) an intact pore (Figure 1O–T) in order to measure global Ca2+ signals. TPC2-A1-N and TPC2-A1-P evoked Ca2+ signals in cells expressing intracellular TPC2GCaMP but not TPC2GCaMP/L265P. Again the responses to TPC2-A1-P were modest compared to TPC2-A1-N.
Systematic structure modifications were performed on both screening hits. Surprisingly, none of the modified versions of TPC-A1-N or TPC2-A1-P showed significantly increased efficacies (Figure 1—figure supplements 4 and 5). TPC2-A1-P analogues missing the trifluoromethoxy residue were completely inactive (Figure 1—figure supplement 6).
Collectively, our data show that TPC2-A1-N and TPC2-A1-P activate TPC2 with differential effects on Ca2+ mobilization.
TPC2 agonists differentially evoke Na+ currents in isolated endo-lysosomes
To determine more directly whether TPC2-A1-N and TPC2-A1-P activate TPC2, endo-lysosomal patch-clamp experiments were performed (Figure 2). TPC2-A1-N elicited currents using Na+ as the major permeant ion, in vacuolin-enlarged endo-lysosomes isolated from TPC2-expressing cells (Figure 2A). The currents were inhibited by ATP as reported previously (Cang et al., 2013). In contrast, no activation was found in cells expressing TPC1 (Figure 1—figure supplement 7). Endo-lysosomes isolated from cells expressing a gain-of-function variant of TPC2 (TPC2M484L) (Chao et al., 2017) showed larger currents compared to the wild-type isoform upon application of TPC2-A1-N (Figure 2B). TPC2-A1-P also evoked currents in endo-lysosomes isolated from cells expressing TPC2 and TPC2M484L (Figure 2C and D). As with TPC2-A1-N, the currents were potentiated by the gain-of-function variant (Figure 2D). Surprisingly, the currents evoked by TPC2-A1-P were significantly larger than those evoked by TPC2-A1-N (Figure 2E–G) in both wild-type and gain-of-function variant. Full concentration-effect relationships for the plateau response in endo-lysosomal patch-clamp experiments indicated EC50 values of 0.6 μM for both TPC2-A1-N and TPC2-A1-P (Figure 2—figure supplement 1A).
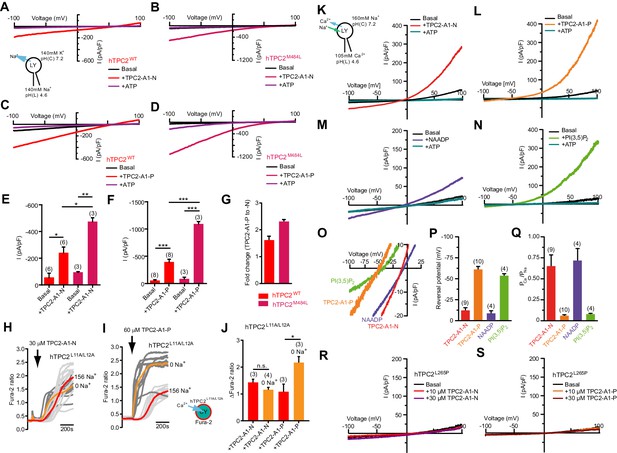
TPC2 agonists alter Ca2+/Na+ permeability of TPC2.
(A–B) Representative TPC2-A1-N-evoked currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing human TPC2 (hTPC2) (A) or a gain-of-function variant, (hTPC2M484L) (B). Currents were obtained before and after addition of 10 µM TPC2-A1-N in the absence or presence of 1 mM ATP. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV. (C–D) Similar to A and B except that TPC2-A1-P (10 µM) was used. (E–G) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV for endo-lysosomes expressing TPC2 or TPC2M484L for TPC2-A1-N (E) and TPC2-A1-P (F), and the fold-change in current evoked by TPC2-A1-P versus TPC2-A1-N in the two variants (G). Red bars are WT and maroon bars are mutant. An unpaired t-test was applied. *p<0.05, **p<0.01, and ***p<0.001 (n > 10 endo-lysosomes per condition). (H–I) Ca2+ signals evoked by TPC2-A1-N (H) or TPC2-A1-P (I) in Fura-2-loaded HEK293 cells stably expressing hTPC2L11A/L12A. Recordings (mean responses from three to four independent experiments) were obtained in standard or Na+-free extracellular solution. (J) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM). An unpaired t-test was applied. *p<0.05. (K–N) Agonist-evoked cation currents from enlarged endo-lysosomes isolated from HEK293 cells stably expressing human TPC2 under bi-ionic conditions: 160 mM Na+ in the cytosol (bath) and 105 mM Ca2+ in the lumen (pipette). Representative current-voltage curves before and after stimulation with either 10 μM TPC2-A1-N (K), 10 μM TPC2-A1-P (L), 50 nM NAADP (4 recordings out of 8 attempts) (M), or 1 μM PI(3,5)P2 (N). ATP (1 mM) was added at the end of each experiment. Recordings were carried out using ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of 0 mV. (O) Expanded views of K-N, showing the reversal potentials (Erev) of currents evoked by the indicated agonist. (P–Q) Statistical analyses of Erev (P) and the calculated relative cationic permeability ratios (PCa/PNa) (Q). Shown are mean values ± SEM. (R–S) Representative TPC2-A1-N- (R) or TPC2-A1-P- (S) evoked currents from endo-lysosomes isolated from HEK293 cells transiently expressing hTPC2L265P under bi-ionic conditions.
-
Figure 2—source data 1
TPC2 agonists alter Ca/Na permeability of TPC2.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig2-data1-v3.xlsx
Increased Na+ currents in the face of reduced Ca2+ signals (Figure 1) upon TPC2-A1-P activation prompted us to examine the effects of Na+ removal on intracellular Ca2+ elevation. For these experiments, we used cells stably expressing TPC2L11A/L12A and agonist concentrations that evoked similar responses. As shown in Figure 2H and J, the amplitude of Ca2+ signals evoked by TPC2-A1-N were unaffected by removal of Na+ from the extracellular solution, although the rate of rise was increased. In contrast, both the amplitude and rate of rise of the responses to TPC2-A1-P were markedly enhanced (Figure 2I and J). These data indicate that TPC2 is activated by TPC2-A1-N and TPC2-A1-P but that its permeability to Na+ differs upon activation.
TPC2 agonists alter Ca2+/Na+ permeability of TPC2
The above analyses raised the intriguing possibility that the selectivity of TPC2 to Ca2+ and Na+ might not be fixed. To directly test this, we measured cation conductance evoked by the two compounds under bi-ionic conditions (luminal side: 105 mM Ca2+, pH 4.6, cytosolic side: 160 mM Na+, pH 7.2) (Figure 2K and L). For these experiments, we used cells stably expressing TPC2. Based on the reversal potential (Erev), the PCa/PNa permeability ratio of TPC2-A1-N-evoked currents (ITPC2-A1-N) was 0.65 ± 0.13 (Figure 2O–Q). In marked contrast, Erev values for TPC2-A1-P-evoked currents (ITPC2-A1-P) were more negative such that the PCa/PNa permeability ratio was 0.04 ± 0.01 (Figure 2O–Q) and thus Na+-selective. These ratios are highly reminiscent of those obtained for endogenous NAADP-mediated currents in planar patch-clamp analyses (Ruas et al., 2015) and for recombinant TPC2 activated by PI(3,5)P2 measured using endo-lysosomal patch-clamp experiments where the channel appeared insensitive to NAADP (Wang et al., 2012). We thus examined the effects of NAADP and PI(3,5)P2 in parallel. NAADP (50 nM) evoked robust currents with an Erev similar to TPC2-A1-N (−8.5 mV for NAADP versus −12 mV for TPC2-A1-N). The PCa/PNa for NAADP (0.73 ± 0.14) was thus similar to TPC2-A1-N (Figure 2M–Q). In contrast, the Erev for PI(3,5)P2 (−53 mV) was similar to TPC2-A1-P (−61 mV) and the previously reported Erev for PI(3,5)P2 (−68 mV) (Wang et al., 2012), corresponding to a PCa/PNa ratio of 0.08 ± 0.01 (Figure 2M–Q). No significant outward Na+ or inward Ca2+ conductances were observed in endo-lysosomes isolated from HEK293 cells expressing ‘pore-dead’ TPC2L265P in the presence of TPC2-A1-N or TPC2-A1-P (Figure 2R–S and Figure 2—figure supplement 1B–C). Moreover, the agonists induced similar shifts in selectivity using luminal solution containing 114 mM Na+ and 30 mM Ca2+ and a bath solution containing 160 mM Na+ (Figure 2—figure supplement 1D–F) confirming that the measured PCa/PNa permeability ratio is independent of the luminal Ca2+ concentration. These data indicate that TPC2-A1-N and TPC2-A1-P not only differentially affect current amplitudes but also influence the PCa/PNa permeability ratio (Figure 2Q). TPC2-A1-N-activated TPC2 shows a higher relative Ca2+ permeability, similar to NAADP-activated TPC2. In contrast, TPC2-A1-P-activated TPC2 shows a lower relative Ca2+ permeability, similar to PI(3,5)P2-activated TPC2. Ion permeation through TPC2 is thus ligand-dependent providing an explanation for conflicting evidence that TPC2 is a NAADP-activated Ca2+ release channel (Brailoiu et al., 2010; Grimm et al., 2014; Pitt et al., 2010; Ruas et al., 2015; Schieder et al., 2010) or a PI(3,5)P2 gated Na+ channel (Boccaccio et al., 2014; Guo et al., 2017; Wang et al., 2012).
TPC2 agonists activate TPC2 through distinct sites
The PI(3,5)P2 binding site in TPC2 has recently been identified (She et al., 2019). To gain mechanistic insight into agonist action, we mutated K204 which is required for activation of TPC2 by PI(3,5)P2 (Kirsch et al., 2018; She et al., 2019) and examined its effect on responses to TPC2-A1-N and TPC2-A1-P. Intracellular Ca2+ elevation mediated by TPC2-A1-N was similar in cells expressing plasma membrane targeted TPC2 with and without the K204A mutation (Figure 3A and C). In contrast, the responses to TPC2-A1-P were reduced approximately two-fold (Figure 3B and C). Similar selective inhibition was observed in GCaMP assays (Figure 3G). Thus, whereas responses to TPC2-A1-N were comparable for the wild-type and mutant channel, the responses to TPC2-A1-P were reduced by the mutation. Furthermore, currents evoked by TPC2-A1-N were largely unaffected by the K204A mutation (Figure 3H and J). In contrast, those to TPC2-A1-P were reduced (Figure 3I and J). Collectively, these data suggest K204 is required for activation of TPC2 by TPC2-A1-P and PI(3,5)P2 but not TPC2-A1-N. Molecular determinants for agonist action in TPC2 thus differ.
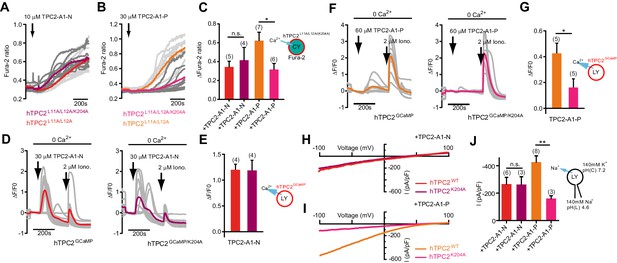
TPC2 agonists activate TPC2 through distinct sites.
(A–B) Representative Ca2+ signals recorded from Fura-2-loaded HeLa cells (n = 10, each) stimulated with the indicated concentration of TPC2-A1-N or -P. Cells were transiently transfected with plasma membrane targeted human TPC2 (hTPC2L11A/L12A) or the PI(3,5)P2-insensitive mutant (TPC2L11A/L12A/K204A). (C) Statistical analysis of the maximal change in Fura-2 ratio (mean ± SEM) with the number of independent transfections in parentheses. An unpaired t-test was applied. *p<0.05. (D–F) Representative Ca2+ signals recorded from HeLa cells (n = 10, each) transiently transfected with GCaMP6(s) fused to wild-type TPC2 (TPC2GCaMP) or the PI(3,5)P2-insensitive mutant (TPC2GCaMP/K204A). Cells were sequentially stimulated with TPC2-A1-N or -P and the Ca2+ ionophore ionomycin in the absence of extracellular Ca2+. (E–G) Statistical analysis of the change in fluorescence intensity (mean ± SEM) of experiments as shown in D and F. An unpaired t-test was applied. *p<0.05. (H–I) Representative TPC2-A1-N or -P-evoked currents from enlarged endo-lysosomes isolated from HEK293 cells transiently expressing human TPC2 (hTPC2) or the PI(3,5)P2-insensitive mutant, (hTPC2K204A). Currents were obtained before and after addition of 10 µM TPC2-A1-N or -P. Recordings were carried out using standard bath and pipette solutions and applying ramp protocols (−100 mV to +100 mV over 500 ms) every 5 s at a holding potential of −60 mV. (J) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV for endo-lysosomes expressing TPC2 or TPC2K204A using TPC2-A1-N or TPC2-A1-P to activate, respectively. *p<0.05 and **p<0.01, using one-way ANOVA followed by Tukey’s post hoc test.
-
Figure 3—source data 1
TPC2 agonists activate TPC2 through distinct sites.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig3-data1-v3.xlsx
TPC2 agonists differentially affect lysosomal pH
Lysosomal function and integrity critically depend on the pH in the lysosomal lumen. Previously, it has been reported that NAADP induced pH changes in the lumen of acidic Ca2+ stores, leading to an alkalinization (Cosker et al., 2010; Morgan and Galione, 2007) but the role of TPC2 in regulating lysosomal pH is conflicting (Ambrosio et al., 2016; Cang et al., 2013; Grimm et al., 2014; Lin et al., 2015; Ruas et al., 2015). We therefore assessed the acute effects of TPC2-A1-N and TPC2-A1-P on lysosomal pH using a recently described novel lysosomal pH sensor, pH-Lemon-GPI (Burgstaller et al., 2019; Figure 4A–K and Video 1). Results from high-resolution array confocal laser scanning microscopy revealed that TPC2-A1-N, but not TPC2-A1-P, increased pH of single vesicles in cells expressing wild-type TPC2 (Figure 4A–B and Video 1). In contrast, no pH changes were observed in cells expressing TPC2L265P. Time-lapse wide-field fluorescence microscopy showed that activation of endogenous TPC2 with TPC2-A1-N modestly increased the lysosomal pH in untransfected cells in a time-dependent manner (Figure 4C and I). The pH response was markedly potentiated in cells expressing wild-type TPC2 but not the pore-mutant (Figure 4D,E and I). The response in the former was approximately half that evoked by direct alkalization with NaN3/NH4Cl (Figure 4K). Again, vesicular pH was largely unaffected by TPC2-A1-P (Figure 4F–H and J). Time-lapse imaging additionally revealed that vesicle motility was strongly impaired by TPC2-A1-N but not TPC2-A1-P (Video 2). The effect of TPC2-A1-N was reversible and appeared to temporally correlate with the increase in vesicular pH (Video 1). In endo-lysosomal patch-clamp experiments, we probed the proton permeability of TPC2 and found that in contrast to TPC2-A1-P, TPC2-A1-N rendered the channel proton-permeable, thus providing a direct channel-dependent mechanism leading to alkalinization (Figure 4L–N). Proton permeability did not substantially change our estimates of relative Ca2+ and Na+ permeability as PCa/PNa values were similar when proton currents were subtracted from currents obtained under bi-ionic conditions (0.44 for TPC2-A1-N and 0.06 for TPC2-A1-P). In sum, these findings demonstrate that TPC2 activation is coupled to lysosomal pH and motility in an agonist-dependent manner.
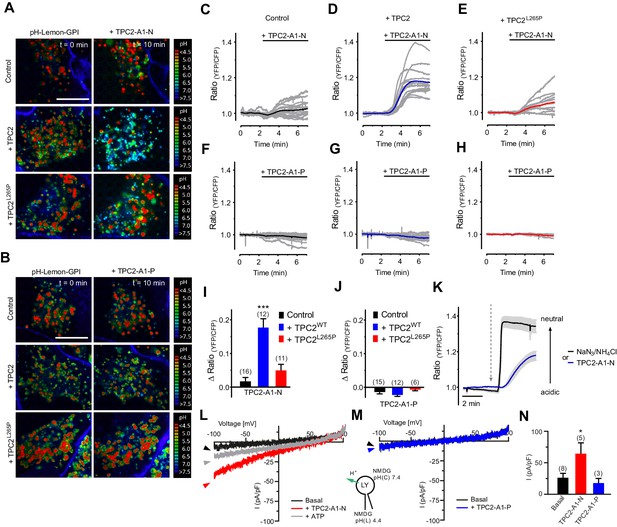
TPC2 agonists differentially affect lysosomal pH and proton conductance.
(A) Representative pseudo colored ratio images (mTurquoise2/EYFP) of vesicle targeted pH-Lemon-GPI in control HeLa cells (upper panel, n = 4/141 cells), HeLa cells positive for wild-type TPC2-mCherry (middle panel, c = 3/70 cells) and HeLa cells positive for TPC2L265P-mCherry (lower panel, n = 3/74 cells) in the absence (t = 0 min; left images) and upon treatment for 10 min with 10 µM TPC2-A1-N (t = 10 min; right images). Scale bar = 5 µm. (B) Representative pseudo colored ratio images (mTurquoise2/EYFP) of vesicle targeted pH-Lemon-GPI in control HeLa cells (upper panel, n = 3/121 cells), HeLa cells positive for wild-type TPC2-mCherry (middle panel, n = 3/59 cells) and HeLa cells positive for TPC2L265P-mCherry (lower panel, n = 3/71 cells) in the absence (t = 0 min; left images) and upon treatment for 10 min with 10 µM TPC2-A1-P (t = 10 min; right images). Scale bar = 5 µm. (C–E) Normalized single cell responses (grey curves) and the respective average response (colored curves) of pH-Lemon-GPI upon treatment with 10 µM TPC2-A1-N in a region of high vesicle density. Shown are ratio curves of pH-Lemon-GPI expressed in control HeLa cells (n = 16 cells; C), in HeLa cells co-expressing wild-type TPC2-mCherry (n = 12 cells; D), and HeLa cells co-expressing TPC2L265P-mCherry (n = 11 cells; E). (F–H) Experiments as in C-E for TPC2-A1-P. Shown are ratio curves of pH-Lemon-GPI expressed in control HeLa cells (n = 15 cells; F), in HeLa cells co-expressing wild-type TPC2-mCherry (n = 12 cells; G), and HeLa cells co-expressing TPC2L265P-mCherry (n = 6 cells, (H). (I–J) Columns represent delta ratio values of pH-Lemon-GPI at min six from curves as shown in panels C-E and F-H, respectively. ***p<0.001 using unpaired student’s t-test. (K) Average ratio signals over time of pH-Lemon-GPI in HeLa cells expressing wild-type TPC2-mCherry in response to either 0.5% NaN3 and 50 mM NH4Cl (black average curve ± SEM, n = 12 cells) or 10 µM TPC2-A1-N (blue average curve ± SEM, n = 12 cells). Cells were treated with compounds as indicated. (L–M) Representative agonist-evoked inward H+ currents from enlarged endo-lysosomes using hTPC2-expressing HEK293 cells. Both bath and pipette solutions contained 1 mM HCl, 5 mM HEPES, 5 mM MES, 150 mM NMDG, pH as indicated, adjusted with MSA. Ramp protocol (−100 mV to +100 mV in 500 ms, holding voltage = 0 mV) was used. Currents were obtained before and after addition of 10 μM TPC2-A1-N (L) or TPC2-A1-P (M). ATP (1 mM) was added at the end of the experiment to block TPC2 (L). (N) Statistical analysis of current densities (mean ± SEM) recorded at −100 mV (inward H+ conductance; H+ from luminal to cytosolic site) as shown in L and M. *p<0.05, using one-way ANOVA followed by Bonferroni’s post hoc test.
-
Figure 4—source data 1
Effect of TPC2-A1-N on vesicular pH.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig4-data1-v3.xlsx
TPC2-A1-N effectively increases vesicular pH in HeLa cells.
Dynamic pseudo-colored ratio (mTurquoise2/EYFP) video of HeLa cells stably expressing pH-Lemon-GPI and co-expressing TPC2 WT upon addition of TPC2-A1-N at the time-point indicated. Pseudo-colored ratio scale with estimated pH values is shown on the right. Scale bar represents 10 µm.
Vesicle movement is impaired upon treatment with TPC2-A1-N, but not with TPC-A1-P.
HeLa cells stably expressing pH-Lemon-GPI and co-expressing either TPC2 WT (upper left panel and lower left panel) or TPC2L265P (upper right panel and lower right panel) were analyzed. Shown are the mTurquoise2 fluorescence signals of pH-Lemon over time upon treatment with TPC2-A1-N (upper panels) and TPC2-A1-P (lower panels) at the time-points indicated. Time-lapse stacks were acquired using wide-field microscopy. Scale bar represents 10 µm.
TPC2 agonists differentially affect lysosomal exocytosis
To further probe the physiological relevance of TPC2 activation by the agonists, we assessed the effect of TPC2-A1-N and TPC2-A1-P on lysosomal exocytosis (Figure 5). Lysosomal exocytosis is involved in a plethora of physiological and pathophysiological processes including release of lysosomal enzymes and inflammatory mediators, clearance of lysosomal storage material and pathogens, plasma membrane repair and cancer progression (Lopez-Castejon and Brough, 2011; Machado et al., 2015; Miao et al., 2015; Xu and Ren, 2015). The Ca2+-dependence of this process has been mostly ascribed to activation of TRPML1 (Xu and Ren, 2015). Here, we used murine macrophages which express high endogenous levels of TPC2 (Cang et al., 2013). To assess lysosomal exocytosis, we quantified translocation of LAMP1 to the cell surface. As shown in Figure 5A–E, TPC2-A1-N was without effect on lysosomal exocytosis. This was not due to lack of efficacy of TPC2-A1-N on the murine channel because Ca2+ measurements with mouse TPC2 fused to GCaMP6 (MmTPC2GCaMP) confirmed that TPC2-A1-N evoked larger responses than TPC2-A1-P (Figure 5—figure supplement 1A) similar to the human channel (Figure 1). In contrast, TPC2-A1-P evoked robust lysosomal exocytosis in a time- and concentration-dependent manner (Figure 5A–E), providing a corollary to the effects of the agonists on lysosomal pH. To further corroborate that TPC2-A1-P triggers TPC2-dependent vesicle fusion in primary macrophages, electrophysiological measurements of membrane capacitance via the whole-cell patch-clamp technique were performed (Figure 5F–H). These data showed that TPC2-A1-P caused a modest increase in cell size. In the final set of experiments, we examined the effects of TPC2 knockout on agonist responses. As shown in Figure 5—figure supplement 1, both TPC2-A1-N and TPC2-A1-P evoked measurable endogenous TPC2-like currents in macrophages from wild-type animals. The amplitudes of the currents were larger for TPC2-A1-P than TPC2-A1-N, again recapitulating agonist effects on human TPC2 (Figure 2). Importantly, currents were reduced in macrophages derived from TPC2 KO cells (Figure 5—figure supplement 1) and the effects of TPC2-A1-P on lysosomal exocytosis and cell size were abolished (Figure 5A–E). TPC2 knockout however failed to affect lysosomal exocytosis in response to ionomycin attesting to specificity (Figure 5A–E). Collectively, these data further identify agonist-selective effects of TPC2 on lysosomal physiology in an endogenous setting.
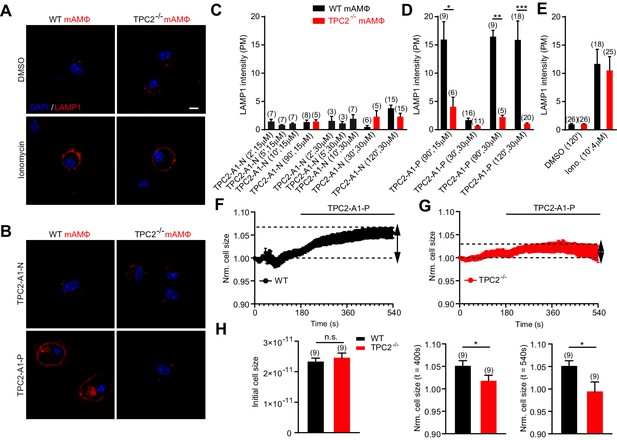
TPC2 agonists differentially affect lysosomal exocytosis.
(A–B) Representative images of LAMP1 translocation assay using murine wild-type and TPC2 KO alveolar macrophages. Shown are results obtained after 120 min treatment with either DMSO, TPC2-A1-P (30 μM) or TPC2-A1-N (30 μM), or 10 min treatment with ionomycin (4 μM). (C–E) Statistical analysis of experiments as shown in A and B at different compound concentrations and incubation times as indicated. Shown are mean values ± SEM. **p<0.01, and ***p<0.001, using two-way ANOVA followed by Bonferroni’s post hoc test. (F–G) Electrophysiological measurements of membrane capacitance via the whole-cell patch-clamp technique as an estimate of cell size (in picofarad [pF]) were used to record fusion of vesicles of primary alveolar macrophages isolated from WT (black, n = 9, F) or TPC2 KO (red, n = 9, G) mice. The normalized, averaged cell size was plotted versus time of the experiment. TPC2-A1-P (30 µM) was applied at 180 s until the end of the experiment indicated by the black bar. Data are shown as mean ± SEM. (H) Bar graphs show mean cell sizes at different time points. The initial cell size was measured immediately after whole cell break in pF and was used for normalization (left panel). Statistical analysis at 400 s (middle panel) and at 540 s (right panel). *p<0.05, unpaired student’s t-test.
-
Figure 5—source data 1
TPC2 agonists differentially affect lysosomal exocytosis.
- https://cdn.elifesciences.org/articles/54712/elife-54712-fig5-data1-v3.xlsx
Discussion
We demonstrate that the ion selectivity of TPC2 is not fixed but rather agonist-dependent. To the best of our knowledge, TPC2 is a unique example of an ion channel that conducts different ions in response to different activating ligands. The novel lipophilic, membrane permeable isoform-selective small molecule agonists of TPC2 (TPC2-A1-N and TPC2-A1-P) characterized herein mimic the physiological actions of NAADP and PI(3,5)P2 most likely through independent binding sites. As such, our data reconcile diametrically opposed opinion regarding activation of TPC2 by endogenous cues and the biophysical nature of the ensuing ion flux. Whether TPC1 also switches its ion selectivity remains to be established. A previous bilayer study found that PI(3,5)P2 did not gate TPC1 but induced modest shifts in relative permeability to different cations when TPC1 was activated by NAADP (Pitt et al., 2014).
Physiologically, we demonstrate inverse effects of the agonists on key lysosomal activities. While TPC2-A1-N increases the pH in the lysosomal lumen in a TPC2-dependent manner, TPC2-A1-P has no significant effect on lysosomal pH. An alkalinizing effect of TPC2-A1-N is in accordance with effects described previously for NAADP, which likewise induces alkalinization (Cosker et al., 2010; Morgan and Galione, 2007) and may be related to agonist-specific effects on proton permeability. The acute nature of these experiments in live cells, made possible by the lipophilicity of TPC2-A1-N, circumvents possible compensatory effects of TPC2 knockout which may have confounded steady-state analyses in previous studies (Cang et al., 2013; Grimm et al., 2014; Ruas et al., 2015). In contrast, TPC2-A1-P but not TPC2-A1-N promoted lysosomal exocytosis. The lack of effect of TPC2-A1-N is surprising given the Ca2+-dependence of lysosomal exocytosis but may relate to its inhibitory effect on lysosomal motility. Boosting lysosomal exocytosis is gaining traction as a strategy to combat disease (Lopez-Castejon and Brough, 2011; Machado et al., 2015; Miao et al., 2015; Xu and Ren, 2015). Selective activation of TPC2 in ‘PI(3,5)P2-mode’ with TPC2-A1-P offers novel scope to achieve this.
In sum, TPC2 can mediate very different physiological and possibly pathophysiological effects depending on how it is activated. TPC2 therefore emerges as an integrator of adenine nucleotide- and phosphoinositide-based messengers with an intrinsic ability to signal through switchable ionic signatures.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | HEK 293 | DSMZ | ACC 305 | |
| Cell line (Homo-sapiens) | HeLa S3 | ATCC | CCL-2.2 | |
| Cell line (Homo-sapiens) | HeLa cells stably expressing pH-Lemon-GPI | This paper | Generated by selective antibiotic G418 disulfate salt from Sigma Aldrich (Cat#A1720). After 4 weeks of cultivation in800 µg/mL, positive cells were selected by FACS analysis. | |
| Cell line (Homo-sapiens) | HEK 293 stably expressing TPC2L11A/L12A -RFP | This paper | Generated by selective antibiotic neomycin from Invitrogen (Cat#10486–025) following the guideline V790-20/V795-20 (Invitrogen) | |
| Cell line (Homo-sapiens) | HEK 293 stably expressing CLN3L253A/I254A -RFP | This paper | Generated by selective antibiotic neomycin from Invitrogen (Cat#10486–025) following the guideline V790-20/V795-20 (Invitrogen) | |
| Recombinant DNA reagent | HsTRPML1-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTRPML2-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTRPML3-YFP | (Grimm et al., 2010) PMID:20189104 | ||
| Recombinant DNA reagent | HsTPC2-YFP | (Chao et al., 2017) PMID:28923947 | ||
| Recombinant DNA reagent | HsTPC2M484L-YFP | (Chao et al., 2017) PMID:28923947 | ||
| Recombinant DNA reagent | HsTPC2L11/12A-YFP | This paper | Generated by site-directed mutagenesis from WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2L11/12A/L265P-YFP | This paper | Generated by site-directed mutagenesis of WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2L11/12A/K204A-YFP | This paper | Generated by site-directed mutagenesis of WT plasmid published by Chao et al. (2017) | |
| Recombinant DNA reagent | HsTPC2-GCaMP6s | This paper | Generated by subcloning (Chao et al., 2017) WT TPC2 construct into Addgene vector #40753 | |
| Recombinant DNA reagent | HsTPC2L265P-GCaMP6s | This paper | Generated by site-directed mutagenesis of WT GCaMP6s plasmid | |
| Recombinant DNA reagent | HsTPC2K204A-GCaMP6s | This paper | Generated by site-directed mutagenesis of WT GCaMP6s plasmid | |
| Recombinant DNA reagent | MmTPC2-GCaMP6s | This paper | Generated by subcloning GCaMP6s from Addgene vector #40753 into WT MmTPC2 vector | |
| Recombinant DNA reagent | HsTPC2L11/12A-GFP | (Brailoiu et al., 2010) PMID:20880839 | ||
| Recombinant DNA reagent | HsTPC2L11/12A-RFP | (Brailoiu et al., 2010) PMID:20880839 | ||
| Recombinant DNA reagent | HsTPC2L11/12A/L265P-GFP | This paper | Generated by site-directed mutagenesis of HsTPC2L11/12A-GFP (plasmid) | |
| Recombinant DNA reagent | TRPML1ΔNC-GFP | (Yamaguchi et al., 2011) PMID:21540176 | ||
| Recombinant DNA reagent | HsTPC2L11/12A/K204A-GFP | (She et al., 2019) PMID:30860481 | ||
| Recombinant DNA reagent | MmTPC1-YFP | (Zong et al., 2009) PMID:19557428 | Generated by subcloning into TOPO pcDNA3.1-YFP, Addgene vector (#13033) | |
| Biological sample | Macrophages from TPC2 KO mouse | (Grimm et al., 2014) PMID:25144390 | ||
| Antibody | LAMP1 antibody | Santa Cruz Biotechnology | sc19992 | 1:200 |
| Transfection reagent | PolyJet | SignaGen Laboratories | SL100688 | |
| Transfection reagent | TurboFect | Thermo Fisher | R0531 | |
| Transfection reagent | Lipofectamine 2000 | Thermo Fisher | 11668 | |
| Commercial assay or kit | Fluo-4 AM, cell permeant | Thermo Fisher | F14202 | |
| Commercial assay or kit | Fura-2 AM, cell permeant | Thermo Fisher | F1221 | |
| Chemical compound, drug | NAADP | Tocris | 3905 | |
| Chemical compound, drug | PI(3,5)P2 | Echelon Biosciences | P-3508 | |
| Chemical compound, drug | Ionomycin | Sigma Aldrich and Cayman Chemical | I-0634 and 11932 | |
| Chemical compound, drug | Tetrandrine | Sigma Aldrich and Santa Cruz Biotechnology | T2695 and sc201492A | |
| Chemical compound, drug | Raloxifene | Cayman Chemical | 10011620 | |
| Chemical compound, drug | Fluphenazine | Sigma Aldrich | F4765 | |
| Chemical compound, drug | ML-SA1 | Merck | 648493 | |
| Chemical compound, drug | ATP | Sigma Aldrich | A9187 | |
| Chemical compound, drug | Sodiumazide (NaN3) | Sigma Aldrich | 09718 | |
| Chemical compound, drug | Ammoniumchloride (NH4Cl) | Sigma Aldrich | S2002 | |
| Software, algorithm | Origin8 | OriginLab | ||
| Software, algorithm | GraphPad Prism | GraphPad Software Inc | ||
| Other | Glass Bottom Dish 35 mm | ibidi | 81218 | |
| Other | Perfusion Chamber PC30 | Next Generation Fluorescence Imaging | PC30 (www.ngfi.eu) | Perfusion chamber used with gravity based perfusion system (NGFI, Graz, Austria) |
| Other | µ-Slide 8 Well | ibidi | 80826 |
High-throughput screening
Request a detailed protocolThe high-throughput screen was performed using a custom-made fluorescence imaging plate reader built into a robotic liquid handling station (Freedom Evo 150, Tecan, Männedorf, Switzerland) as previously described (Urban et al., 2016). HEK293 cells were used that stably expressed RFP fusion proteins of human TPC2 (C-terminally tagged) or human CLN3 (N-terminally tagged) rerouted to the plasma membrane. Targeting was achieved by mutation of the endo-lysosomal targeting motifs (hTPC2L11A/L12A, hCLN3L253A/I254A). Mutants were generated by site-directed mutagenesis using the QuikChange II XL protocol (Agilent), according to the manufacturer’s instructions. Stable HEK293 cell lines were generated using 400 μg/mL geneticin (G418, Sigma). If G418-resistant foci were not identified after 3–4 days, the concentration of G418 was increased to 800 μg/mL. After 2–3 weeks cells were picked from G418-resistant foci and colonies were expanded in six well plates. RFP expression was assessed using confocal microscopy when cells were >50% confluent. Colonies with more than 95% RFP positive cells were selected, grown to >90% confluency, split and further expanded. For HTS experiments, cells were cultured at 37°C with 5% of CO2 in Dulbecco’s modified Eagle medium (Thermo Fisher), supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany), 2 mM L-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 400–800 µg/mL G418. Cells were seeded on black-walled, clear-bottom 384-well plates (Greiner, Germany) and incubated with Fluo-4/AM (4 µM; Life Technologies, Eugene, OR) for 30 min at 37°C, washed and resuspended in a HEPES-buffered solution 1 (HBS1) comprising 132 mM NaCl, 6 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM HEPES, and 5.5 mM D-glucose (pH was adjusted to 7.4 with NaOH). For primary screening, individual compounds from Roche libraries (Xplore libraries X30 and X50, Roche, Basel, CH) were diluted in HBS1 to a working concentration of 100 µM. After recording the baseline for 30 s, compounds were injected to a final concentration of 10 µM. Recording continued for 180 s per quadrant (total 750 s). If high intensities were measured in both cell lines, the compounds were deemed false positives and excluded. Concentration-effect relationships were plotted using GraphPad Prism five and fitted to the Hill equation. The identity of the cell lines used has been authenticated by STR profiling. No mycoplasma contamination has been reported.
Ca2+ imaging
Request a detailed protocolSingle cell Ca2+ imaging experiments were performed using Fura-2. HeLa cells and HEK293 cells were cultured at 37°C with 5% of CO2 in Dulbecco’s modified Eagle medium (Gibco), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cells were plated onto poly-L-lysine (sigma)-coated glass coverslips, grown overnight and transiently transfected for 18–24 hr with plasmids using lipofectamine 2000 (Invitrogen) or TurboFect (Thermo Fisher) according to the manufacturer’s instructions. For Ca2+ influx experiments, cells were transfected with human TPC2 (C-terminally tagged with GFP or RFP) (Brailoiu et al., 2010) or TRPML1 (N-terminally-tagged with GFP (Yamaguchi et al., 2011) or C-terminally tagged with YFP). They were targeted to the plasma membrane by mutation/deletion of the endo-lysosomal targeting motifs. For Ca2+ imaging experiments with TRPML2 or TRPML3 (both C-terminally tagged with YFP Grimm et al., 2010), the latter two sufficiently locating to the plasma membrane when overexpressed. A pore-dead mutant of plasma membrane, GFP-tagged TPC2 (hTPC2L11A/L12A/L265P) was generated by site-directed mutagenesis. Transfected cells were loaded for 1 hr at room temperature with Fura-2 AM (2.5 µM) and 0.005% (v/v) pluronic acid (both from Invitrogen) in HEPES-buffered solution 2 (HBS2) comprising 1.25 mM KH2PO4, 2 mM CaCl2, 2 mM MgSO4, 3 mM KCl, 156 mM NaCl, 10 mM D-glucose and 10 mM HEPES (adjusted to pH 7.4 with HCl). After loading, cells were washed three times in HBS2, and mounted in an imaging chamber. All recordings were performed in HBS2. Ca2+ imaging experiments were also performed in HEK293 cells stably expressing C-terminally RFP-tagged hTPC2L11A/L12A (see above). These cells were plated, loaded with Fura-2 and recorded as with HeLa cells, except for that in indicated experiments, NaCl in HBS2 was replaced with NMDG. For lysosomal Ca2+ release experiments, HeLa cells were transfected with human TPC2, C-terminally tagged with GCaMP6(s) (hTPC2GCaMP). The hTPC2 sequence was amplified by PCR from the pcDNA3.1 plasmid encoding YFP-tagged hTPC2 as described previously (Chao et al., 2017), using a forward primer carrying a NheI restriction site followed by a Kozak sequence, and a sequence binding the first 17 base-pairs of hTPC2: TATGCTAGCGCCACCATGGCGGAACCCCAGGC. The reverse primer also contained a NheI restriction site, preceded by a GSG-linker coding sequence, and the final 20 base-pairs of hTPC2, excluding the TPC2 stop codon: ATAGCTAGCACCAGAACCCCTGCACAGCCACAGGTG. The PCR product was cloned into the GCaMP6(s) (Addgene ID 40753) vector by insertion into a NheI site, yielding plasmids encoding 6xHis tag-HsTPC2-GSG-Xpress tag-GCaMP6 fusion proteins. The ligated plasmid was sequenced by Sanger sequencing using standard CMV forward and pEGFP reverse primers, to confirm the insertion took place as desired. A pore dead mutant (hTPC2GCaMP/L265P) was generated by site-directed mutagenesis using the QuikChange II XL protocol (Agilent), according to the manufacturer’s instructions, using the forward primer sequence GAGTCTCTGACTTCCCCCCTGGTGCTGCTGAC (reverse primer reverse complement of the former). All recordings were performed in nominally Ca2+-free HBS2. Images were acquired every 3 s at 20X (Fura-2) or 40X magnification using a cooled coupled device camera (TILL photonics) attached to an Olympus IX71 inverted fluorescence microscope fitted with a monochromator light source. Fura-2 was excited at 340 nm/380 nm, and GCaMP6(s) was excited at 470 nm. Emitted fluorescence was captured using 440 nm or 515 nm long-pass filters, respectively.
Endo-lysosomal patch-clamp experiments
Request a detailed protocolManual whole-endo-lysosomal patch-clamp recordings were performed as described previously (Chen et al., 2017). HEK293 cells were plated onto poly-L-lysine (Sigma)-coated glass coverslips, grown over night and transiently transfected for 17–25 hr with plasmids using TurboFect (Thermo Fisher) according to the manufacturer’s instructions. Cells expressing wild-type (hTPC2) and a gain-of-function variant (hTPC2M484L) of human TPC2 tagged at their C-termini with YFP were used (Chao et al., 2017). Cells were treated with either vacuolin or YM201636 (1 μM and 800 nM overnight, respectively) to enlarge endo-lysosomes. Currents were recorded using an EPC-10 patch-clamp amplifier (HEKA, Lambrecht, Germany) and PatchMaster acquisition software (HEKA). Data were digitized at 40 kHz and filtered at 2.8 kHz. Fast and slow capacitive transients were cancelled by the compensation circuit of the EPC-10 amplifier. Glass pipettes for recording were polished and had a resistance of 4–8 MΩ. For all experiments, salt-agar bridges were used to connect the reference Ag-AgCl wire to the bath solution to minimize voltage offsets. Liquid junction potential was corrected as described (Chen et al., 2017). For the application of agonists, cytoplasmic solution was completely exchanged. Unless otherwise stated, the cytoplasmic solution comprised 140 mM K-MSA, 5 mM KOH, 4 mM NaCl, 0.39 mM CaCl2, 1 mM EGTA and 10 mM HEPES (pH was adjusted with KOH to 7.2). Luminal solution comprised 140 mM Na-MSA, 5 mM K-MSA, 2 mM Ca-MSA, 1 mM CaCl2, 10 mM HEPES and 10 mM MES (pH was adjusted to 4.6 with MSA). 500 ms voltage ramps from −100 to +100 mV were applied every 5 s, holding potential at 0 mV. The current amplitudes at −100 mV were extracted from individual ramp current recordings. For MmTPC1 measurements a one step protocol was applied (+140 mV over 2 s, holding potential of −70 mV) and the cytoplasmic solution contained 140 mM Na-gluconate, 5 mM NaOH, 4 mM KCl, 2 mM MgCl2, 0.39 mM CaCl2, 1 mM EGTA and mM 10 HEPES (pH 7.2). The luminal solution contained 140 mM Na-MSA, 5 mM K-MSA, 2 mM Ca-MSA, 1 mM CaCl2, 10 mM HEPES and 10 mM MES (pH was adjusted to 4.6 with MSA). All statistical analyses were done using Origin8 software.
For analyses under bi-ionic conditions, HEK293 cells stably expressing hTPC2 tagged at its C-terminus with YFP were used. The cytoplasmic solution comprised 160 mM NaCl and 5 mM HEPES (pH was adjusted with NaOH to 7.2) and the luminal solution comprised 105 mM CaCl2, 5 mM HEPES and 5 mM MES (pH was adjusted to 4.6 with MSA). The permeability ratio (PCa/PNa) was calculated according to Fatt and Ginsborg (1958):
for bi-ionic test (Figure 2Q and Figure 2—figure supplement 1F), and equation according to Eq. 13.47 from Jackson (2006),
for internal and external solutions containing the same monovalent and divalent cations (Figure 2—figure supplement 1D–F). PCa = Ca2+ permeability; PNa = Na+ permeability; γCa = Ca2+ activity coefficient (0.52); γNa = Na+ activity coefficient (0.75); [Ca]o = concentration of Ca2+ in the lumen; [Na]i = concentration of Na+ in the cytosol; [Na]o = concentration of Na+ in the lumen; Erev = reversal potential; F, R = standard thermodynamic constants; T = temperature.
Isolation of murine alveolar macrophages
Request a detailed protocolFor preparation of primary alveolar macrophages, mice were deeply anesthetized and euthanized by exsanguination. The trachea was exposed and cannulated by inserting a 20-gauge catheter (B. Braun). Cells were harvested by eight consecutive lung lavages with 1 mL of DPBS (Dulbeccos’s Phosphate-Buffered Saline) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% antibiotics. For experimentation, alveolar macrophages were directly seeded onto 12 mm glass cover slips and used within 5 days after preparation.
Lysosomal exocytosis experiments
Request a detailed protocolAlveolar macrophages (3 × 104), isolated from wild-type and TPC2 KO mice (described in Grimm et al., 2014) were seeded on 8-well plates (Ibidi) and cultured overnight. Cells were washed once with Minimum Essential Media (MEM) supplemented with 10 mM HEPES and then treated with TPC2-A1-N or TPC2-A1-P as indicated. Ionomycin (4 µM for 10 min) was used as positive control. Following treatment, cells were incubated with an anti-LAMP1 antibody (1:200, SantaCruz) in MEM supplemented with 10 mM HEPES and 1% BSA for 20 min on ice. Cells were then fixed with 2.6% PFA (Thermo Fisher) for 20 min and incubated with Alexa Fluor 488 conjugated secondary antibody (Thermo Fisher) for 1 hr in PBS containing 1% BSA. Nuclei were stained with DAPI. Confocal images were acquired using an LSM 880 microscope (Zeiss) with 40X magnification.
Capacitance measurements
Request a detailed protocolVesicle fusion of wild-type and TPC2-deficient primary alveolar macrophages was analyzed by measuring cell membrane capacitance using the patch-clamp technique as previously described (Zierler et al., 2016). In brief, vesicle fusion was recorded using the automated capacitance cancellation function of the EPC-10 (HEKA, Lambrecht, Germany). Measurements were performed in a tight seal whole-cell configuration at room temperature. Membrane capacitance values directly captured after breaking the seal between the membrane and the glass pipette were used as a reference for the initial cell size. Further recorded capacitance values were normalized to this initial determined capacitance. Extracellular solution contained: 140 mM NaCl, 1 mM CaCl2, 2.8 mM KCl, 2 mM MgCl2, 10 mM HEPES-NaOH, 11 mM glucose (pH 7.2, 300 mosmol/L). Internal solution contained: 120 mM potassium glutamate, 8 mM NaCl, 1 mM MgCl2, 10 mM HEPES-NaOH, 0.1 mM GTP (pH 7.2, 280 mosmol/L). At 180 s TPC2-A1-P (30 µM, diluted in external solution) was applied via an application pipette.
Lysosomal pH measurements
Request a detailed protocolCell culture and transfection: DMEM (Sigma Aldrich) containing 10% fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2.5 µg/mL Fungizone (Thermo Fisher) was used to grow HeLa cells (all obtained from ATCC, Guernsey, UK). Transfection of cells in six-well (Greiner-Bio-One, Kremsmünster, Austria) was performed using PolyJet transfection reagent (SignaGen Laboratories, Rockville). HeLa cells stably expressing pH-Lemon-GPI (NGFI, Graz, Austria) were generated by selection with 800 µg/mL of G418 (Sigma Aldrich, St. Louis, USA) and FACS sorting for CFP (excitation at 405 nm). Wide-field fluorescence microscopy experiments were performed using an OLYMPUS IX73 inverted microscope (OLYMPUS, Vienna, Austria) using a 40X objective (UApo N 340, 40X/1.35 Oil, ∞/0.17/FN22, OLYMPUS). Illumination was performed using an OMICRON LedHUB High-Power LED Light Engine, equipped with a 455 nm and 505 nm LED light source (OMICRON electronics, Vienna, Austria), and 430 nm and 500 nm excitation filters (AHF Analysentechnik, Tübingen, Germany), respectively. Images were captured at a binning of 2 using a Retiga R1 CCD camera (TELEDYNE QIMAGING, Surrey, Canada) and emissions were separated using an optical beam splitter (DV2, Photometrics, Arizona). Array Confocal microscopy was performed using an array confocal laser scanning microscope (ACLSM) built on a fully automatic inverse microscope (Axio Observer.Z1, Zeiss, Göttingen, Germany) using a 100x objective (Plan-Apochromat 100x, 1.4 Oil M27). Excitation was performed using laser light of diode lasers (Visitron Systems): CFP of pH-lemon was excited at 445 nm and 505 nm respectively. Emitted light was acquired with emission filters ET480/40 m for CFP and ET525/50 m for EYFP (Chroma Technologies, VT). A Photometrics CCD camera (CoolSnap HQ2) was used to capture all images at a binning of 1. Device control and image acquisition was performed using VisiView image acquisition and control software (Visitron Systems, Puchheim, Germany) for both devices. Images were processed using MetaMorph analysis software (Molecular Devices, San Jose). Data and statistical analyses were done using GraphPad Prism Software.
Synthesis of the compounds
Request a detailed protocolAll chemicals used were of analytical grade and were obtained from abcr (Karlsruhe, Germany), Fischer Scientific (Schwerte, Germany), Sigma-Aldrich (now Merck, Darmstadt, Germany), TCI (Eschborn, Germany) or Th. Geyer (Renningen, Germany). HPLC grade and dry solvents were purchased from VWR (Darmstadt, Germany) or Sigma-Aldrich, all other solvents were purified by distillation. Hydrophobic phase separation filters (MN 617 WA, 125 mm) were purchased from Macherey Nagel (Düren, Germany). All reactions were monitored by thin-layer chromatography (TLC) using pre-coated plastic sheets POLYGRAM SIL G/UV254 from Macherey-Nagel and detected by irradiation with UV light (254 nm or 366 nm). Flash column chromatography (FCC) was performed on Merck silica gel Si 60 (0.015–0.040 mm).
NMR spectra (1H, 13C, DEPT, COSY, HSQC/HMQC, HMBC) were recorded at 23°C on an Avance III 400 MHz Bruker BioSpin or Avance III 500 MHz Bruker BioSpin instrument, unless otherwise specified. Chemical shifts δ are stated in parts per million (ppm) and are calibrated using residual protic solvents as an internal reference for proton (CDCl3: δ = 7.26 ppm, (CD3)2SO: δ = 2.50 ppm) and for carbon the central carbon resonance of the solvent (CDCl3: δ = 77.16 ppm, (CD3)2SO: δ = 39.52 ppm). Multiplicity is defined as s = singlet, d = doublet, t = triplet, q = quartet, sext = sextet, m = multiplet. NMR spectra were analyzed with NMR software MestReNova, version 12.0.1–20560 (Mestrelab Research S.L.). High-resolution mass spectra were performed by the LMU Mass Spectrometry Service applying a Thermo Finnigan MAT 95 or Joel MStation Sektorfeld instrument at a core temperature of 250°C and 70 eV for EI or a Thermo Finnigan LTQ FT Ultra Fourier Transform Ion Cyclotron Resonance device at 250°C for ESI. IR spectra were recorded on a Perkin Elmer FT-IR Paragon 1000 instrument as neat materials. Absorption bands were reported in wave number (cm−1) with ATR PRO450-S. Melting points were determined by the open tube capillary method on a Büchi melting point B-540 apparatus and are uncorrected. Microwave-assisted reactions were carried out in a Discover (S-Class Plus) SP microwave reactor (CEM GmbH, Kamp-Lintfort, Germany). HPLC purities were determined using an HP Agilent 1100 HPLC with a diode array detector and an Agilent Poroshell column (120 EC-C18; 3.0 × 100 mm; 2.7 micron) with acetonitrile/water as eluent (60:40 acetonitrile/water + 0.1% formic acid).
Preparation of the TPC2-A1-N series
General procedure A – Amide coupling
Request a detailed protocolAccording to Sjogren et al. (1991) the appropriate aniline (1.0 eq.) and 2-cyanoacetic acid (1.0 eq.) were dissolved in DMF and cooled to 0°C. DCC (1.0 eq.) was added portion wise. The mixture was warmed up to rt over 1 hr and subsequently diluted with hexanes/EtOAc (1:1). Precipitates were removed by filtration and the filtrate was extracted once with 1 M aq. HCl and thrice with EtOAc. The combined organic layers were washed with sat. aq. NaCl solution, dried over Na2SO4, filtered and concentrated in vacuo. Recrystallization from EtOH yielded the desired amides.
General procedure B – synthesis of N-aryl cyanoacetamides
Request a detailed protocolAccording to Sjogren et al. (1991) the appropriate amides received from general procedure A (1.0 eq.) were dissolved in dry THF, the solution was cooled to 0°C and NaH (dispersion in paraffin, 60%, 2.3 eq.) was added. After stirring for 15 min, the appropriate benzoyl chloride (1.1 eq.) was added. The mixture was stirred at 0°C for 1 hr and then cautiously treated with 1 M HCl. The precipitate was collected by filtration, washed with ice water and cold EtOH and recrystallized from toluene to give the desired cyanoacetamides.
If the appropriate benzoyl chloride was not commercially available, it was prepared by refluxing the appropriate benzoic acid (1.1 eq.) in SOCl2 (55 eq.) for 1 hr and concentrating in vacuo. The resulting acid chloride was immediately transferred to the reaction.
Preparation of the TPC2-A1-P series
General procedure C – Paal-Knorr pyrrole synthesis
Request a detailed protocolFollowing a general procedure published by Kang et al. (2010) the appropriate β-ketoester (1.1 eq.) was dissolved in dry THF and cooled to 0°C, before NaH (dispersion in paraffin, 60%, 1.5 eq.) was added portion wise. After the suspension was stirred for 30 min, a solution of appropriate halogenated acetophenone (1.0 eq.) and KI (1.0 eq.) in dry THF was added dropwise. The reaction mixture was allowed to warm up to rt over 2 hr, then poured on water and extracted three times with diethyl ether. The combined organic phases were washed with sat. aq. NaHCO3 solution, dried over Na2SO4, filtered and concentrated in vacuo. The residue was dissolved in acetic acid and the appropriate primary amine (2.0 eq.) was added dropwise. The reaction mixture was stirred at 80°C for 18 hr. The solvent was removed in vacuo, the residue disperged in water and extracted three times with diethyl ether. The collected organic phases were washed with sat. aq. NaHCO3 solution, dried over Na2SO4 and concentrated in vacuo. Purification was accomplished by FCC and recrystallization from EtOH if not otherwise specified.
General procedure D – alkaline deprotection of the pyrrolecarboxylic esters
Request a detailed protocolLiOH (10 eq.) was added to a solution of the appropriate ester (1.0 eq.) in dioxane/H2O (5:1) and the reaction mixture was stirred in a closed vessel under microwave irradiation (pmax = 8 bar, Pmax = 200 W, Tmax = 180°C) for 1–18 hr. The suspension was diluted with water to thrice original volume and aq. 2 M HCl was added dropwise under vigorous stirring until the mixture was strongly acidic. The formed precipitate was collected by filtration, washed with water and dried. If necessary the acids were recrystallized from EtOH to yield the pure products.
Synthesis of TPC2-A1-N and analogs
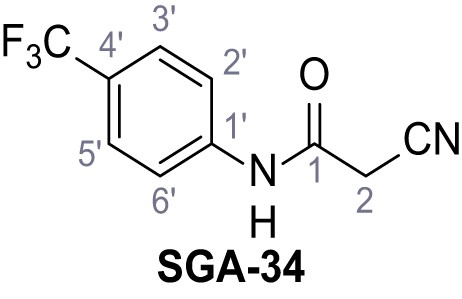
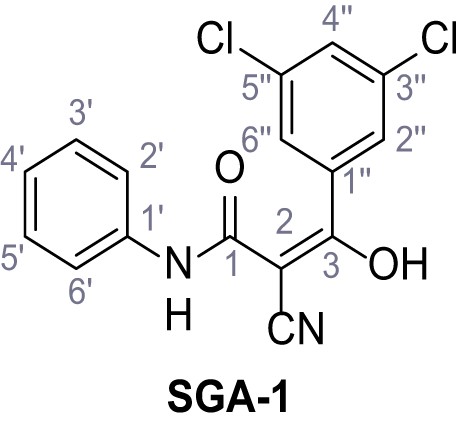
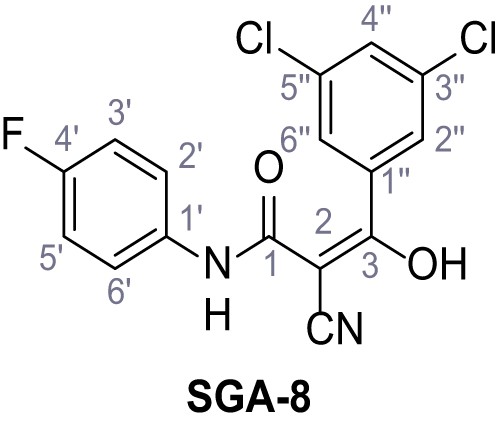
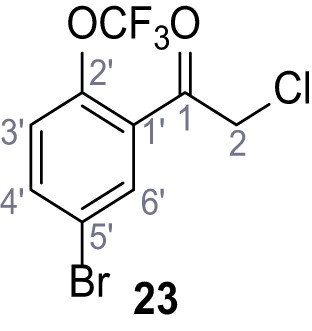
Request a detailed protocolAccording to general procedure A, 4-(trifluoromethyl)aniline (812 µL, 6.47 mmol, 1.1 eq.), 2-cyanoacetic acid (500 mg, 5.88 mmol, 1.0 eq.) and DCC (1.27 g, 6.17 mmol, 1.1 eq.) in DMF (7.0 mL) were used to yield amide SGA-34 as colorless crystals (983 mg, 4.31 mmol, 73%). Analytical data are in accordance with literature (Davies et al., 2009; Sjogren et al., 1991). Rf = 0.14 (4:1 hexanes/acetone). m.p.: 195°C [(Sjogren et al., 1991): 191–193°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.65 (s, 1H, NH), 7.75 (d, J = 8.8 Hz, 2H, 3’-H, 5’-H), 7.70 (d, J = 8.8 Hz, 2H, 2’-H, 6’-H), 3.95 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.9 (C-1), 141.9 (C-1’), 126.3 (q, JCF = 3.7 Hz, C-3’, C-5’), 124.4 (q, JCF = 271.4 Hz, CF3), 124.0 (q, JCF = 32.0 Hz, C-4’), 119.2 (C-2’, C-6’), 115.7 (CN), 27.0 (C-2). IR (ATR) max/cm−1=3287, 3221, 3147, 1681, 1612, 1557, 1319, 1110, 1065, 849, 835. HRMS (ESI): calcd. for C10H6F3N2O (M-H)- 227.04377; found 227.04371. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
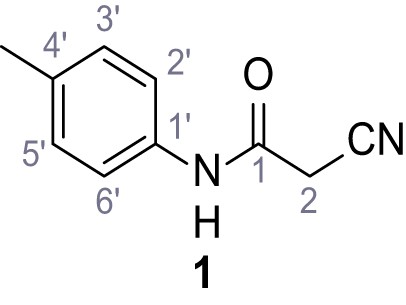
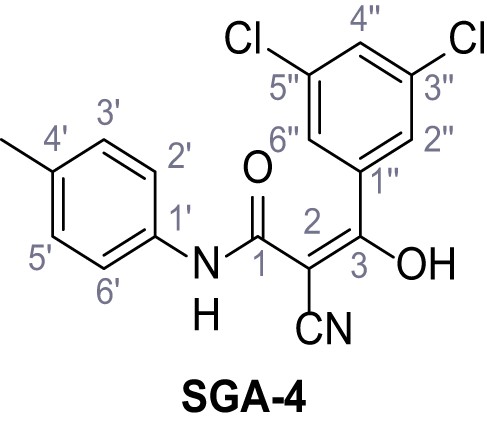
According to general procedure A, p-toluidine (712 µL, 6.47 mmol, 1.1 eq.), 2-cyanoacetic acid (500 mg, 5.88 mmol, 1.0 eq.) and DCC (1.27 g, 6.17 mmol, 1.1 eq.) in DMF (7.0 mL) were used to yield amide one as colorless crystals (728 mg, 4.18 mmol, 71%). Analytical data are in accordance with literature (Yuan et al., 2019). Rf = 0.14 (4:1 hexanes/acetone). m.p.: 184°C [(Yuan et al., 2019): 186°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.19 (s, 1H, NH), 7.47–7.36 (m, 2H, 2’-H, 6’-H), 7.13 (d, J = 8.2 Hz, 2H, 3’-H, 5’-H), 3.86 (s, 2H, 2 hr), 2.25 (s, 3H, CH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 160.7 (CO), 135.9 (C-1’), 132.9 (C-4’), 129.3 (C-3’, C-5’), 119.2 (C-2’, C-6’), 116.0 (CN), 26.6 (C-2), 20.4 (CH3). IR (ATR) max/cm−1=3267, 3207, 3137, 1660, 1613, 1552, 1510, 819. HRMS (ESI): calcd. for C10H9N2O (M-H)- 173.07204; found 173.07194. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
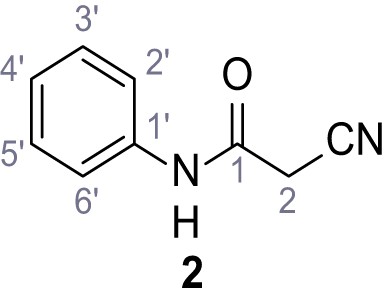
According to general procedure A, aniline (1.96 mL, 21.5 mmol, 1.0 eq.), 2-cyanoacetic acid (2.01 g, 23.6 mmol, 1.1 eq.) and DCC (4.87 g, 23.6 mmol, 1.1 eq.) in DMF (20 mL) were used to yield amide two as colorless crystals (2.60 g, 16.2 mmol, 76%). Analytical data are in accordance with literature (Yuan et al., 2019). Rf = 0.12 (4:1 hexanes/acetone). m.p.: 202°C [(Yuan et al., 2019): 172°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.28 (s, 1H, NH), 7.54 (dt, J = 8.7, 1.6 Hz, 2H, 2’-H, 6’-H), 7.39–7.29 (m, 2H, 3’-H, 5’-H), 7.15–7.04 (m, 1H, 4’-H), 3.89 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 161.0 (CO), 138.4 (C-1’), 128.9 (C-3’, C-5’), 123.9 (C-4’), 119.2 (C-2’, C-6’), 115.9 (CN), 26.7 (C-2). IR (ATR) max/cm−1=3265, 3207, 3143, 3099, 3052, 1653, 1620, 1557, 1299, 943, 761, 696. HRMS (ESI): calcd. for C9H7N2O (M-H)- 159.05639; found 159.05628. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
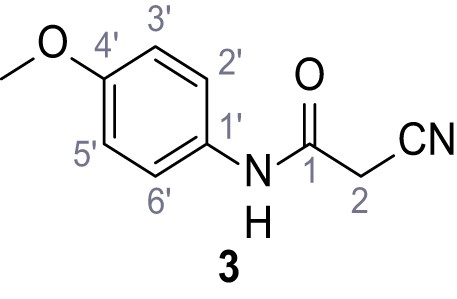
According to general procedure A, p-anisidine (2.36 mL, 20.3 mmol, 1.0 eq.), 2-cyanoacetic acid (19.0 g, 22.3 mmol, 1.1 eq.) and DCC (4.61 g, 22.3 mmol, 1.1 eq.) in DMF (20 mL) were used to yield amide three as pale blue crystals (1.58 g, 8.31 mmol, 41%). Analytical data are in accordance with literature (Yuan et al., 2019). Rf = 0.10 (4:1 hexanes/acetone). m.p.: 137°C [(Yuan et al., 2019): 176°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.14 (s, 1H, NH), 7.53–7.38 (m, 2H, 2’-H, 6’-H), 7.00–6.83 (m, 2H, 3’-H, 5’-H), 3.84 (s, 2H, 2 hr), 3.72 (s, 3H, OCH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 160.4 (CO), 155.6 (C-4’), 131.4 (C-1’), 120.8 (C-2’, C-6’), 116.0 (CN), 114.0 (C-3’, C-5’), 55.2 (OCH3), 26.5 (C-2). IR (ATR) max/cm−1=3299.3150, 1655, 1608, 1557, 1511, 1251, 1032, 828. HRMS (ESI): calcd. for C10H9N2O2 (M-H)- 189.06695; found 189.06688. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
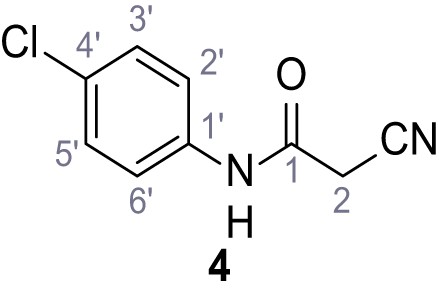
According to general procedure A, 4-chloroaniline (682 µL, 23.0 mmol, 1.0 eq.), 2-cyanoacetic acid (2.16 g, 25.4 mmol, 1.1 eq.) and DCC (5.23 g, 25.4 mmol, 1.1 eq.) in DMF (20 mL) were used to yield amide four as colorless crystals (3.46 g, 17.8 mmol, 77%). Analytical data are in accordance with literature (Yuan et al., 2019). Rf = 0.14 (3:2 hexanes/acetone). m.p.: 207°C [(Yuan et al., 2019): 179°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.42 (s, 1H, NH), 7.62–7.51 (m, 2H, 2’-H, 6’-H), 7.50–7.27 (m, 2H, 3’-H, 5’-H), 3.91 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 161.2 (CO), 137.3 (C-1’), 128.8 (C-3’, C-5’), 127.5 (C-4’), 120.8 (C-2’, C-6’), 115.8 (CN), 26.8 (C-2). IR (ATR) max/cm−1=3264, 3200, 3132, 3083, 1664, 1610, 1548, 1491, 832. HRMS (ESI): calcd. for C9H635ClN2O (M-H)- 193.01741; found 193.01750. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
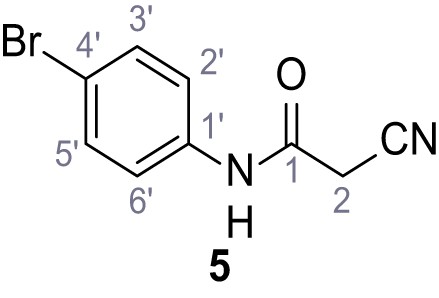
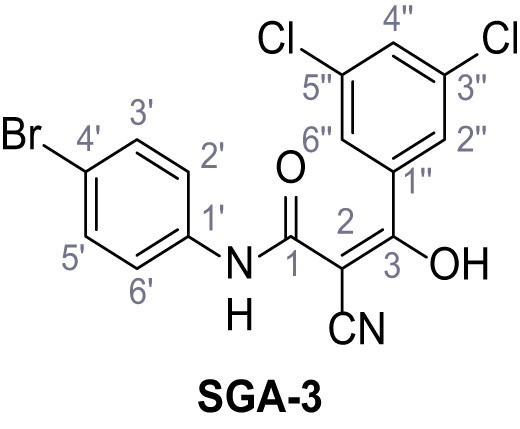
According to general procedure A, 4-bromoaniline (2.00 mL, 17.1 mmol, 1.0 eq.), 2-cyanoacetic acid (1.60 g, 18.8 mmol, 1.1 eq.) and DCC (3.88 g, 18.8 mmol, 1.1 eq.) in DMF (20 mL) were used to yield amide five as colorless crystals (1.62 g, 6.76 mmol, 40%). Analytical data are in accordance with literature (Yuan et al., 2019). Rf = 0.14 (4:1 hexanes/acetone). m.p.: 186°C [(Yuan et al., 2019): 185°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.42 (s, 1H, NH), 7.52 (s, 4H, 2’-H, 3’-H, 5’-H, 6’-H), 3.90 (s, 2H, 2 hr).13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.2 (CO), 137.7 (C-1‘), 131.7 (C-2‘, C-6’ or C-3’, C-5’), 121.2 (C-2‘, C-6’ or C-3’, C-5’), 115.8 (C-4’), 115.5 (CN), 26.8 (C-2). IR (ATR) max/cm−1=3322, 2927, 2849, 1608, 1547, 1245, 828. HRMS (ESI): calcd. for C9H679BrN2O (M-H)- 236.96690; found 236.96692. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
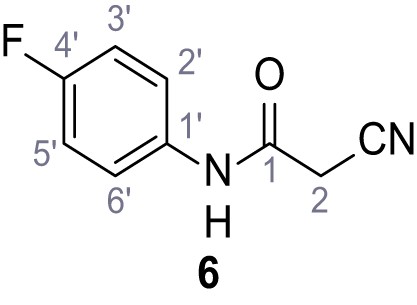
According to general procedure A, 4-fluoroaniline (2.16 mL, 22.5 mmol, 1.0 eq.), 2-cyanoacetic acid (1.91 g, 22.5 mmol, 1.0 eq.) and DCC (4.64 g, 22.5 mmol, 1.0 eq.) in DMF (20 mL) were used to yield amide six as colorless crystals (3.00 g, 16.9 mmol, 75%). Analytical data are in accordance with literature (Ammar et al., 2006). Rf = 0.11 (4:1 hexanes/acetone). m.p.: 179°C [(Ammar et al., 2006): 158–160°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.34 (s, 1H, NH), 7.66–7.46 (m, 2H, 2’-H, 6’-H), 7.27–7.09 (m, 2H, 3’-H, 5’-H), 3.89 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.0 (CO), 158.3 (d, JCF = 240.5 Hz, C-4’), 134.7 (d, JCF = 2.7 Hz, C-1’), 121.1 (d, JCF = 7.9 Hz, C-2’, C-6’), 115.9 (CN), 115.5 (d, JCF = 22.3 Hz, C-3’, C-5’), 26.6 (C-2). IR (ATR) max/cm−1=3274, 3166, 3107, 1662, 1623, 1566, 1505, 834. HRMS (ESI): calcd. for C9H6FN2O (M-H)- 177.04696; found 177.04687. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
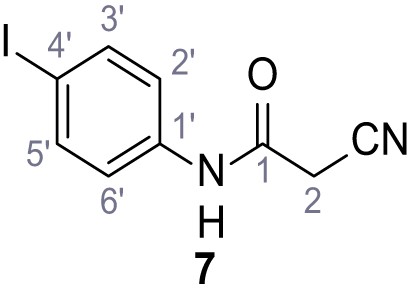
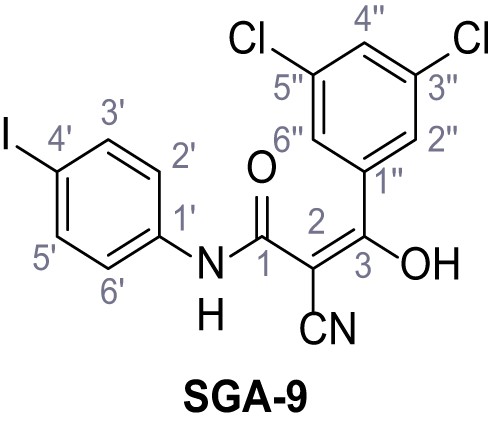
According to general procedure A, 4-iodoaniline (4.50 g, 20.5 mmol, 1.0 eq.), 2-cyanoacetic acid (17.5 g, 20.5 mmol, 1.0 eq.) and DCC (4.24 g, 20.5 mmol, 1.0 eq.) in DMF (20 mL) were used to yield amide seven as pale blue crystals (4.50 g, 15.7 mmol, 77%). The compound is literature known, but no analytical data are available (Sjogren et al., 1991). Rf = 0.11 (4:1 hexanes/acetone). m.p.: 218°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.38 (s, 1H, NH), 7.78–7.60 (m, 2H, 3’-H, 5’-H), 7.49–7.29 (m, 2H, 2’-H, 6’-H), 3.90 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.2 (CO), 138.2 (C-1’), 137.6 (C-3’, C-5’), 121.4 (C-2’, C-6’), 115.8 (CN), 87.6 (C-4’), 26.8 (C-2). IR (ATR) max/cm−1=3265, 3188, 3113, 3078, 1666, 1543, 1391, 1299, 823. HRMS (ESI): calcd. for C9H6IN2O (M-H)- 284.95303; found 284.95302. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
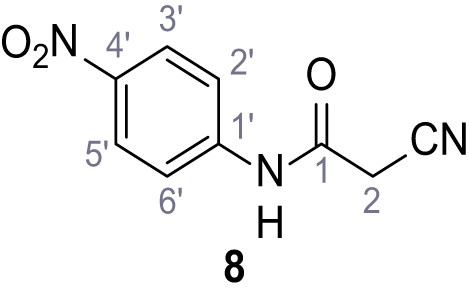
According to general procedure A, 4-nitroaniline (696 µL, 7.24 mmol, 1.0 eq.), 2-cyanoacetic acid (616 mg, 7.24 mmol, 1.0 eq.) and DCC (1.49 g, 7.24 mmol, 1.0 eq.) in DMF (20 mL) were used to yield amide eight as yellow solid (918 mg, 4.47 mmol, 62%). Analytical data are in accordance with literature (Sohn et al., 2017). Rf = 0.11 (4:1 hexanes/acetone). m.p.: 218°C [(Sohn et al., 2017): 220°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.88 (s, 1H, NH), 8.35–8.15 (m, 2H, 3’-H, 5’-H), 7.93–7.71 (m, 2H, 2’-H, 6’-H), 4.00 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 162.2 (CO), 144.4 (C-1’), 142.7 (C-4’), 125.1 (C-3’, C-5’), 119.0 (C-2’, C-6’), 115.5 (CN), 27.2 (C-2). IR (ATR) max/cm−1=3287, 1673, 1562, 1503, 1336, 1259, 860, 748. HRMS (ESI): calcd. for C9H6N3O3 (M-H)- 204.04146; found 204.04146. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
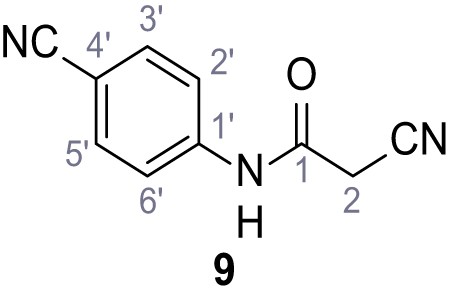
According to general procedure A, 4-aminobenzonitrile (1.00 g, 8.46 mmol, 1.0 eq.), 2-cyanoacetic acid (7.20 g, 8.46 mmol, 1.0 eq.) and DCC (1.75 g, 8.46 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide nine as yellow solid (1.22 g, 6.57 mmol, 78%). The compound is literature known, but no analytical data are available (Sjogren et al., 1991). Rf = 0.08 (4:1 hexanes/acetone). m.p.: 201°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.72 (s, 1H, NH), 7.82–7.78 (m, 2H, 3’-H, 5’-H), 7.74–7.70 (m, 2H, 2’-H, 6’-H), 3.97 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 162.0 (CO), 142.5 (C-1’), 133.5 (C-3’, C-5’), 119.3 (C-2’, C-6’), 118.9 (C-4’), 115.6 (CH2CN), 105.7 (CN), 27.1 (C-2). IR (ATR) max/cm−1=3268, 3194, 3118, 2229, 1599, 1538, 1504, 845. HRMS (ESI): calcd. for C10H6N3O (M-H)- 184.05164; found 184.05161. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
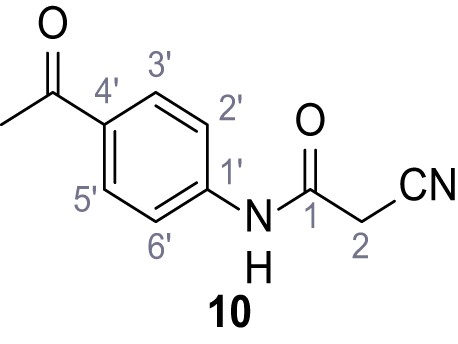
According to general procedure A, 4-aminoacetophenone (1.30 mL, 7.40 mmol, 1.0 eq.), 2-cyanoacetic acid (629 mg, 7.40 mmol, 1.0 eq.) and DCC (1.53 g, 7.40 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 10 as yellow crystals (802 mg, 3.97 mmol, 54%). Analytical data are in accordance with literature (Metwally et al., 2017). Rf = 0.37 (3:2 hexanes/acetone). m.p.: 194°C [(Metwally et al., 2017): 225°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.63 (s, 1H, NH), 7.99–7.91 (m, 2H, 3’-H, 5’-H), 7.74–7.62 (m, 2H, 2’-H, 6’-H), 3.96 (s, 2H, 2 hr), 2.53 (s, 3H, CH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 196.5 (COCH3), 161.7 (HNCO), 142.6 (C-1‘), 132.2 (C-4‘), 129.6 (C-3‘, C-5‘), 118.5 (C-2‘, C-6‘), 115.7 (CN), 27.0 (C-2), 26.5 (CH3). IR (ATR) max/cm−1=3286, 2250, 1695, 1651, 1599, 1536, 1279, 1249, 833, 720. HRMS (ESI): calcd. for C11H9N2O2 (M-H)- 201.06695; found 201.06694. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
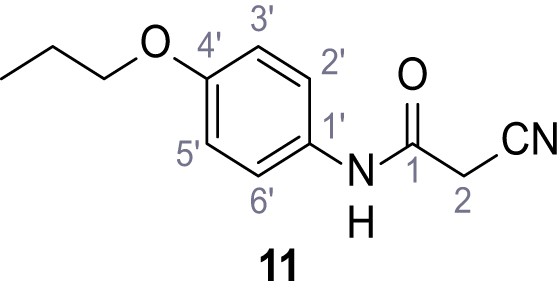
According to general procedure A, 4-propoxyaniline (679 µL, 4.49 mmol, 1.0 eq.), 2-cyanoacetic acid (382 mg, 4.49 mmol, 1.0 eq.) and DCC (927 mg, 4.49 mmol, 1.0 eq.) in DMF (2.0 mL) were used to yield amide 11 as colorless crystals (604 mg, 2.77 mmol, 62%). Rf = 0.24 (95:5 CH2Cl2/EtOH). m.p.: 183°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.13 (s, 1H, NH), 7.51–7.38 (m, 2H, 2’-H, 6’-H), 6.98–6.83 (m, 2H, 3’-H, 5’-H), 3.88 (t, J = 7.1 Hz, 2H, CH2CH2CH3), 3.84 (s, 2H, 2 hr), 1.70 (sext, J = 7.1 Hz, 2H, CH2CH2CH3), 0.96 (t, J = 7.1 Hz, 3H, CH2CH2CH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 160.4 (CO), 155.1 (C-4’), 131.4 (C-1’), 120.8 (C-2’, C-6’), 116.0 (CN), 114.5 (C-3’, C-5’), 69.0 (CH2CH2CH3), 26.5 (C-2), 22.0 (CH2CH2CH3), 10.4 (CH2CH2CH3). IR (ATR) max/cm−1=3283, 3096, 1607, 1559, 1508, 1239, 828, 570. HRMS (ESI): calcd. for C12H13N2O2 (M-H)- 217.09825; found 217.09832. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
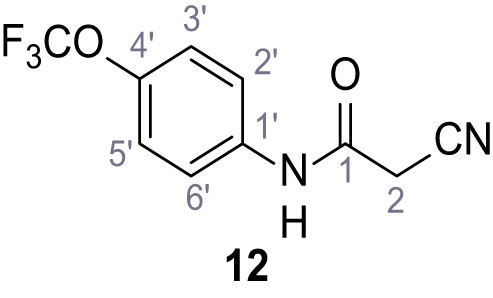
According to general procedure A, 4-(trifluoromethoxy)aniline (939 µL, 7.00 mmol, 1.0 eq.), 2-cyanoacetic acid (595 mg, 7.00 mmol, 1.0 eq.) and DCC (1.44 g, 7.00 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 12 as colorless solid (1.23 g, 5.03 mmol, 72%). The compound is literature known, but no analytical data are available (Sjogren et al., 1991). Rf = 0.39 (95:5 CH2Cl2/EtOH). m.p.: 154°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.49 (s, 1H, NH), 7.74–7.55 (m, 2H, 2’-H, 6’-H), 7.47–7.26 (m, 2H (3’-H, 5’-H), 3.92 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.3 (CO), 144.0 (C-4‘), 137.5 (C-1‘), 121.8 (C-3’, C-5’), 120.7 (C-2’, C-6’), 120.1 (q, JCF = 255.7 Hz, OCF3), 115.8 (CN), 26.8 (C-2). IR (ATR) max/cm−1=3278, 2975, 1667, 1616, 1557, 1508, 1277, 1205, 1171. HRMS (ESI): calcd. for C10H6F3N2O2 (M-H)- 243.03869; found 243.03869. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
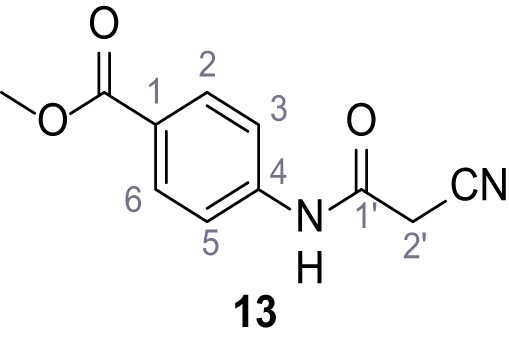
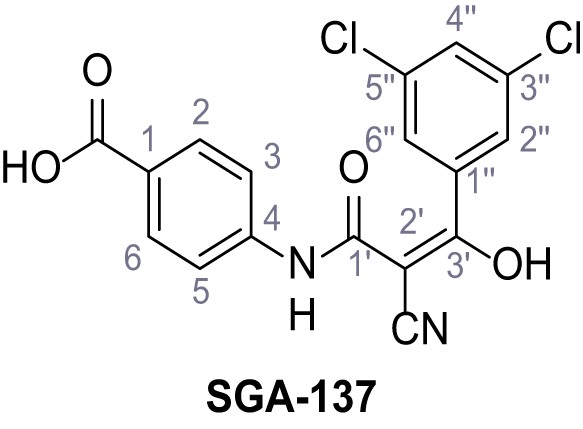
According to general procedure A, methyl 4-aminobenzoate (3.00 g, 19.5 mmol, 1.0 eq.), 2-cyanoacetic acid (1.66 g, 19.5 mmol, 1.0 eq.) and DCC (4.01 g, 19.5 mmol, 1.0 eq.) in DMF (15 mL) were used to yield amide 13 as colorless solid (2.76 g, 12.6 mmol, 65%). The compound is literature known, but no analytical data are available (Sjogren et al., 1991). Rf = 0.44 (3:2 hexanes/acetone). m.p.: 162°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.63 (s, 1H, NH), 8.01–7.87 (m, 2H, 2 hr, 6 hr), 7.76–7.61 (m, 2H, 3 hr, 5 hr), 3.96 (s, 2H, 2 hr), 3.82 (s, 3H, CH3). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 165.7 (COOCH3), 161.7 (HNCO), 142.7 (C-4), 130.4 (C-2, C-6), 124.6 (C-1), 118.6 (C-3, C-5), 115.7 (CN), 52.0 (CH3), 27.0 (C-2). IR (ATR) max/cm−1=2809, 1722, 1608, 1558, 1507, 1431, 1274, 1110, 757. HRMS (ESI): calcd. for C11H9N2O3 (M-H)- 217.06187; found 217.06187. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
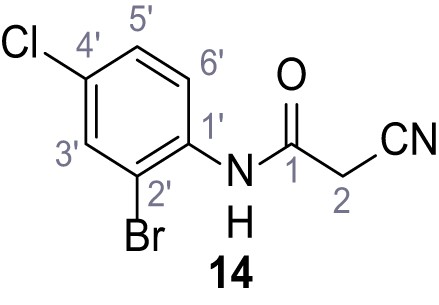
According to general procedure A, 2-bromo-4-chloroaniline (1.00 g, 4.84 mmol, 1.0 eq.), 2-cyanoacetic acid (412 mg, 4.84 mmol, 1.0 eq.) and DCC (999 mg, 4.84 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 14 as colorless crystals (919 mg, 3.36 mmol, 69%). Rf = 0.22 (3:2 hexanes/acetone). m.p.: 157°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 9.97 (s, 1H, NH), 7.83 (d, J = 2.4 Hz, 1H, 3’-H), 7.63 (d, J = 8.7 Hz, 1H, 6’-H), 7.49 (dd, J = 8.7, 2.4 Hz, 1H, 5’-H), 3.98 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.8 (CO), 134.7 (C-1’), 132.0 (C-3’), 130.7 (C-4’), 128.2 (C-5’), 128.0 (C-6’), 118.5 (C-2’), 115.7 (CN), 26.1 (C-2). IR (ATR) max/cm−1=3281, 2258, 1666, 1577, 1530, 1470, 1284, 822. HRMS (ESI): calcd. for C9H579Br35ClN2O (M-H)- 270.92793; found 270.92809. Purity (HPLC): >93% (λ = 210 nm), >93% (λ = 254 nm).
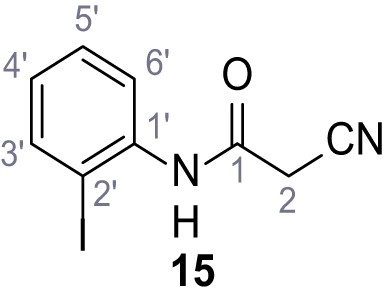
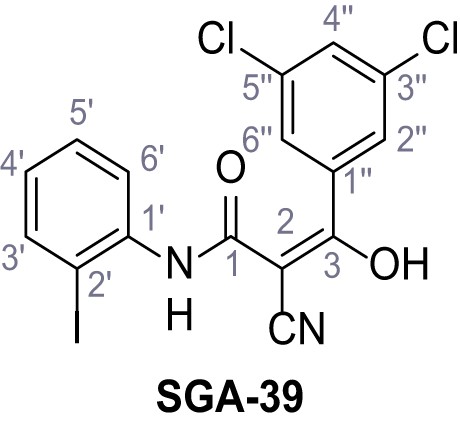
According to general procedure A, 2-iodoaniline (1.00 g, 4.57 mmol, 1.0 eq.), 2-cyanoacetic acid (388 mg, 4.57 mmol, 1.0 eq.) and DCC (942 mg, 4.57 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 15 as brown crystals (913 g, 3.19 mmol, 70%). Rf = 0.16 (4:1 hexanes/acetone). m.p.: 161°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 9.88 (s, 1H, NH), 7.90 (d, J = 7.6 Hz, 1H, 3’-H), 7.46–7.38 (m, 2H, 5’-H, 6’-H), 7.03 (ddd, J = 8.6, 5.4, 3.6 Hz, 1H, 4’-H), 3.93 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.4 (CO), 139.1 (C-3’), 138.7 (C-1’), 128.8 (C-5’), 128.3 (C-4’), 127.4 (C-6’), 115.8 (CN), 96.4 (C-2’), 26.0 (C-2). IR (ATR) max/cm−1=3252, 2263, 1658, 1577, 1542, 1433, 1015, 758, 768. HRMS (ESI): calcd. for C9H6IN2O (M-H)- 284.95303; found 284.95295. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
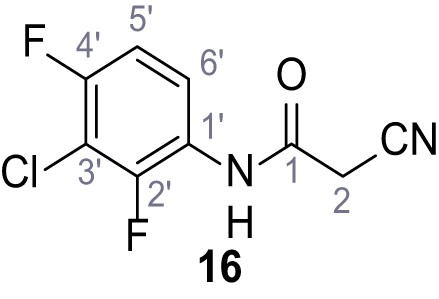
According to general procedure A, 3-chloro-2,4-difluoroaniline (1.06 g, 6.48 mmol, 1.0 eq.), 2-cyanoacetic acid (551 mg, 6.48 mmol, 1.0 eq.) and DCC (1.34 g, 6.48 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 16 as colorless crystals (1.01 g, 4.39 mmol, 68%). Rf = 0.16 (4:1 hexanes/acetone). m.p.: 144°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.32 (s, 1H, NH), 7.79 (td, J = 9.0, 5.8 Hz, 1H, 6’-H), 7.33 (td, J = 9.0, 2.1 Hz, 1H, 5’-H), 3.99 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 162.0 (CO), 154.8 (dd, JCF = 246.5, 2.0 Hz, C-2’ or C-4’), 150.4 (dd, JCF = 250.4, 3.3 Hz, C-2’ or C-4’), 123.4 (dd, JCF = 8.8, 2.4 Hz, C-6’), 123.0 (dd, JCF = 11.8, 3.5 Hz, C-1’), 115.7 (CN), 111.9 (dd, JCF = 21.4, 3.8 Hz, C-5’), 108.7 (dd, JCF = 21.9, 19.7 Hz, C-3’), 26.3 (C-2). IR (ATR) max/cm−1=3274.2935, 2264, 1681, 1551, 1488, 1443, 1012, 831, 628. HRMS (ESI): calcd. for C9H435ClF2N2O (M-H)- 228.99857; found 228.99850. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
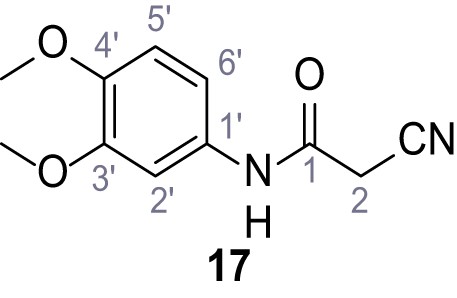
According to general procedure A, 3,4-dimethoxyaniline (1.00 g, 6.53 mmol, 1.0 eq.), 2-cyanoacetic acid (555 mg, 6.53 mmol, 1.0 eq.) and DCC (1.35 g, 6.53 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 17 as violet crystals (1.10 g, 4.99 mmol, 76%). The compound is literature known, but no analytical data are available (Edraki et al., 2016). Rf = 0.05 (4:1 hexanes/acetone). m.p.: 174°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.15 (s, 1H, NH), 7.21 (d, J = 2.4 Hz, 1H, 2’-H), 7.04 (dd, J = 8.7, 2.4 Hz, 1H, 6’-H), 6.90 (d, J = 8.7 Hz, 1H, 5’-H), 3.84 (s, 2H, 2 hr), 3.79–3.66 (m, 6H, 2x OCH3). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 160.5 (CO), 148.6 (C-3’), 145.3 (C-4’), 131.9 (C-1’), 116.0 (CN), 112.0 (C-5’), 111.2 (C-6’), 104.3 (C-2’), 55.7 (OCH3), 55.4 (OCH3), 26.6 (C-2). IR (ATR) max/cm−1=3273, 2914, 2256, 1660, 1513, 1239, 1132, 1020, 837. HRMS (ESI): calcd. for C11H11N2O3 (M-H)- 219.07752; found 219.07751. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
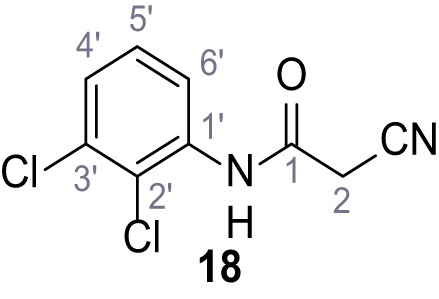
According to general procedure A, 2,3-dichloroaniline (745 µL, 6.30 mmol, 1.0 eq.), 2-cyanoacetic acid (536 mg, 6.30 mmol, 1.0 eq.) and DCC (1.30 g, 6.30 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 18 as colorless crystals (414 mg, 1.81 mmol, 29%). Rf = 0.21 (4:1 hexanes/acetone). m.p.: 176°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.11 (s, 1H, NH), 7.69 (dd, J = 8.1, 1.3 Hz, 1H, 4’-H or 6’-H), 7.51 (dd, J = 8.1, 1.3 Hz, 1H, 4’-H or 6’-H), 7.38 (t, J = 8.1 Hz, 1H, 5’-H), 4.02 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 161.9 (CO), 136.1 (C-1’ or C-3’), 132.0 (C-1’ or C-3’), 128.2 (C-5’), 127.3 (C-4’ or C-6’), 125.3 (C-2’), 124.8 (C-4’ or C-6’), 115.7 (CN), 26.3 (C-2). IR (ATR) max/cm−1=3287, 2253, 1666, 1580, 1527, 1415, 1338, 1182, 953, 788. HRMS (ESI): calcd. for C9H535Cl2N2O (M-H)- 226.97844; found 226.97859. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
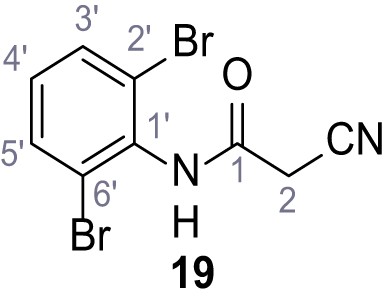
According to general procedure A, 2,6-dibromoaniline (1.00 g, 4.00 mmol, 1.0 eq.), 2-cyanoacetic acid (340 mg, 4.00 mmol, 1.0 eq.) and DCC (825 mg, 4.00 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 19 as colorless crystals (514 mg, 1.62 mmol, 40%). Rf = 0.48 (3:2 hexanes/acetone). m.p.: 187°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.37 (s, 1H, NH), 7.74 (d, J = 8.1 Hz, 2H, 3’-H, 5’-H), 7.22 (t, J = 8.1 Hz, 1H, 4’-H), 3.93 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 161.1 (CO), 134.7 (C-1‘), 132.4 (C-3‘, C-5‘), 130.7 (C-4‘), 123.9 (C-2‘, C-6‘), 115.6 (CN), 25.4 (C-2). IR (ATR) max/cm−1=3326, 2926, 2851, 1626, 1568, 1539, 1242, 642. HRMS (ESI): calcd. for C9H579Br2N2O (M-H)- 314.87741; found 314.87761. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure A, 3,5-bis(trifluoromethyl)aniline (1.09 mL, 7.00 mmol, 1.0 eq.), 2-cyanoacetic acid (595 mg, 7.00 mmol, 1.0 eq.) and DCC (1.44 g, 7.00 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 20 as colorless crystals (1.68 g, 5.67 mmol, 81%). The compound is literature known, but no analytical data are available (Shah et al., 2018). Rf = 0.43 (95:5 CH2Cl2/EtOH). m.p.: 141°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 10.96 (s, 1H, NH), 8.28–8.12 (m, 2H, 2’-H, 6’-H), 7.90–7.77 (m, 1H, 4’-H), 4.00 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 162.4 (CO), 140.2 (C-1’), 130.9 (q, JCF = 32.9 Hz, C-3’, C-5’), 123.1 (q, JCF = 272.7 Hz, CF3), 119.2–118.7 (m, C-2’, C-6’), 116.9–116.5 (m, C-4’), 115.4 (CN), 27.1 (C-2). IR (ATR) max/cm−1=3313, 1695, 1572, 1471, 1381, 1272, 1132, 889, 703, 681. HRMS (ESI): calcd. for C11H5F6N2O (M-H)- 295.03116; found 295.03127. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
According to general procedure A, N-methyl-4-(trifluoromethyl)aniline (403 µL, 2.85 mmol, 1.0 eq.), 2-cyanoacetic acid (243 mg, 2.85 mmol, 1.0 eq.) and DCC (589 mg, 2.85 mmol, 1.0 eq.) in DMF (10 mL) were used to yield amide 21 as colorless solid (513 mg, 2.12 mmol, 74%). Analytical data are in accordance with literature (Kobayashi and Harayama, 2009). Rf = 0.58 (95:5 CH2Cl2/EtOH). m.p.: 70°C [(Kobayashi and Harayama, 2009): 66–68°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 7.77 (d, J = 8.6 Hz, 2H, 3’-H, 5’-H), 7.40 (d, J = 8.2 Hz, 2H, 2’-H, 6’-H), 3.34 (s, 3H, CH3), 3.23 (s, 2H, 2 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 161.4 (CO), 145.6 (C-1’), 131.7 (C-4’), 127.9 (C-2’, C-6’ or C-3’, C-5’), 127.8 (C-2’, C-6’ or C-3’, C-5’), 123.5 (q, J = 271.4 Hz, CF3), 113.7 (CN), 38.1 (CH3), 25.6 (C-2). IR (ATR) max/cm−1=3152, 2355, 1657, 1611, 1322, 1122, 1103, 1065, 848. HRMS (ESI): calcd. for C11H8F3N2O (M-H)- 241.05942; found 241.05939. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure A, 4-(trifluoromethyl)benzylamine (2.50 mL, 17.5 mmol, 1.0 eq.), 2-cyanoacetic acid (1.49 g, 17.5 mmol, 1.0 eq.) and DCC (3.62 g, 17.5 mmol, 1.0 eq.) in DMF (15 mL) were used to yield amide 22 as colorless solid (299 mg, 1.23 mmol, 7%). Analytical data are in accordance with literature (Guo et al., 2011). Rf = 0.46 (3:2 hexanes/acetone). m.p.: 113°C [(Guo et al., 2011): 128°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 8.83 (t, J = 5.7 Hz, 1H, NH), 7.74–7.66 (m, 2H, 3’-H, 5’-H), 7.54–7.44 (m, 2H, 2’-H, 6’-H), 4.38 (d, J = 5.7 Hz, 2H, CH2), 3.73 (s, 2H, 2 hr). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 162.5 (CO), 143.6 (C-1’), 128.0 (C-2’, C-6’), 127.7 (q, JCF = 31.7 Hz, C-4’), 125.2 (q, JCF = 3.7 Hz, C-3’, C-5’). 124.3 (q, JCF = 272.1 Hz, CF3), 116.1 (CN), 42.2 (CH2), 25.3 (C-2). IR (ATR) max/cm−1=3316, 2937, 2364, 1734, 1664, 1547, 1325, 1152, 1107, 1066. HRMS (ESI): calcd. for C11H8F3N2O (M-H)- 241.05942; found 241.05949. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
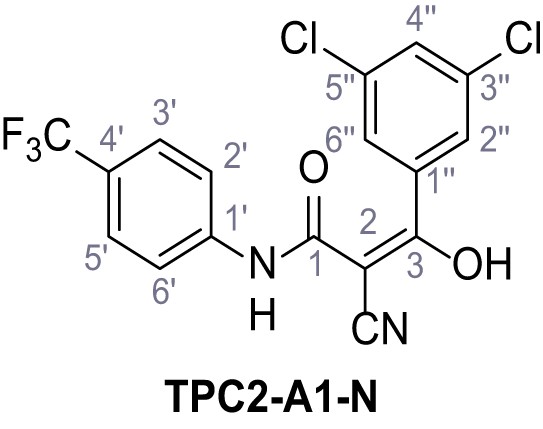
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – TPC2-A1-N.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to yield TPC2-A1-N as colorless crystals (276 mg, 0.688 mmol, 69%). Rf = 0.62 (4:1 hexanes/acetone). m.p.: 202°C [(Sjogren et al., 1991): 208–210°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.36 (s, 1H, NH), 7.79–7.76 (m, 2H, 2’-H, 6’-H), 7.65 (t, J = 1.9 Hz, 1H, 4’’-H), 7.62–7.60 (m, 2H, 3’-H, 5’-H), 7.59 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.7 (C-3), 166.5 (C-1), 144.9 (C-1’’), 143.6 (C-1’), 133.5 (C-3’’, C-5’’), 128.6 (C-4’’), 126.2–125.9 (m, C-3’, C-5’ and C-2’’, C-6’’), 124.6 (q, JCF = 286.1 Hz, CF3), 123.3 (C-2), 121.8 (q, JCF = 31.8 Hz, C-4’), 118.7 (C-2’, C-6’), 77.7 (CN). IR (ATR) max/cm−1=3293, 2213, 1538, 1409, 1320, 1268, 1244, 1167, 1106, 1070, 837, 810, 660, 591. HRMS (ESI): calcd. for C17H835Cl2F3N2O2 (M-H)- 398.99204; found 398.99202. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and benzoyl chloride (128 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-10 as colorless crystals (185 mg, 0.557 mmol, 56%). Analytical data are in accordance with literature (Davies et al., 2009). Rf = 0.28 (3:2 hexanes/acetone). m.p.: 242°C [(Davies et al., 2009): 245–247°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.12 (s, 1H, NH), 7.84–7.74 (m, 2H, 2’-H, 6’-H), 7.70–7.58 (m, 4H, 3’-H, 5’-H, Ph), 7.48–7.39 (m, 3H, Ph). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 185.7 (C-3), 167.3 (C-1), 143.1 (C-1‘), 139.7 (qPh), 135.0 (C-2), 130.0 (Ph), 127.8 (Ph), 127.6 (Ph), 126.0 (q, JCF = 3.8 Hz, C-3’, C-5’), 125.0 (d, JCF = 270.6 Hz, CF3), 122.3 (d, JCF = 35.2 Hz, C-4’), 119.4 (C-2’, C-6’), 77.8 (CN). IR (ATR) max/cm−1=3283, 2216, 1592, 1550, 1309, 1109, 1067, 840, 694. HRMS (ESI): calcd. for C17H10F3N2O2 (M-H)- 331.06999; found 331.06985. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
2-Cyano-3-(3,5-dinitrophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-11.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dinitrobenzoyl chloride (254 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-11 as red crystals (124 mg, 0.294 mmol, 29%). Rf = 0.21 (3:2 hexanes/acetone). m.p.: 240°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.32 (s, 1H, NH), 10.00 (s, 1H, OH), 8.89–8.80 (m, 3H, 2’’-H, 4’’-H, 6’’-H), 7.84–7.75 (m, 2H, 2’-H, 6’-H), 7.67–7.58 (m, 2H, 3’-H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 180.5 (C-3), 166.1 (C-1), 147.6 (C-3‘‘, C-5‘‘), 144.4 (C-1’’), 143.5 (C-1‘), 127.6 (C-2‘‘, C-6‘‘),126.1 (q, JCF = 3.5 Hz, C-3‘, C-5‘),124.2 (q, JCF = 271.0 Hz, CF3), 123.3 (C-2), 121.8 (q, JCF = 31.7 Hz, C-4’), 119.0 (C-4’’), 118.6 (C-2’, C-6’), 77.7 (CN). IR (ATR) max/cm−1=3262, 3093, 2223, 1539, 1342, 1317, 1115, 1067, 841, 730, 703, 687. HRMS (ESI): calcd. for C17H8F3N4O6 (M-H)- 421.04014; found 421.04021. Purity (HPLC):>96% (λ = 210 nm), >96% (λ = 254 nm).
2-Cyano-3-(4-nitrophenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-15.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 4-nitrobenzoyl chloride (204 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-15 as yellow crystals (276 mg, 0.688 mmol, 69%). Rf = 0.19 (3:2 hexanes/acetone). m.p.: 217°C [(Sjogren et al., 1991): 211–214°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.39 (s, 1H, NH), 8.28–8.19 (m, 2H, 3’’-H, 5’’-H), 7.84–7.80 (m, 2H, 2’’-H, 6’’-H), 7.78 (d, J = 8.6 Hz, 2H, 2’-H, 6’-H), 7.60 (d, J = 8.6 Hz, 2H, 3’’-H, 5’’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 184.3 (C-3), 166.4 (C-1), 148.1 (C-1’’), 147.5 (C-4’’), 143.7 (C-1’), 128.6 (C-2’’, C-6’’), 126.1 (q, JCF = 3.6 Hz, C-3’, C-5’), 124.6 (q, JCF = 270.9 Hz, CF3), 123.2 (C-2), 123.1 (C-3’’, C-5’’), 121.6 (q, JCF = 32.0 Hz, C-4’), 118.6 (C-2’, C-6’), 77.9 (CN). IR (ATR) max/cm−1=3307, 2219, 1551, 1320, 1111, 1069, 844, 750, 700. HRMS (ESI): calcd. for C17H9F3N3O4 (M-H)- 376.05506; found 376.05509. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
3-(4-Chlorophenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-16.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 4-chlorobenzoyl chloride (141 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-16 as colorless crystals (222 mg, 0.605 mmol, 61%). Rf = 0.32 (3:2 hexanes/acetone). m.p.: 220°C [(Sjogren et al., 1991): 218–220°C]. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.31 (s, 1H, NH), 7.81–7.75 (m, 2H, 2’-H, 6’-H), 7.68–7.63 (m, 2H, 2’’-H, 6’’-H), 7.63–7.58 (m, 2H, 3’-H, 5’-H), 7.49–7.44 (m, 2H, 3’’-H, 5’’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 184.8 (C-3), 167.0 (C-1), 143.5 (C-1’), 139.6 (C-1’’), 134.1 (C-4’’), 129.4 (C-2’’, C-6’’), 127.8 (C-3’’, C-5’’), 126.0 (q, JCF = 3.6 Hz, C-3’, C-5’), 124.6 (q, JCF = 271.0 Hz, CF3), 123.0 (C-2), 121.8 (q, JCF = 31.0 Hz, C-4’), 118.9 (C-2’, C-6’), 77.5 (CN). IR (ATR) max/cm−1=3282.2215, 1587, 1240, 1302, 1129, 1113, 1097, 839. HRMS (ESI): calcd. for C17H935ClF3N2O2 (M-H)- 365.03101; found 365.03108. Purity (HPLC): >96% (λ = 210 nm), >96% (λ = 254 nm).
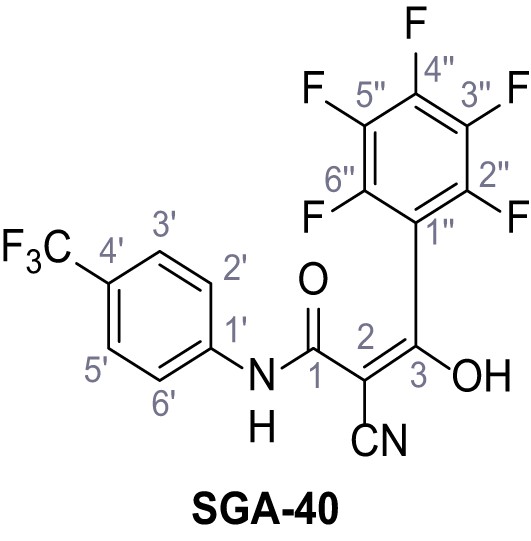
2-Cyano-3-hydroxy-3-(2,3,4,5,6-pentafluorophenyl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-40.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 2,3,4,5,6-pentafluorobenzoyl chloride (158 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-40 as colorless crystals (236 mg, 0.560 mmol, 56%). Rf = 0.28 (3:2 hexanes/acetone). m.p.: 161°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.70 (s, 1H, NH), 7.81–7.72 (m, 2H, 2’-H, 6’-H), 7.67–7.56 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 172.6 (C-3), 164.9 (C-1), 143.9–140.9 (m, C-2’’, C-6’’ or C-3’’, C-5’’), 143.3 (C-1’), 141.9–138.9 (m, C-4’’), 138.5–135.1 (m, C-2’’, C-6’’ or C-3’’, C-5’’), 126.1 (q, JCF = 3.6 Hz, C-3’, C-5’), 124.6 (q, JCF = 271.0 Hz, CF3), 122.0 (q, JCF = 31.9 Hz, C-4’), 117.5–117.0 (m, C-1’’), 121.7 (C-2’, C-6’), 118.7 (C-2), 81.8 (CN). IR (ATR) max/cm−1=2230, 1590, 1543, 1524, 1497, 1323, 1116, 1000, 839. HRMS (ESI): calcd. for C17H5F8N2O2 (M-H)- 421.02288; found 421.02337. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
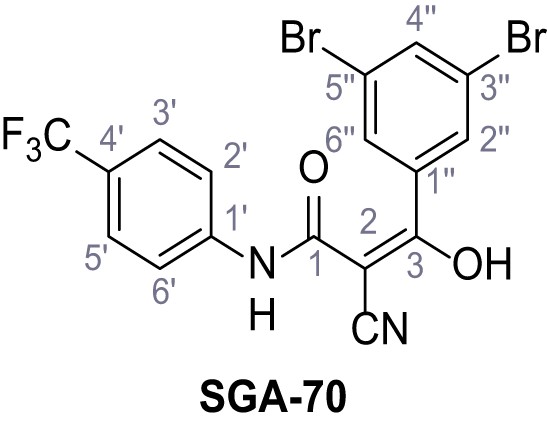
2-Cyano-3-(3,5-dibromophenyl)-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-70.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dibromobenzoic acid (308 mg, 1.10 mmol, 1.1 eq.; converted into the corresponding aryl chloride) were used to give SGA-70 as light yellow crystals (206 mg, 0.420 mmol, 42%). Rf = 0.32 (3:2 hexanes/acetone). m.p.: 211°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.32 (s, 1H, NH), 8.96 (s, 1H, OH), 7.91–7.81 (m, 1H, 4’’-H), 7.78–7.72 (m, 4H, 2’-H, 6’-H, 2’’-H, 6’’-H), 7.65–7.55 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.5 (C-3), 166.4 (C-1), 145.5 (C-1’’), 143.6 (C-1’), 133.7 (C-4’’), 129.2 (C-2’’, C-6’’), 126.0 (q, JCF = 3.5 Hz, C-3’, C-5’), 123.9 (q, JCF = 271.2 Hz, CF3), 123.3 (C-2), 121.8 (C-3’’, C-5’’), 121.6 (q, JCF = 31.9 Hz, C-4’), 118.5 (C-2’, C-6’), 77.4 (CN). IR (ATR) max/cm−1=2218, 1594, 1538, 1315, 1166, 1109, 1068, 838, 750. HRMS (ESI): calcd. for C17H879Br2F3N2O2 (M-H)- 486.89101; found 486.89128. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
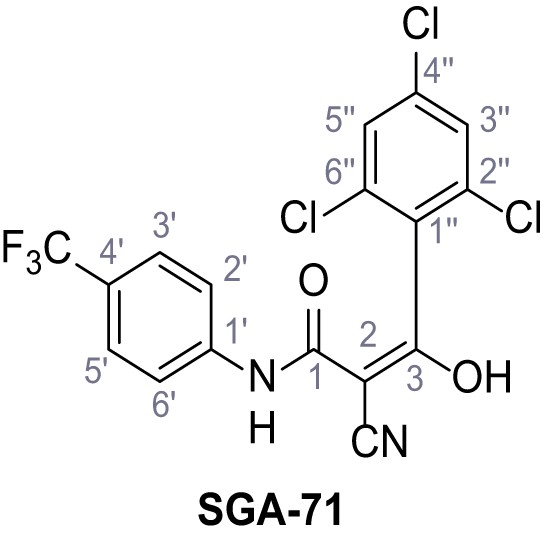
2-Cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-71.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 2,4,6-trichlorobenzoyl chloride (172 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-71 as colorless crystals (158 mg, 0.363 mmol, 36%). Rf = 0.14 (3:2 hexanes/acetone). m.p.: 220°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 11.92 (s, 1H, NH), 7.79–7.74 (m, 2H, 2’-H, 6’-H), 7.66 (s, 2H, 3’’-H, 5’’-H), 7.63–7.58 (m, 2H, 3’-H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 180.7 (C-3), 165.7 (C-1), 143.6 (C-1’), 139.5 (C-1’’), 133.1 (C-4’’), 132.0 (C-2’’, C-6’’), 127.8 (C-3’’, C-5’’), 126.1 (q, JCF = 3.6 Hz, C-3’, C-5’), 124.6 (q, JCF = 271.1 Hz, CF3), 121.9 (C-2), 121.6 (q, JCF = 31.9 Hz, C-4’), 118.4 (C-2’, C-6’), 79.5 (CN). IR (ATR) max/cm−1=2230, 1598, 1541, 1318, 1116, 841. HRMS (ESI): calcd. for C17H735Cl3F3N2O2 (M-H)- 432.95307; found 432.95394. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
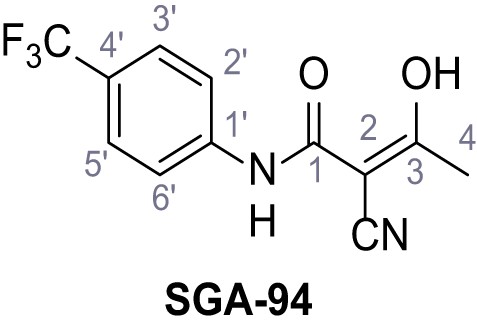
2-Cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)but-2-enamide (Teriflunomide) – SGA-94.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and acetyl chloride (78.5 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-94 as colorless crystals (206 mg, 0.761 mmol, 76%). Analytical data are in accordance with literature (Métro et al., 2012). Rf = 0.65 (3:2 hexanes/acetone). m.p.: 224°C [(Métro et al., 2012): 230–232°C]. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.28 (s, 1H, OH), 10.91 (s, 1H, NH), 7.81–7.72 (m, 2H, 2’-H, 6’-H), 7.70–7.61 (m, 2H, 3’-H, 5’-H), 2.25 (s, 3H, 4 hr). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 187.1 (C-3), 166.4 (C-1), 141.9 (C-1’), 125.9 (q, JCF = 3.7 Hz, C-3’, C-5’), 124.4 (q, JCF = 271.3 Hz, CF3), 123.5 (q, JCF = 32.1 Hz, C-4’), 120.7 (C-2’, C-6’), 118.9 (C-2), 80.5 (CN), 23.5 (C-4). IR (ATR) max/cm−1=2335, 2214, 1551, 1319, 1154, 1113, 840, 679. HRMS (ESI): calcd. for C12H8F3N2O2 (M-H)- 269.05434; found 269.05423. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
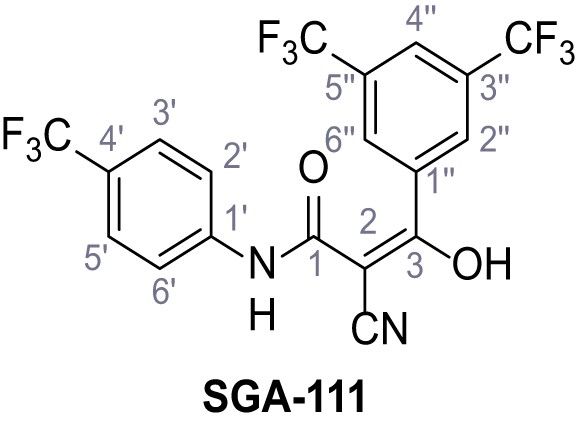
3-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-111.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-bis(trifluoromethyl)benzoyl chloride (199 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-111 as colorless crystals (357 mg, 0.762 mmol, 76%). Rf = 0.43 (3:2 hexanes/acetone). m.p.: 230°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.36 (s, 1H, NH), (m, 3H, 2’’-H, 6’’-H, 4’’-H), 7.87–7.71 (m, 2H, 2’-H, 6’-H), 7.69–7.51 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.4 (C-3), 166.3 (C-1), 144.0 (C-1’’), 143.6 (C-1’), 129.8 (q, JCF = 32.8 Hz, C-3’’, C-5’’), 128.1 (q, JCF = 4.7 Hz, C-2’’, C-6’’), 126.1 (q, JCF = 3.6 Hz, C-3’, C-5’), 126.0 (q, JCF = 257.2 Hz, m-CF3), 123.4 (C-2), 123.3 (q, JCF = 271.6 Hz, p-CF3), 122.7 (q, JCF = 4.0 Hz, C-4’’), 121.7 (q, JCF = 31.9 Hz, C-4’), 118.5 (C-2’, C-6’), 77.7 (CN). IR (ATR) max/cm−1=3289, 2218, 1349, 1323, 1284, 1186, 1139, 1114, 836. HRMS (ESI): calcd. for C19H8F9N2O2 (M-H)- 467.04475; found 467.04496. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
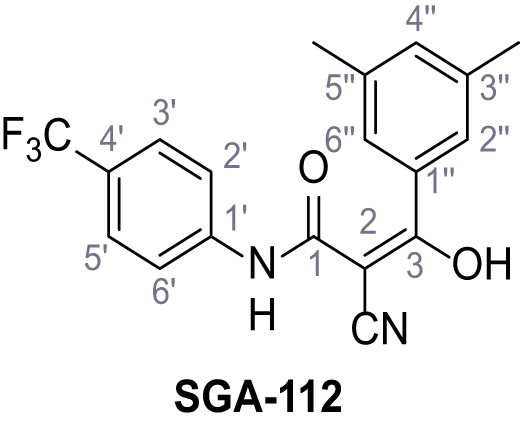
2-Cyano-3-(3,5-dimethylphenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-112.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dimethylbenzoyl chloride (163 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-112 as colorless crystals (175 mg, 0.486 mmol, 49%). Rf = 0.48 (3:2 hexanes/acetone). m.p.: 187°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.08 (s, 1H, NH), 7.85–7.74 (m, 2H, 2’-H, 6’-H), 7.66–7.58 (m, 2H, 3’-H, 5’-H), 7.25 (s, 2H, 2’’-H, 6’’-H), 7.09 (s, 1H,4’’-H), 2.30 (s, 6H, CH3). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 186.5 (C-3), 167.8 (C-1), 143.6 (C-1’), 140.1 (C-1’’), 137.2 (C-3’’, C-5’’), 131.8 (C-4’’), 126.4 (q, JCF = 3.8 Hz, C-3’, C-5’), 125.8 (C-2’’, C-6’’), 125.3 (q, JCF = 270.8 Hz, CF3), 122.6 (q, JCF = 31.9 Hz, C-4’), 121.8 (C-2), 119.9 (C-2’, C-6’), 78.2 (CN), 21.4 (CH3). IR (ATR) max/cm−1=3270, 2218, 1526.1319, 1247, 1157, 1110, 1066, 839. HRMS (ESI): calcd. for C19H14F3N2O2 (M-H)- 359.10129; found 359.10142. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
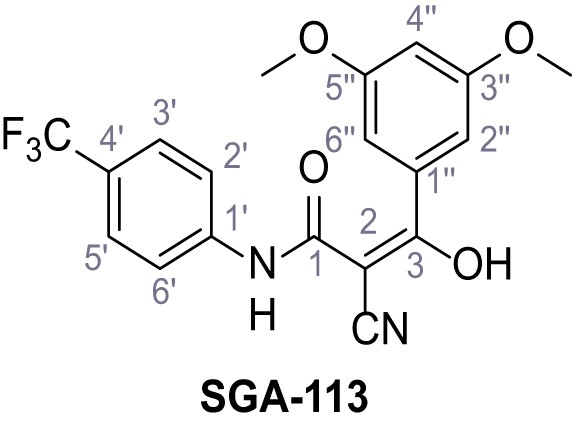
2-Cyano-3-(3,5-dimethoxyphenyl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-113.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dimethoxybenzoyl chloride (221 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-113 as colorless crystals (309 mg, 0.789 mmol, 79%). Rf = 0.54 (3:2 hexanes/acetone). m.p.: 203°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.99 (s, 1H, NH), 7.73–7.63 (m, 4H, 2’-H, 6’-H, 3’-H, 5’-H), 7.12 (d, J = 2.3 Hz, 2H, 2’’-H, 6’’-H), 6.69 (t, J = 2.3 Hz, 1H, 4’’-H), 3.85 (s, 6H, OCH3). 13C NMR (101 MHz, CDCl3) δ/ppm = 184.1 (C-3), 168.7 (C-1), 161.0 (C-3’’, C-5’’), 139.1 (C-1’), 133.9 (C-1’’), 127.8 (q, JCF = 33.2 Hz, C-4’), 126.7 (q, JCF = 3.8 Hz, C-3’, C-5’), 123.9 (q, JCF = 270.1 Hz, CF3), 121.0 (C-2’, C-6’), 117.5 (C-2), 106.4 (C-2’’, C-6’’), 106.0 (C-4’’), 78.5 (CN), 55.8 (OCH3). IR (ATR) max/cm−1=3297, 2215, 1550, 1324, 1208, 1156, 1095, 1067, 834. HRMS (ESI): calcd. for C19H14F3N2O4 (M-H)- 391.09112; found 391.09140. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
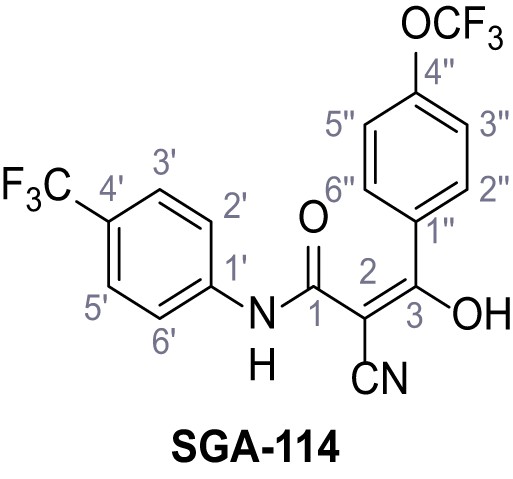
2-Cyano-3-hydroxy-3-(4-(trifluoromethoxy)phenyl)-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-114.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 4-(trifluoromethoxy)benzoyl chloride (173 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-114 as colorless crystals (200 mg, 0.481 mmol, 48%). Rf = 0.58 (3:2 hexanes/acetone). m.p.: 198°C [(Sjogren et al., 1991): 188–190°C]. 1H NMR (500 MHz, CDCl3) δ/ppm = 8.12–8.05 (m, 2H, 2’’-H, 6’’-H), 7.96 (s, 1H, NH), 7.78–7.62 (m, 4H, 2’-H, 6’-H, 3’-H, 5’-H), 7.42–7.32 (m, 2H, 3’’-H, 5’’-H). 13C NMR (126 MHz, CDCl3) δ/ppm = 182.6 (C-3), 168.5 (C-1), 152.8 (C-4’’), 139.0 (C-1’), 130.7 (C-2’’, C-6’’), 130.5 (C-1’’), 128.0 (q, JCF = 33.0 Hz, C-4’), 126.7 (q, JCF = 3.7 Hz, C-3’, C-5’), 123.9 (q, JCF = 271.7 Hz, CF3), 121.0 (C-2’, C-6’), 120.8 (C-3’’, C-5’’), 120.6 (q, JCF = 259.4 Hz, OCF3), 117.3 (C-2), 78.6 (CN). IR (ATR) max/cm−1=3285, 2215, 1597, 1551, 1505, 1268, 1168, 1128, 839. HRMS (ESI): calcd. for C18H9F6N2O3 (M-H)- 415.05228; found 415.05225. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
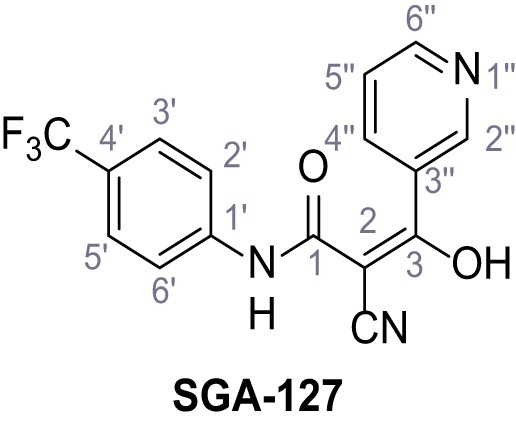
2-Cyano-3-hydroxy-3-(pyridin-3-yl)-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-127.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and nicotinoyl chloride hydrochloride (196 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-127 as orange solid (169 mg, 0.506 mmol, 51%). Rf = 0.00 (3:2 hexanes/EtOAc). m.p.: 235°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.22 (s, 1H, NH), 9.12 (s, 1H, 2’’-H), 8.88 (d, J = 5.6 Hz, 1H, 6’’-H), 8.70 (d, J = 8.0 Hz, 1H, 4’’-H), 8.04 (dd, J = 8.0, 5.6 Hz, 1H, 5’’-H), 7.83–7.72 (m, 2H, 2’-H, 6’-H), 7.67–7.56 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 180.0 (C-3), 165.9 (C-1), 143.5 (C-1’), 143.3 (C-6’’), 143.0 (C-4’’), 141.9 (C-2’’), 139.9 (C-3’’), 126.2 (C-5’’), 126.1 (q, JCF = 3.6 Hz, C-3’, C-5’), 124.2 (q, JCF = 270.7 Hz, CF3), 123.3 (C-2), 121.8 (q, JCF = 32.0 Hz, C-4’), 118.5 (C-2’, C-6’), 78.5 (CN). IR (ATR) max/cm−1=2356, 2191, 1533, 1317, 1105, 1060, 849, 698. HRMS (ESI): calcd. for C16H9F3N3O2 (M-H)- 332.06523; found 332.06517. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
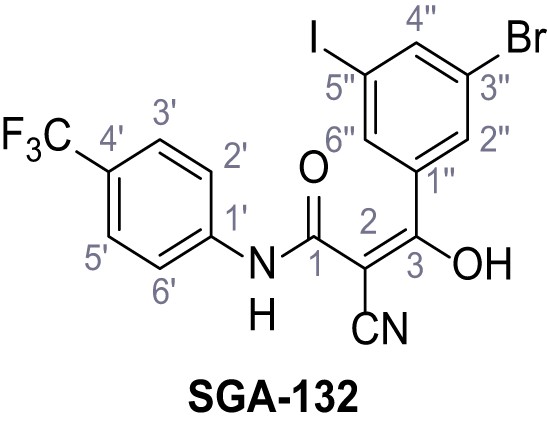
3-(3-Bromo-5-iodophenyl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-132.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3-bromo-5-iodobenzoic acid (360 mg, 1.10 mmol, 1.1 eq.; converted into the corresponding aryl chloride) were used to give SGA-132 as colorless crystals (332 mg, 0.618 mmol, 62%). Rf = 0.38 (3:2 hexanes/EtOAc). m.p.: 207°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.31 (s, 1H, NH), 7.97 (s, 1H, 4’’-H), 7.89 (s, 1H, 2’’-H or 6’’-H), 7.78–7.72 (m, 3H, 2’-H, 6’-H and 2’’-H or 6’’-H), 7.63–7.55 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.6 (C-3), 166.4 (C-1), 145.4 (C-1’’), 143.6 (C-1’), 139.1 (C-4’’), 135.0 (C-2’’ or C-6’’), 129.5 (C-2’’ or C-6’’), 126.0 (q, JCF = 3.8 Hz, C-3’, C-5’), 123.9 (q, JCF = 263.8 Hz, CF3), 123.3 (C-2), 121.7 (q, JCF = 31.6 Hz, C-4’), 121.6 (C-3’’ or C-5’’), 118.6 (C-2’, C-6’), 95.0 (C-3’’ or C-5’’), 77.4 (CN). IR (ATR) max/cm−1=3276, 2213, 1532, 1318, 1163, 1112, 731. HRMS (ESI): calcd. for C17H879BrIF3N2O2 (M-H)- 534.87714; found 534.87813. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
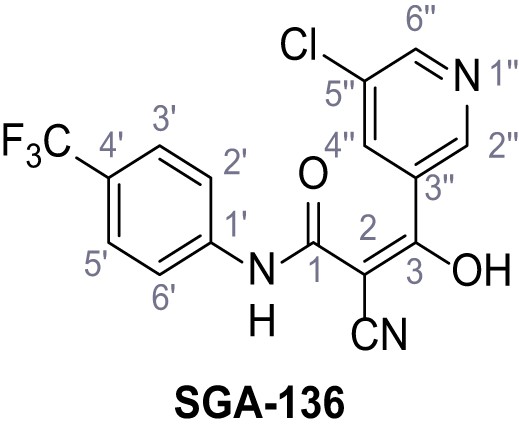
3-(5-Chloropyridin-3-yl)−2-cyano-3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-136.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 5-chloronicotinic acid (173 mg, 1.10 mmol, 1.1 eq.; converted into the corresponding aryl chloride) were used to give SGA-136 as light pink crystals (225 mg, 0.611 mmol, 61%). Rf = 0.15 (3:2 hexanes/acetone). m.p.: 221°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.33 (s, 1H, NH), 8.74 (s, 1H, 2’’-H or 4’’-H), 8.69–8.64 (m, 1H, 6’’-H), 8.11–8.05 (m, 1H, 2’’-H or 4’’-H), 7.81–7.73 (m, 2H, 2’-H, 6’-H), 7.64–7.55 (m, 2H, 3’-H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 181.7 (C-3), 166.2 (C-1), 147.9 (C-6’’), 146.1 (C-2’’ or C-4’’), 143.6 (C-1’), 138.8 (C-3’’ or C-5’’), 134.9 (C-2’’ or C-4’’), 130.5 (C-3’’ or C-5’’), 126.1 (q, JCF = 3.7 Hz, C-3’, C-5’), 124.6 (q, JCF = 271.0 Hz, CF3), 123.5 (C-2), 121.6 (q, JCF = 31.5 Hz, C-4’), 118.5 (C-2’, C-6’), 78.2 (CN). IR (ATR) max/cm−1=3285, 2225, 1539, 1308, 1113, 951, 838. HRMS (ESI): calcd. for C16H835ClF3N3O2 (M-H)- 366.02626; found 366.02652. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
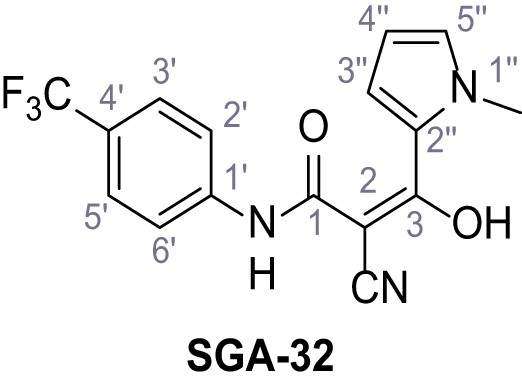
2-Cyano-3-(1-methyl-1H-pyrrol-2-yl)−3-hydroxy-N-(4-(trifluoromethyl)phenyl)acrylamide – SGA-32.
According to general procedure B, amide SGA-34 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 1-methylpyrrole-2-carbonyl chloride (158 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-32 as colorless crystals (221 mg, 0.660 mmol, 66%). Rf = 0.57 (3:2 hexanes/acetone). m.p.: 203°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.86 (s, 1H, NH), 7.69–7.61 (m, 4H, 2’-H, 6’-H, 3’-H, 5’-H), 7.57 (dd, J = 4.3, 1.6 Hz, 1H, 3’’-H), 6.93–6.90 (m, 1H, 4’’-H), 6.27 (dd, J = 4.3, 2.5 Hz, 1H, 5’’-H), 3.93 (s, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ/ppm = 175.6 (C-3), 170.2 (C-1), 139.5 (C-1’), 132.6 (C-5’’), 127.4 (q, JCF = 31.8 Hz, C-4’), 126.6 (q, J = 3.8 Hz, C-3’, C-5’), 124.1 (C-2’’), 124.0 (q, JCF = 271.4 Hz, CF3), 121.3 (C-3’’), 120.7 (C-2’, C-6’), 119.0 (C-2), 110.0 (C-4’’), 72.6 (CN), 38.7 (CH3). IR (ATR) max/cm−1=3273, 2210, 1518, 1379, 1228, 1111, 996, 837, 745. HRMS (ESI): calcd. for C16H11F3N3O2 (M-H)- 334.08088; found 334.08090. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
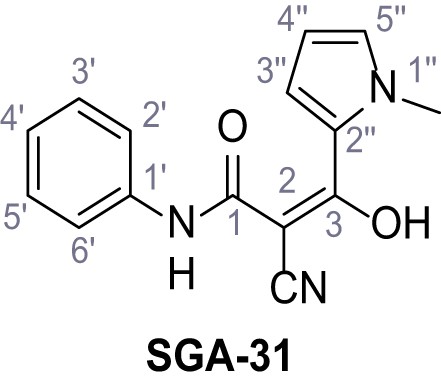
2-Cyano-3-(1-methyl-1H-pyrrol-2-yl)−3-hydroxy-N-phenylacrylamide (Prinomide) – SGA-31.
According to general procedure B, amide 2 (228 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 1-methylpyrrole-2-carbonyl chloride (158 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-31 as colorless crystals (200 mg, 0.747 mmol, 75%). Rf = 0.65 (3:2 hexanes/acetone). m.p.: 171°C [(Walker, 1981): 174–175°C]. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.74 (s, 1H, NH), 7.54 (dd, J = 4.3, 1.6 Hz, 1H, 3’’-H), 7.50 (d, J = 8.0 Hz, 2H, 2’-H, 6’-H), 7.44–7.34 (m, 2H, 3’-H, 5’-H), 7.20 (t, J = 7.4 Hz, 1H, 4’-H), 6.89 (t, J = 1.9 Hz, 1H, 5’’-H), 6.25 (dd, J = 4.3, 2.5 Hz, 1H, 4’’-H), 3.92 (s, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ/ppm = 175.6 (C-3), 170.0 (C-1), 136.2 (C-1’), 132.0 (C-5’’), 129.3 (C-3’, C-5’), 125.7 (C-4’), 124.4 (C-2’’), 121.3 (C-2’, C-6’), 120.7 (C-3’’), 119.1 (C-2), 109.7 (C-4’’), 72.6 (CN), 38.5 (CH3). IR (ATR) max/cm−1=3293, 2210, 1526, 1382, 1231, 751, 736, 687. HRMS (ESI): calcd. for C15H12N3O2 (M-H)- 266.09350; found 266.09370. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 2 (160 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-1 as yellow solid (227 mg, 0.681 mmol, 68%). Rf = 0.67 (3:2 hexanes/acetone). m.p.: 200°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 11.95 (s, 1H, NH), 7.61 (t, J = 1.9 Hz, 1H, 4’’-H), 7.56 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.53 (dd, J = 8.5, 1.0 Hz, 2H, 2’-H, 6’-H), 7.27–7.22 (m, 2H, 3’-H, 5’-H), 6.96–6.90 (m, 1H, 4’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.2 (C-3), 166.1 (C-1), 145.3 (C-1’’), 140.1 (C-1’), 133.4 (C-3’’, C-5’’), 128.7 (C-3’, C-5’), 128.3 (C-4’’), 126.1 (C-2’’, C-6’’), 123.6 (C-2), 121.7 (C-4’), 118.8 (C-2’, C-6’), 77.4 (CN). IR (ATR) max/cm−1=3294, 2212, 1579, 1445, 810, 748, 683. HRMS (ESI): calcd. for C16H935Cl2N2O2 (M-H)- 331.00466; found 331.00465. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 1 (174 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-4 as yellow crystals (185 mg, 0.534 mmol, 53%). Rf = 0.28 (3:2 hexanes/acetone). m.p.: 214°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.82 (s, 1H, NH), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.56 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.46–7.39 (m, 2H, 2’-H, 6’-H), 7.09–7.02 (m, 2H, 3’-H, 5’-H), 2.24 (s, 3H, CH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.0 (C-3), 166.0 (C-1), 145.1 (C-1’’), 137.4 (C-1’), 133.4 (C-3’’, C-5’’), 130.6 (C-4’), 129.1 (C-3’, C-5’), 128.3 (C-4’’), 126.1 (C-2’’, C-6’’), 123.5 (C-2), 119.0 (C-2’, C-6’), 77.5 (CN), 20.4 (CH3). IR (ATR) max/cm−1=3299, 2212, 1543, 1518.866, 806, 654. HRMS (ESI): calcd. for C17H1135Cl2N2O2 (M-H)- 345.02031; found 345.02052. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
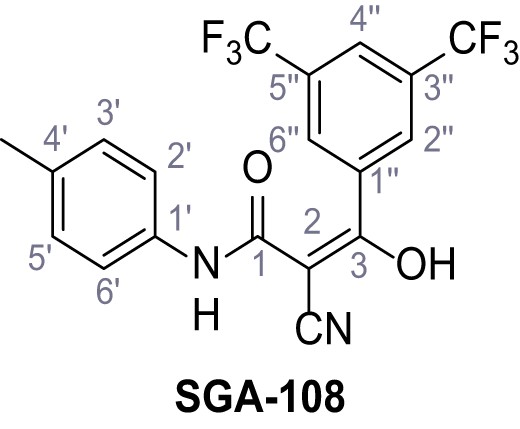
3-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-hydroxy-N-(p-tolyl)acrylamide – SGA-108.
According to general procedure B, amide 1 (174 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-bis(trifluoromethyl)benzoyl chloride (199 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-108 as light yellow crystals (208 mg, 0.502 mmol, 50%). Rf = 0.28 (3:2 hexanes/acetone). m.p.: 183°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 8.45 (s, 2H, 2’’-H, 6’’-H), 8.10 (s, 1H, 4’’-H), 7.86 (s, 1H, NH), 7.43–7.37 (m, 2H, 2’-H, 6’-H), 7.25–7.20 (m, 2H, 3’-H, 5’-H), 2.37 (s, 3H, CH3). 13C NMR (126 MHz, CDCl3) δ/ppm = 180.4 (C-3), 167.9 (C-1), 136.7 (C-4’), 134.7 (C-1’’), 132.7 (C-1’), 132.6 (q, JCF = 34.2 Hz, C-3’’, C-5’’), 130.1 (C-3’, C-5’), 128.6 (q, JCF = 3.1 Hz, C-2’’, C-6’’), 126.3 (q, JCF = 3.7 Hz, C-4’’), 122.8 (q, JCF = 272.9 Hz, CF3), 121.8 (C-2’, C-6’), 116.5 (C-2), 79.7 (CN), 21.2 (CH3). IR (ATR) max/cm−1=3276, 2213, 1538, 1278, 1129, 811, 681. HRMS (ESI): calcd. for C19H11F6N2O2 (M-H)- 413.07302; found 413.07301. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
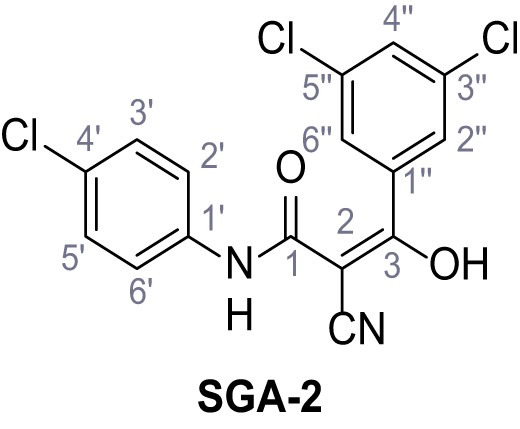
N-(4-Chlorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-2.
According to general procedure B, amide 4 (195 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-2 as yellow solid (256 mg, 0.695 mmol, 70%). Rf = 0.71 (3:2 hexanes/acetone). m.p.: 230°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.12 (s, 2H, NH), 7.62 (t, J = 1.9 Hz, 1H, 4’’-H), 7.60–7.57 (m, 2H, 2’-H, 6’-H), 7.55 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.30–7.26 (m, 2H, 3’-H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.5 (C-3), 166.1 (C-1), 145.4 (C-1’’), 139.1 (C-1’), 133.4 (C-3’’, C-5’’), 128.6 (C-3’, C-5’), 128.3 (C-4’’), 126.0 (C-2’’, C-6’’), 125.0 (C-4’), 123.6 (C-2), 120.2 (C-2’, C-6’), 77.2 (CN). IR (ATR) max/cm−1=3305, 2212, 1545, 1495, 1506, 1316, 1098, 809. HRMS (ESI): calcd. for C16H835Cl3N2O2 (M-H)- 364.96568; found 364.96593. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 5 (239 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-3 as yellow crystals (289 mg, 0.702 mmol, 70%). Rf = 0.73 (3:2 hexanes/acetone). m.p.: 237°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.09 (s, 1H, NH), 7.62 (t, J = 1.9 Hz, 1H, 4’’-H), 7.58–7.51 (m, 4H, 2’-H, 6’-H, 2’’-H, 6’’-H), 7.44–7.38 (m, 2H, 3’-H, 5’-H).13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.4 (C-3), 166.1 (C-1), 145.2 (C-1’’), 139.4 (C-1’), 133.4 (C-3’’, C-5’’), 131.4 (C-3’, C-5’), 128.3 (C-4’’), 126.0 (C-2’’, C-6’’), 122.6 (C-2), 120.7 (C-2’, C-6’), 112.9 (C-4’), 77.4 (CN). IR (ATR) max/cm−1=3309, 2211, 1540, 1493, 1403, 1316, 1290, 1012, 809. HRMS (ESI): calcd. for C16H879Br35Cl2N2O2 (M-H)- 408.91517; found 408.91581. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 5 (239 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dibromobenzoic acid (308 mg, 1.10 mmol, 1.1 eq.; converted into the corresponding aryl chloride) were used to give SGA-75 as colorless crystals (222 mg, 0.443 mmol, 44%). Rf = 0.20 (3:2 hexanes/acetone). m.p.: 233°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.05 (s, 1H, NH), 7.86 (t, J = 1.8 Hz, 1H, 4’-H), 7.73 (d, J = 1.8 Hz, 2H, 2’’-H, 6’’-H), 7.57–7.50 (m, 2H, 2’-H, 6’-H), 7.46–7.37 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.2 (C-3), 166.1 (C-1), 145.5 (C-1’’), 139.4 (C-1’), 133.7 (C-4’’), 131.5 (C-3’, C-5’), 129.2 (C-2’’, C-6’’), 122.7 (C-2), 121.8 (C-3’’, C-5’’), 120.8 (C-2’, C-6’), 113.0 (C-4’), 77.4 (CN). IR (ATR) max/cm−1=3314, 2214, 1596, 1543, 865, 817, 748, 658. HRMS (ESI): calcd. for C16H879Br3N2O2 (M-H)- 496.81414; found 496.81449. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
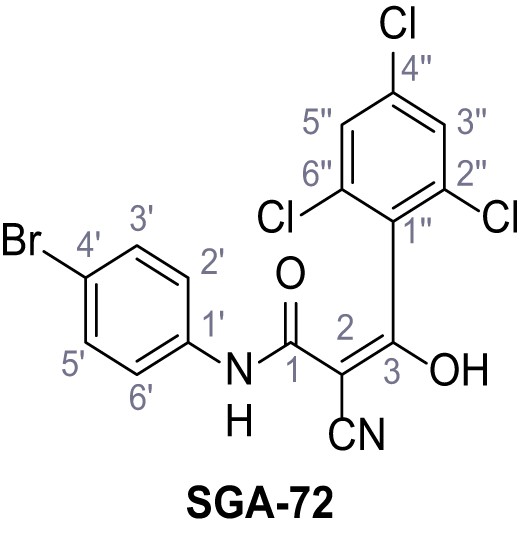
N-(4-Bromophenyl)−2-cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)acrylamide – SGA-72.
According to general procedure B, amide 5 (239 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 2,4,6-trichlorobenzoyl chloride (172 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-72 as colorless solid (227 mg, 0.507 mmol, 51%). Rf = 0.14 (3:2 hexanes/acetone). m.p.: 191°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.69 (s, 1H, NH), 7.65 (s, 2H, 3’’-H, 5’’-H), 7.58–7.52 (m, 2H, 2’-H, 6’-H), 7.44–7.38 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 180.3 (C-3), 165.4 (C-1), 139.6 (C-1’ or C-1’’), 139.4 (C-1’ or C-1’’), 133.0 (C-4’’), 132.0 (C-2’’, C-6’’), 131.5 (C-3’, C-5’), 127.8 (C-3’’, C-5’’), 122.1 (C-2), 120.5 (C-2’, C-6’), 112.9 (C-4’), 79.5 (CN). IR (ATR) max/cm−1=2238, 1588, 1539, 1488, 1307, 856, 818. HRMS (ESI): calcd. for C16H779Br35Cl3N2O2 (M-H)- 442.87620; found 442.87747. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 6 (178 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-8 as white crystals (220 mg, 0.626 mmol, 63%). Rf = 0.69 (3:2 hexanes/acetone). m.p.: 225°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.92 (s, 1H, NH), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.60–7.53 (m, 4H, 2’-H, 6’-H, 2’’-H, 6’’-H), 7.12–7.03 (m, 2H, 3’-H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.2 (C-3), 166.1 (C-1), 157.3 (d, JCF = 236.9 Hz, C-4’), 144.9 (C-1’’), 136.3 (d, JCF = 2.4 Hz, C-1’), 133.4 (C-3’’, C-5’’), 128.4 (C-4’’), 126.1 (C-2’’, C-6’’), 123.3 (C-2), 120.5 (d, JCF = 7.6 Hz, C-2’, C-6’), 115.2 (d, JCF = 22.0 Hz, C-3’, C-5’), 77.4 (CN). IR (ATR) max/cm−1=3296, 2212, 1739, 1549, 1506, 1210, 823, 809, 645. HRMS (ESI): calcd. for C16H835Cl2FN2O2 (M-H)- 348.99523; found 348.99526. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 7 (286 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-9 as yellow crystals (279 mg, 0.608 mmol, 61%). Rf = 0.71 (3:2 hexanes/acetone). m.p.: 238°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.04 (s, 1H, NH), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.59–7.53 (m, 4H, 3’-H, 5’-H, 2’’-H, 6’’-H), 7.44–7.38 (m, 2H, 2’-H, 6’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.4 (C-3), 166.2 (C-1), 145.1 (C-1’’), 139.8 (C-1’), 137.3 (C-3’, C-5’ or C-2’’, C-6’’), 133.4 (C-3’’, C-5’’), 128.4 (C-4’’), 126.1 (C-3’, C-5’ or C-2’’, C-6’’), 123.3 (C-2), 121.1 (C-2’, C-6’), 84.5 (C-4’), 77.5 (CN). IR (ATR) max/cm−1=3303, 2217, 1592, 1523, 1485, 1314, 817, 806, 658. HRMS (ESI): calcd. for C16H835Cl2IN2O2 (M-H)- 456.90130; found 456.90014. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
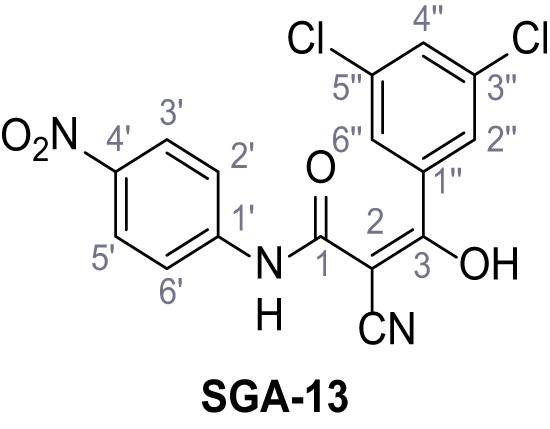
2-Cyano-3-(3,5-dichlorophenyl)-N-(4-nitrophenyl)−3-hydroxyacrylamide – SGA-13.
According to general procedure B, amide 8 (205 mg, 1.00 mmol, 1.0 eq.) in dry THF (16 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-13 as yellow solid (208 mg, 0.550 mmol, 55%). Rf = 0.82 (3:2 hexanes/acetone). m.p.: 246°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.64 (s, 1H, NH), 8.19–8.13 (m, 2H, 3’-H, 5’-H), 7.82–7.77 (m, 2H, 2’-H, 6’-H), 7.64 (t, J = 1.9 Hz, 1H, 4’’-H), 7.57 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 181.6 (C-3), 166.9 (C-1), 149.4 (C-1’’), 147.0 (C-1’), 141.3 (C-4’), 133.9 (C-3’’, C-5’’), 129.0 (C-4’’), 126.5 (C-2’’, C-6’’), 125.7 (C-3’, C-5’), 121.8 (C-2), 118.6 (C-2’, C-6’), 77.0 (CN). IR (ATR) max/cm−1=3314, 2209, 1568, 1546, 1514, 1498, 1340, 1309, 847, 813, 656. HRMS (ESI): calcd. for C16H835Cl2N3O4 (M-H)- 375.98973; found 375.98970. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
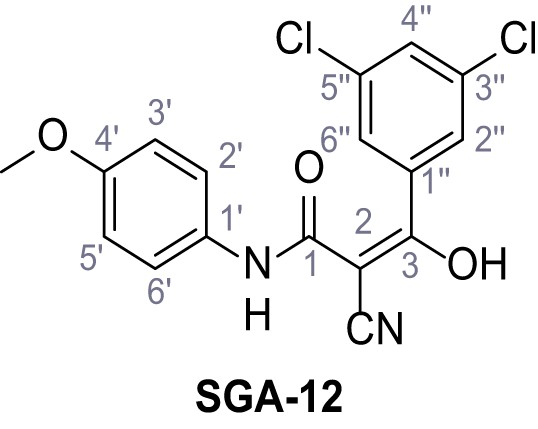
2-Cyano-3-(3,5-dichlorophenyl)-N-(4-methoxyphenyl)−3-hydroxyacrylamide – SGA-12.
According to general procedure B, amide 3 (190 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-12 as yellow solid (220 mg, 0.606 mmol, 61%). Rf = 0.76 (3:2 hexanes/acetone). m.p.: 207°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.36 (s, 1H, NH), 9.87 (s, 1H, OH), 7.71 (t, J = 1.8 Hz, 1H, 4’-H), 7.65 (d, J = 1.8 Hz, 2H, 2’’-H, 6’’-H), 7.49–7.41 (m, 2H, 2’-H, 6’-H), 6.90–6.85 (m, 2H, 3’-H, 5’-H), 3.72 (s, 3H, OCH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 181.6 (C-3), 166.4 (C-1), 155.2 (C-4’), 142.6 (C-1’’), 133.7 (C-3’’, C-5’’), 131.9 (C-1’), 129.3 (C-4’’), 126.3 (C-2’’, C-6’’), 121.7 (C-2’, C-6’), 121.4 (C-2), 113.9 (C-3’, C-5’), 78.1 (CN), 55.2 (OCH3). IR (ATR) max/cm−1=3296, 2211, 1601, 1467, 1441, 1297, 1251, 1032, 764. HRMS (ESI): calcd. for C17H1135Cl2N2O3 (M-H)- 361.01522; found 361.01516. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
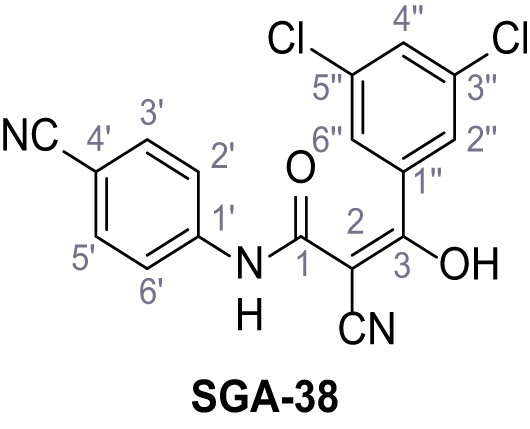
2-Cyano-N-(4-cyanophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-38.
According to general procedure B, amide 9 (185 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-38 as yellow solid (137 mg, 0.382 mmol, 38%). Rf = 0.12 (3:2 hexanes/acetone). m.p.: 236°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.42 (s, 1H, NH), 7.75–7.72 (m, 2H, 3’-H, 5’-H), 7.71–7.67 (m, 2H, 2’-H, 6’-H), 7.64 (t, J = 1.9 Hz, 1H, 4’’-H), 7.57 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 183.0 (C-3), 166.4 (C-1), 145.1 (C-1’’), 144.3 (C-1’), 133.5 (C-3’’, C-5’’), 133.3 (C-3’, C-5’), 128.5 (C-4’’), 126.0 (C-2’’, C-6’’), 123.2 (C-4’), 119.4 (C-2), 118.8 (C-2’, C-6’), 103.1 (4’-CN), 77.4 (2-CN). IR (ATR) max/cm−1=3321, 2360, 2340, 1533, 839, 655. HRMS (ESI): calcd. for C17H835Cl2N3O2 (M-H)- 355.99991; found 356.00057. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
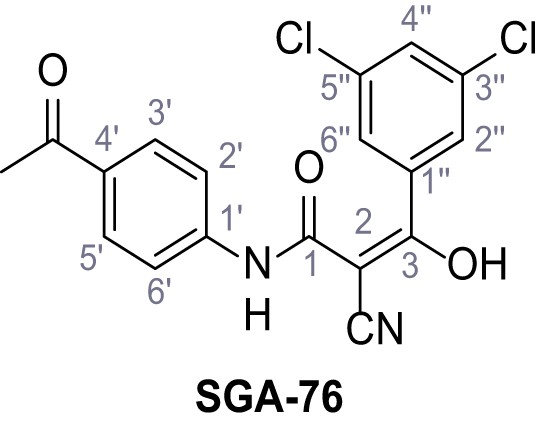
N-(4-Acetylphenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-76.
According to general procedure B, amide 10 (202 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-76 as off white solid (296 mg, 0.789 mmol, 79%). Rf = 0.08 (3:2 hexanes/acetone). m.p.: 209°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.32 (s, 1H, NH), 7.91–7.86 (m, 2H, 3’-H, 5’-H), 7.71–7.66 (m, 2H, 2’-H, 6’-H), 7.64 (t, J = 1.9 Hz, 1H, 4’’-H), 7.58 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 2.51 (s, 3H, CH3, collapses with DMSO). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 196.3 (CO), 182.8 (C-3), 166.3 (C-1), 145.1 (C-1’’), 144.5 (C-1’), 133.5 (C-3’’, C-5’’), 130.4 (C-4’), 129.7 (C-3’, C-5’), 128.5 (C-4’’), 126.1 (C-2’’, C-6’’), 123.3 (C-2), 117.9 (C-2’, C-6’), 77.6 (CN), 26.3 (CH3). IR (ATR) max/cm−1=3304, 2207, 1682, 1596, 1544, 1355, 1272, 809. HRMS (ESI): calcd. for C18H1135Cl2N2O3 (M-H)- 373.01522; found 373.01580. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
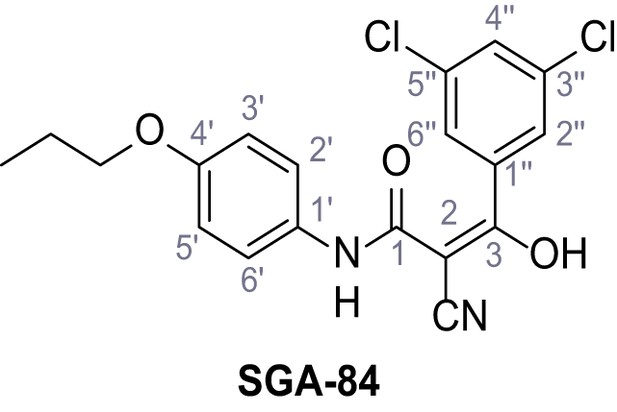
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-propoxyphenyl)acrylamide – SGA-84.
According to general procedure B, amide 11 (218 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-84 as yellow solid (245 mg, 0.626 mmol, 63%). Rf = 0.15 (3:2 hexanes/acetone + 2% TEA). m.p.: 184°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 11.36 (s, 1H, NH), 9.30 (s, 1H, OH), 7.71 (t, J = 1.7 Hz, 1H, 4’’-H), 7.67–7.62 (m, 2H, 2’’-H, 6’’-H), 7.49–7.40 (m, 2H, 3’-H, 5’-H), 6.90–6.82 (m, 2H, 2’-H, 6’-H), 3.88 (t, J = 6.5 Hz, 2H, CH2CH2CH3), 1.71 (sext, J = 7.2 Hz, 2H, CH2CH2CH3), 0.97 (t, J = 7.4 Hz, 3H, CH2CH2CH3). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 181.6 (C-3), 166.4 (C-1), 154.6 (C-4’), 142.7 (C-1’’), 133.7 (C-3’’, C-5’’), 131.9 (C-1’), 129.3 (C-4’’), 126.3 (C-2’’, C-6’’), 121.6 (C-3’, C-5’), 121.5 (C-2), 114.5 (C-2’, C-6’), 78.0 (CN), 69.1 (CH2CH2CH3), 22.1 (CH2CH2CH3), 10.4 (CH2CH2CH3). IR (ATR) max/cm−1=3304, 2208, 1601, 1550, 1511, 1249, 1235, 822, 811. HRMS (ESI): calcd. for C19H1535Cl2N2O3 (M-H)- 389.04652; found 389.04631. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
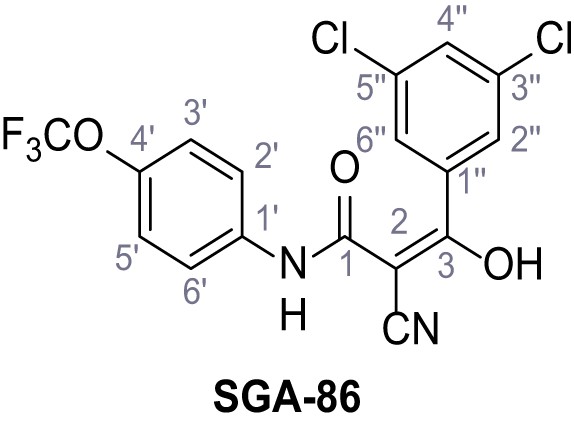
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethoxy)phenyl)acrylamide – SGA-86.
According to general procedure B, amide 12 (244 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-86 as colorless solid (344 mg, 0.825 mmol, 83%). Rf = 0.53 (3:2 hexanes/acetone). m.p.: 195°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.02 (s, 1H, NH), 9.41 (s, 1H, OH), 7.69–7.64 (m, 3H, 2’-H, 6’-H, 4’’-H), 7.59 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.28–7.23 (m, 2H, 3’-H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.3 (C-3), 166.3 (C-1), 144.6 (C-1’’), 142.7 (C-4’), 139.1 (C-1’), 133.5 (C-3’’, C-5’’), 128.6 (C-4’’), 126.1 (C-2’’, C-6’’), 123.0 (C-2), 121.6 (C-3’, C-5’), 120.3 (q, JCF = 255.2 Hz, OCF3), 120.2 (C-2’, C-6’), 77.6 (CN). IR (ATR) max/cm−1=3304, 2218, 1614, 1536, 1506, 1262, 1208, 1164, 661. HRMS (ESI): calcd. for C17H835Cl2F3N2O3 (M-H)- 414.98696; found 414.98676. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
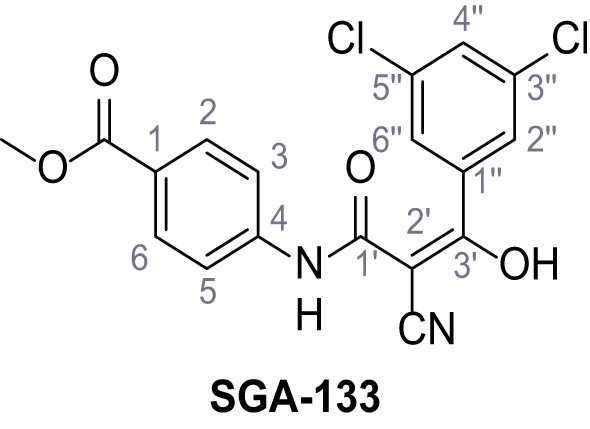
Methyl 4-(2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamido)benzoate – SGA-133.
According to general procedure B, amide 13 (218 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used. The resulting solid was washed with hexanes, EtOH and water to give SGA-133 as colorless solid (331 mg, 0.846 mmol, 85%). Rf = 0.15 (3:2 hexanes/acetone). m.p.: 234°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.33 (s, 1H, NH), 11.41 (s, 1H, OH), 7.90–7.83 (m, 2H, 2 hr, 6 hr), 7.71–7.66 (m, 2H, 3 hr, 5 hr), 7.64 (t, J = 1.9 Hz, 1H, 4’’-H), 7.57 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 3.81 (s, 3H, CH3). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.8 (C-3’), 166.3 (C-1’), 166.0 (CO), 145.1 (C-1’’), 144.6 (C-4), 133.4 (C-3’’, C-5’’), 130.4 (C-2, C-6), 128.5 (C-4’’), 126.1 (C-2’’, C-6’’), 123.3 (C-2’), 122.3 (C-1), 118.1 (C-3, C-5), 77.6 (CN), 51.7 (CH3). IR (ATR) max/cm−1=3304, 3093, 2215, 1727, 1591, 1534, 1415, 1283, 1262, 1112, 810, 766. HRMS (ESI): calcd. for C18H1135Cl2N2O4 (M-H)- 389.01014; found 389.01077. Purity (NMR):>96% (λ = 210 nm),>96% (λ = 254 nm).
Ester SGA-133 (95.0 mg, 0.243 mmol, 1.0 eq.) was dissolved in dioxane/H2O (3:1; 4.0 mL) and LiOH (61.2 mg, 2.43 mmol, 10 eq.) was added. The mixture was stirred at rt for 3 hr, before 1 M aq. HCl (5.0 mL) was added. The precipitate was filtered off, washed with cold water and dried to give SGA-137 as colorless solid (59.6 mg, 0.158 mmol, 65%). Rf = 0.00 (3:2 hexanes/acetone). m.p.: 244°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.30 (s, 1H, NH), 7.87–7.81 (m, 2H, 2 hr, 6 hr), 7.68–7.64 (m, 2H, 3 hr, 5 hr), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.57 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.7 (C-3’), 167.1 (CO), 166.3 (C-1’), 145.2 (C-1’’), 144.2 (C-4), 133.4 (C-3’’, C-5’’), 130.5 (C-2, C-6), 128.4 (C-4’’), 126.1 (C-2’’, C-6’’), 123.5 (C-1), 123.4 (C-2’), 118.0 (C-3, C-5), 77.5 (CN). IR (ATR) max/cm−1=3311, 2215, 1696, 1595, 1550, 1415, 1294, 855, 770. HRMS (ESI): calcd. for C17H935Cl2N2O4 (M-H)- 374.99449; found 374.99480. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
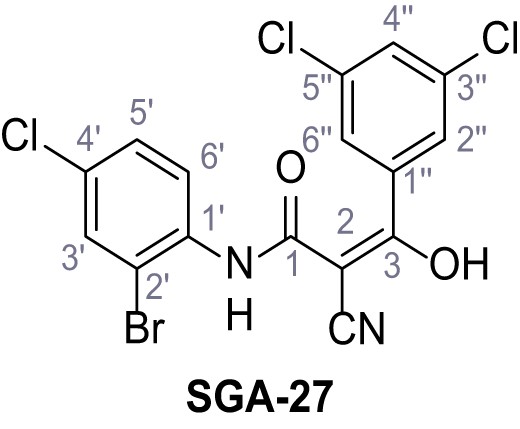
N-(2-Bromo-4-chlorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-27.
According to general procedure B, amide 14 (274 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-27 as colorless solid (330 mg, 0.738 mmol, 74%). Rf = 0.25 (3:2 hexanes/acetone). m.p.: 203°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 12.29 (s, 1H, NH), 8.56 (d, J = 9.0 Hz, 1H, 6’-H), 7.68 (d, J = 2.5 Hz, 1H, 3’-H), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.58 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.36 (dd, J = 9.0, 2.5 Hz, 1H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.7 (C-3), 166.4 (C-1), 145.2 (C-1’’), 137.8 (C-1’), 133.4 (C-3’’, C-5’’), 131.4 (C-6’), 128.4 (C-4’’), 127.8 (C-5’), 126.1 (C-2’’, C-6’’), 125.4 (C-4’), 123.5 (C-2), 122.1 (C-3’), 112.1 (C-2’), 77.3 (CN). IR (ATR) max/cm−1=3366, 3087, 2208, 1556, 1521, 1469, 1360, 1292, 863, 815. HRMS (ESI): calcd. for C16H779Br35Cl3N2O2 (M-H)- 442.87620; found 442.87736. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
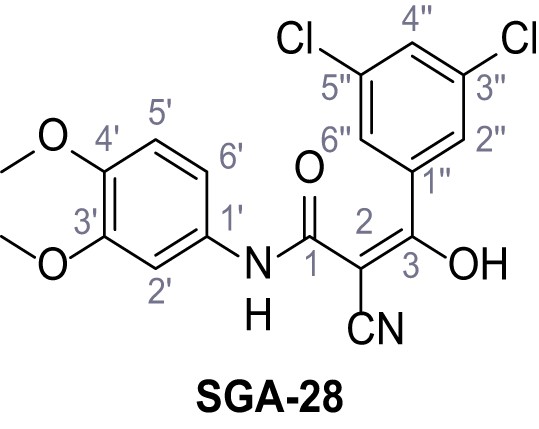
2-Cyano-3-(3,5-dichlorophenyl)-N-(3,4-dimethoxyphenyl)−3-hydroxyacrylamide – SGA-28.
According to general procedure B, amide 17 (220 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-28 as yellow solid (200 mg, 0.508 mmol, 51%). Rf = 0.17 (3:2 hexanes/acetone). m.p.: 195°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 11.52 (s, 1H, NH), 7.68 (t, J = 1.8 Hz, 1H, 4’’-H), 7.62 (d, J = 1.8 Hz, 2H, 2’’-H, 6’’-H), 7.30 (d, J = 2.2 Hz, 1H, 2’-H), 7.01 (dd, J = 8.7, 2.2 Hz, 1H, 6’-H), 6.86 (d, J = 8.7 Hz, 1H, 5’-H), 3.73 (s, 3H, OCH3), 3.71 (s, 3H, OCH3). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 181.7 (C-3), 166.2 (C-1), 148.6 (C-3’), 144.4 (C-4’), 143.6 (C-1’’), 133.6 (C-3’’, C-5’’), 133.0 (C-1’), 128.9 (C-4’’), 126.2 (C-2’’, C-6’’), 122.2 (C-2), 112.3 (C-5’), 111.5 (C-6’), 104.9 (C-2’), 77.9 (CN), 55.8 (OCH3), 55.4 (OCH3). IR (ATR) max/cm−1=3284, 2217, 1608, 1549, 1516, 1238, 1028, 810. HRMS (ESI): calcd. for C18H1335Cl2N2O4 (M-H)- 391.02579; found 391.02608. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure B, amide 15 (286 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-39 as yellow solid (237 mg, 0.515 mmol, 52%). Rf = 0.20 (3:2 hexanes/acetone). m.p.: 171°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 11.87 (s, 1H, NH), 8.27 (dd, J = 8.3, 1.5 Hz, 1H, 6’-H), 7.80 (dd, J = 7.9, 1.5 Hz, 1H, 3’-H), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.59 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.29 (ddd, J = 8.4, 7.3, 1.5 Hz, 1H, 5’-H), 6.80–6.67 (m, 1H, 4’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.2 (C-3), 166.4 (C-1), 145.1 (C-1’’), 141.5 (C-1’), 139.0 (C-3’), 133.4 (C-3’’, C-5’’), 128.4 (C-4’’), 128.3 (C-5’), 126.1 (C-2’’, C-6’’), 124.0 (C-4’), 123.6 (C-2), 122.1 (C-6’), 89.2 (C-2’), 77.2 (CN). IR (ATR) max/cm−1=3337, 2213, 1579, 1537, 1294, 742. HRMS (ESI): calcd. for C16H835Cl2IN2O2 (M-H)- 456.90130; found 456.90095. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
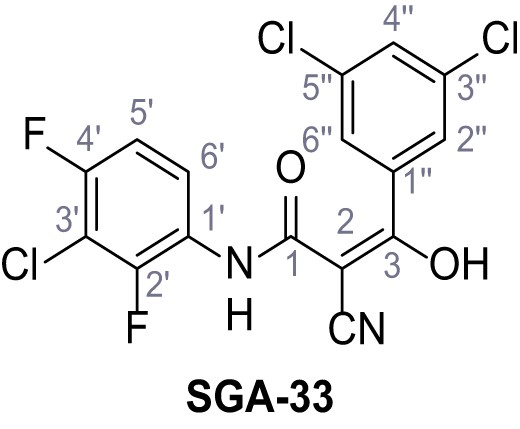
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-33.
According to general procedure B, amide 16 (231 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-33 as colorless crystals (290 mg, 0.719 mmol, 72%). Rf = 0.15 (3:2 hexanes/acetone). m.p.: 193°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.34 (s, 1H, NH), 8.43 (td, J = 9.0, 6.0 Hz, 1H, 6’-H), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.58 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.23 (td, J = 9.2, 2.1 Hz, 1H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.9 (C-3), 166.3 (C-1), 152.1 (d, JCF = 242.4 Hz, C-4’), 147.7 (d, JCF = 245.4 Hz, C-2’), 145.0 (C-1’’), 133.4 (C-3’’, C-5’’), 128.5 (C-4’’), 126.3 (dd, JCF = 10.3, 2.6 Hz, C-1’), 126.1 (C-2’’, C-6’’), 123.2 (C-2), 119.1 (dd, JCF = 7.8, 3.0 Hz, C-6’), 111.5 (dd, JCF = 20.7, 3.6 Hz, C-5’), 107.8 (dd, JCF = 22.2, 2.9 Hz, C-3’), 77.3 (CN). IR (ATR) max/cm−1=3293, 2206, 1539, 1497, 1370, 1289, 1023, 803. HRMS (ESI): calcd. for C16H635Cl3F2N2O2 (M-H)- 400.94684; found 400.94724. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
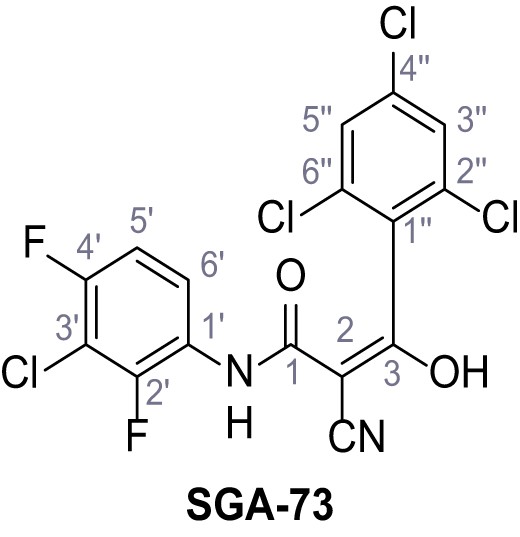
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-hydroxy-3-(2,4,6-trichlorophenyl)acrylamide – SGA-73.
According to general procedure B, amide 16 (231 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 2,4,6-trichlorobenzoyl chloride (172 µL, 1.10 mmol, 1.1 eq.) were used to give SGA-73 as colorless solid (217 mg, 0.496 mmol, 50%). Rf = 0.16 (1:1 hexanes/acetone). m.p.: 199°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 11.93 (s, 1H, NH), 8.45 (td, J = 9.1, 5.9 Hz, 1H, 6’-H), 7.66 (s, 2H, 3’’-H, 5’’-H), 7.23 (td, J = 9.2, 2.0 Hz, 1H, 5’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 180.8 (C-3), 165.6 (C-1), 152.1 (d, JCF = 242.6 Hz, C-4’), 147.5 (d, JCF = 245.6 Hz, C-2’), 139.4 (C-1’’ or C-4’’), 133.1 (C-1’’ or C-4’’), 132.0 (C-2’’, C-6’’), 127.8 (C-3’’, C-5’’), 126.3 (dd, JCF = 10.2, 3.2 Hz, C-1’), 121.8 (C-2), 118.8 (dd, JCF = 7.9, 2.9 Hz, C-6’), 111.5 (dd, JCF = 20.7, 3.5 Hz, C-5’), 107.9 (dd, JCF = 22.0, 19.2 Hz, C-3’), 79.4 (CN). IR (ATR) max/cm−1=3279, 2222, 1626, 1590, 1519, 1484, 1445, 1359, 1275, 1017, 816, 631. HRMS (ESI): calcd. for C16H535Cl4F2N2O2 (M-H)- 434.90787; found 434.90791. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
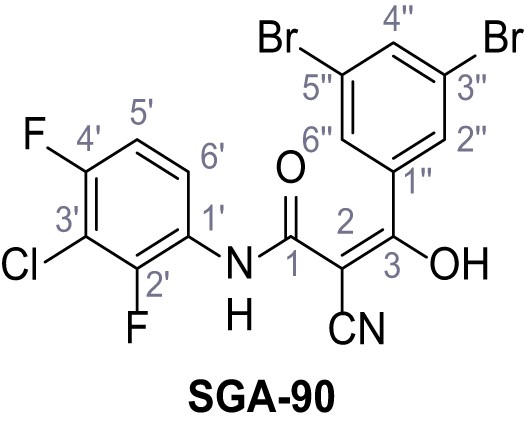
N-(3-Chloro-2,4-difluorophenyl)−2-cyano-3-(3,5-dibromophenyl)−3-hydroxyacrylamide – SGA-90.
According to general procedure B, amide 16 (231 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dibromobenzoic acid (308 mg, 1.10 mmol, 1.1 eq.; converted into the corresponding aryl chloride) were used to give SGA-90 as light yellow solid (374 mg, 0.759 mmol, 76%). Rf = 0.16 (1:1 hexanes/acetone). m.p.: 192°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.33 (s, 1H, NH), 8.43 (td, J = 8.9, 6.0 Hz, 1H, 6’-H), 7.86 (t, J = 1.7 Hz, 1H, 4’’-H), 7.74 (d, J = 1.7 Hz, 2H, 2’’-H, 6’’-H), 7.23 (td, J = 9.3, 2.0 Hz, 1H, 5’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 182.7 (C-3), 166.3 (C-1), 152.1 (d, JCF = 242.4 Hz, C-2’), 147.7 (d, JCF = 242.4 Hz, C-4’), 145.4 (C-1’’), 133.7 (C-4’’), 129.2 (C-2’’, C-6’’), 126.3 (dd, JCF = 10.3, 3.2 Hz, C-1’), 123.2 (C-2), 121.8 (C-3’’, C-5’’), 119.1 (dd, JCF = 7.5, 2.9 Hz, C-6’), 111.5 (dd, JCF = 20.7, 3.6 Hz, C-5’), 108.3–107.7 (m, C-3’), 77.3 (CN). IR (ATR) max/cm−1=3297, 2205, 1586, 1531, 1496, 1022, 803, 750. HRMS (ESI): calcd. for C16H679Br2ClF2N2O2 (M-H)- 488.84581; found 488.84607. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
2-Cyano-N-(2,3-dichlorophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-78.
According to general procedure B, amide 18 (229 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-78 as colorless crystals (344 mg, 0.856 mmol, 86%). Rf = 0.17 (3:2 hexanes/acetone). m.p.: 211°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.49 (s, 1H, NH), 8.58 (dd, J = 8.3, 1.5 Hz, 1H, 6’-H), 7.63 (t, J = 1.9 Hz, 1H, 4’’-H), 7.59 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.27 (t, J = 8.2 Hz, 1H, 5’-H), 7.20 (dd, J = 8.0, 1.5 Hz, 1H, 4’-H). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 182.8 (C-3), 166.4 (C-1), 145.1 (C-1’’), 139.3 (C-1’), 133.4 (C-3’’, C-5’’), 131.4 (C-3’), 128.5 (C-4’’), 128.0 (C-5’), 126.1 (C-2’’, C-6’’), 123.4 (C-2), 122.6 (C-4’), 119.3 (C-2’), 119.1 (C-6’), 77.5 (CN). IR (ATR) max/cm−1=3355, 2203, 1649, 1584, 1539, 1453, 1415, 872, 812, 777. HRMS (ESI): calcd. for C16H735Cl4N2O2 (M-H)- 398.92671; found 398.92789. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
2-Cyano-N-(2,6-dibromophenyl)−3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-77.
According to general procedure B, amide 19 (318 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-77 as colorless crystals (66.0 mg, 0.134 mmol, 13%). Rf = 0.78 (3:2 hexanes/acetone). m.p.: 226°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 10.64 (s, 1H, NH), 8.09–7.90 (m, 3H, 2’’-H, 6’’-H, 4’’-H), 7.79 (d, J = 8.0 Hz, 2H, 3’-H, 5’-H), 7.26 (t, J = 8.0 Hz, 1H, 4’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 162.4 (C-1, C-3), 136.6 (C-1’’), 135.2 (C-1’), 134.6 (C-3’’, C-5’’), 132.3 (C-3’, C-5’), 131.5 (C-4’’), 130.7 (C-4’), 126.4 (C-2’’, C-6’’), 124.4 (C-2’, C-6’), 76.4 (CN), C-2 is missing. IR (ATR) max/cm−1=3206, 2215, 1650.1567, 1516, 1283, 779, 750, 723. HRMS (ESI): calcd. for C16H779Br235Cl2N2O2 (M-H)- 486.82568; found 486.82870. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
N-(3,5-Bis(trifluoromethyl)phenyl)−2-cyano-3-(3,5-dichlorophenyl)−3-hydroxyacrylamide – SGA-85.
According to general procedure B, amide 20 (296 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-85 as colorless solid (320 mg, 0.682 mmol, 68%). Rf = 0.15 (3:2 hexanes/acetone). m.p.: 210°C. 1H NMR (400 MHz, (CD3)2SO) δ/ppm = 12.58 (s, 1H, NH), 8.93 (s, 1H, OH), 8.25 (s, 2H, 2’-H, 6’-H), 7.65 (t, J = 1.9 Hz, 1H, 4’’-H), 7.62–7.56 (m, 3H, 4’-H, 2’’-H, 6’’-H). 13C NMR (101 MHz, (CD3)2SO) δ/ppm = 183.5 (C-3), 167.3 (C-1), 145.4 (C-1’’), 142.3 (C-1’), 134.0 (C-3’’, C-5’’), 131.2 (q, JCF = 32.6 Hz, C-3’, C-5’), 129.1 (C-4’’), 126.5 (C-2’’, C-6’’), 123.7 (q, JCF = 270.8 Hz, CF3), 123.4 (C-2), 119.1–118.8 (m, C-2’, C-6’), 114.8–114.5 (m, C-4’), 77.6 (CN). IR (ATR) max/cm−1=2230, 1637, 1571, 1547, 1375, 1275, 1175, 1128, 810. HRMS (ESI): calcd. for C18H735Cl2F6N2O2 (M-H)- 466.97943; found 466.97933. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-methyl-N-(4-(trifluoromethyl)phenyl)-acrylamide – SGA-115.
According to general procedure B, amide 21 (242 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-115 as colorless solid (198 mg, 0.476 mmol, 48%). Rf = 0.67 (3:2 hexanes/acetone). m.p.: 147°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.80–7.73 (m, 2H, 3’-H, 5’-H), 7.64 (d, J = 1.9 Hz, 2H, 2’’-H, 6’’-H), 7.49 (t, J = 1.9 Hz, 1H, 4’’-H), 7.47–7.41 (m, 2H, 2’-H, 6’-H), 3.48 (s, 3H, CH3). 13C NMR (101 MHz, CDCl3) δ/ppm = 184.2 (C-3), 170.4 (C-1), 145.2 (C-1’), 135.8 (C-1’’), 135.5 (C-3’’, C-5’’), 132.5 (C-4’’), 131.4 (q, JCF = 32.9 Hz, C-4’), 127.8 (C-2’, C-6’), 127.5 (q, J = 3.6 Hz, C-3’, C-5’), 127.2 (C-2’’, C-6’’), 123.7 (q, JCF = 272.6 Hz, CF3), 115.3 (C-2), 78.6 (CN), 39.9 (CH3). IR (ATR) max/cm−1=3074, 2214, 1578, 1540, 1396, 1331, 1165, 1118, 807. HRMS (ESI): calcd. for C18H1035Cl2F3N2O2 (M-H)- 413.00769; found 413.00806. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
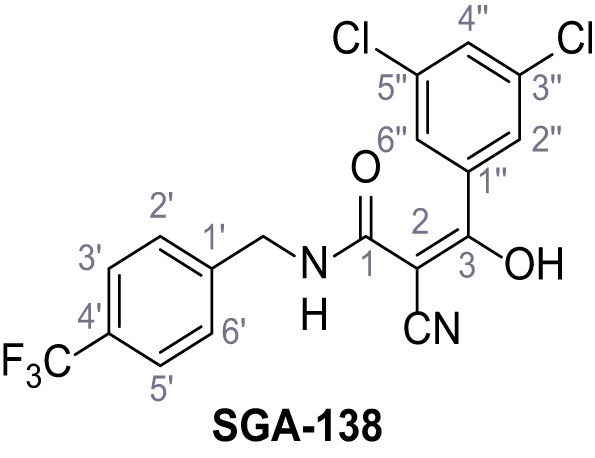
2-Cyano-3-(3,5-dichlorophenyl)−3-hydroxy-N-(4-(trifluoromethyl)benzyl)acrylamide – SGA-138.
According to general procedure B, amide 22 (242 mg, 1.00 mmol, 1.0 eq.) in dry THF (10 mL), NaH (92.0 mg, 2.30 mmol, 2.3 eq.) and 3,5-dichlorobenzoyl chloride (230 mg, 1.10 mmol, 1.1 eq.) were used to give SGA-138 as colorless solid (228 mg, 0.549 mmol, 55%). Rf = 0.70 (3:2 hexanes/acetone). m.p.: 173°C. 1H NMR (500 MHz, (CD3)2SO) δ/ppm = 9.57 (s, 1H, NH), 7.84 (t, J = 1.8 Hz, 1H, 4’’-H), 7.77 (d, J = 1.8 Hz, 2H, 2’’-H, 6’’-H), 7.74–7.69 (m, 2H, 3’-H, 5’-H), 7.57–7.50 (m, 2H, 2’-H, 6’-H), 4.52 (s, 2H, CH2). 13C NMR (126 MHz, (CD3)2SO) δ/ppm = 181.1 (C-3), 169.2 (C-1), 143.5 (C-1’), 138.5 (C-1’’), 134.2 (C-3’’, C-5’’), 130.8 (C-4’’), 128.1 (C-2’, C-6’), 127.7 (q, JCF = 31.7 Hz, C-4’), 126.6 (C-2’’, C-6’’), 125.3 (q, JCF = 3.8 Hz, C-3’, C-5’), 124.3 (q, JCF = 271.9 Hz, CF3), 118.1 (C-2), 78.0 (CN), 42.4 (CH2). IR (ATR) max/cm−1=3326, 2206, 1538, 1324, 1108, 1067, 805. HRMS (ESI): calcd. for C18H1035Cl2F3N2O2 (M-H)- 413.00769; found 413.00780. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
Synthesis of TPC2-A1-P and analogs
Request a detailed protocol4-Bromo-2-iodo-1-(trifluoromethoxy)benzene (610 mg, 1.66 mmol, 1.0 eq.) was dissolved in dry THF (8.0 mL) and cooled to −78°C, then n-BuLi (0.670 mL, 1.66 mmol, 1.0 eq.) was added dropwise. The mixture was stirred for 20 min at −78°C and a solution of 2-chloro-N-methoxy-N-methylacetamide (700 mg, 4.99 mmol, 3.0 eq.) in dry THF (8.0 mL) was added slowly. The mixture was stirred for 1 hr at −78°C and then poured on sat. aq. NH4Cl solution. The mixture was extracted with pentane, the organic layer was washed with sat. aq. NaCl solution, dried using hydrophobic phase separation filter papers and filtered through a short silica column (eluent: pentane). The product was carefully concentrated under ambient pressure to yield a colorless oil (23, 221 mg, 0.696 mmol, 42%). Rf = 0.65 (9:1 hexanes/EtOAc). 1H NMR (500 MHz, CDCl3) δ/ppm = 7.93 (d, J = 2.5 Hz, 1H, 6’-H), 7.71 (dd, J = 8.8, 2.5 Hz, 1H, 4’-H), 7.25–7.22 (m, 1H, 3’-H), 4.61 (s, 2H, 2 hr). 13C NMR (126 MHz, CDCl3) δ/ppm = 190.5 (C-1), 146.2 (C-2’), 137.1 (C-4’), 134.1 (C-6’), 130.8 (C-5’), 122.2 (C-3’), 120.8 (C-1’), 120.3 (q, JCF = 261 Hz, OCF3), 49.0 (C-2). IR (ATR) max/cm−1=1703, 1592, 1480, 1398, 1308, 1252, 1174, 1129, 1088, 822, 664. HRMS (EI): calcd. for C9H579Br35ClF3O2 (M)˙+ 315.9108; found 315.9106. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
2'-(Trifluoromethoxy)acetophenone (779 µL, 4.90 mmol, 1.0 eq.) was dissolved in CH2Cl2 (10 mL), then p-toluenesulfonic acid (86.1 mg, 0.490 mmol, 0.10 eq.) and N-bromosuccinimide (872 mg, 4.90 mmol, 1.0 eq.) were added. The mixture was stirred for 24 hr at rt, then sat. aq. NaCl (10 mL) was added. The aqueous phase was extracted with CH2Cl2 (3 × 10 mL), dried using a hydrophobic filter paper and concentrated in vacuo. Purification by FCC (pentane/EtOAc 9:1) yielded bromoketone 24 (672 mg, 2.87 mmol, 49%) as light brown liquid. Analytical data are in accordance with literature (Saruta et al., 2015). Rf = 0.58 (9:1 hexanes/EtOAc). 1H NMR (400 MHz, CDCl3) δ/ppm = 7.81 (dd, J = 7.8, 1.8 Hz, 1H, 6’-H), 7.60 (ddd, J = 8.3, 7.6, 1.8 Hz, 1H, 4’-H), 7.41 (td, J = 7.6, 1.0 Hz, 1H, 5’-H), 7.37–7.32 (m, 1H, 3’-H), 4.47 (s, 2H, 2 hr). 13C NMR (101 MHz, CDCl3) δ/ppm = 191.5 (C-1), 147.1 (q, J = 1.7 Hz, C-2’), 134.3 (C-4’), 131.6 (C-6’), 129.3 (C-1’), 127.3 (C-5’), 120.6 (C-3’), 120.5 (q, J = 260.1 Hz, OCF3), 35.1 (C-2). IR (ATR) max/cm−1=1698, 1603, 1450, 1295, 1248, 1200, 1160. HRMS (EI): calcd. for C9H679BrF3O2 (M)˙+ 281.9498; found 281.9494. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
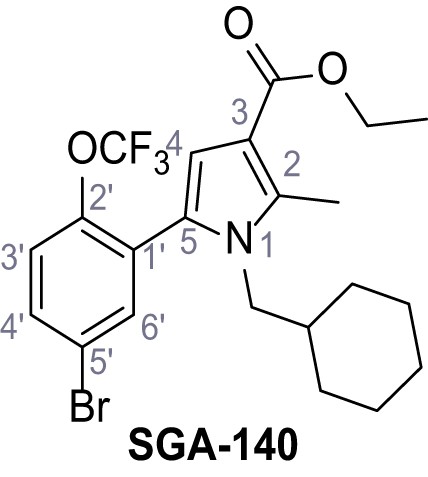
Ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylate.
– SGA-140.
Following general procedure C, ethyl acetoacetate (88.0 µL, 0.693 mmol, 1.1 eq.) in dry THF (3.0 mL), NaH (37.8 mg, 0.945 mmol, 1.5 eq.) and a solution of ketone 23 (200 mg, 0.630 mmol, 1.0 eq.) and KI (209 mg, 1.26 mmol, 2.0 eq.) in dry THF (3.0 mL) were used. Then the residue was dissolved in acetic acid (6.0 mL) and cyclohexanemethanamine (0.160 mL, 1.26 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 99:1) yielded SGA-140 as colorless oil (140 mg, 0.287 mmol, 46%). Rf = 0.30 (9:1 hexanes/EtOAc). 1H NMR (400 MHz, CDCl3) δ/ppm = 7.54–7.50 (m, 2H, 4’-H, 6’-H), 7.20 (ddt, J = 7.6, 3.0, 1.5 Hz, 1H, 3’-H), 6.55 (s, 1H, 4 hr), 4.27 (q, J = 7.1 Hz, 2H, CH2CH3), 3.59 (d, J = 7.1 Hz, 2H, CH2-cy), 2.59 (s, 3H, CH3), 1.60–1.54 (m, 3H, cy), 1.38–1.32 (m, 6H, CH2CH3, cy), 1.07–0.99 (m, 3H, cy), 0.68–0.59 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 165.6 (COOEt), 146.3 (C-2’), 137.6 (C-3), 135.7 (C-6’), 132.4 (C-4’), 129.0 (C-5’), 126.5 (C-5), 122.1 (C-3’), 120.3 (q, JCF = 260.2 Hz, OCF3), 119.9 (C-1’), 112.6 (C-2), 112.2 (C-4), 59.6 (CH2CH3), 50.8 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.7 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=2976, 2925, 2854, 1699, 1254, 1240, 1206, 1190, 1169, 1080, 1064, 774. HRMS (ESI): calcd. for C22H2679BrF3NO3 (M+H)+ 488.10427; found 488.10459. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
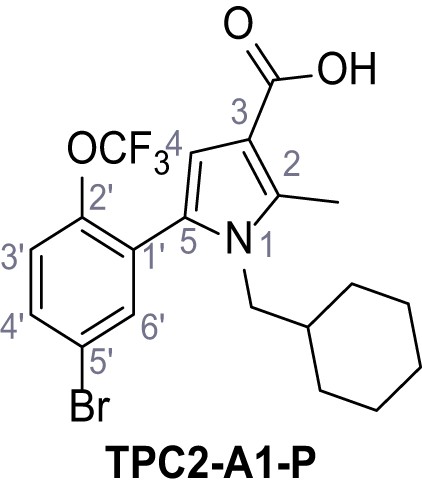
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylic acid – TPC2-A1-P.
According to general procedure D, LiOH (51.6 mg, 2.05 mmol, 10 eq.) and a solution of SGA-140 (100 mg, 0.205 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 2 hr the reaction was completed and recrystallization from EtOH gave TPC2-A1-P as a colorless solid (51.2 mg, 0.111 mmol, 54%). Rf = 0.14 (9:1 hexanes/EtOAc). m.p.: 202°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 11.34 (s, 1H, COOH), 7.56–7.51 (m, 2H, 3’-H, 4’-H), 7.23–7.19 (m, 1H, 6’-H), 6.61 (s, 1H, 4 hr), 3.60 (d, J = 7.1 Hz, 2H, CH2-cy), 2.60 (s, 3H, CH3), 1.62–1.56 (m, 3H, cy), 1.40–1.33 (m, 3H, cy), 1.09–1.01 (m, 3H, cy), 0.68–0.61 (m, 2H, cy). 13C NMR (126 MHz, CDCl3) δ/ppm = 170.0 (COOH), 146.4 (C-2’), 138.8 (C-2), 135.7 (C-6’), 132.6 (C-4’), 128.8 (C-5’), 126.8 (C-5), 122.2 (C-3’), 120.3 (q, JCF = 259.3 Hz, OCF3), 119.9 (C-1’), 112.9 (C-4), 111.6 (C-3), 50.9 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.7 (cy), 12.3 (CH3). IR (ATR) max/cm−1=2961, 2924, 2875, 2853, 2359, 2342, 1667, 1266, 1243, 1212, 1198, 1171, 925, 779, 658. HRMS (ESI): calcd. for C20H2079BrF3NO3 (M-H)- 458.05841; found 458.05889. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
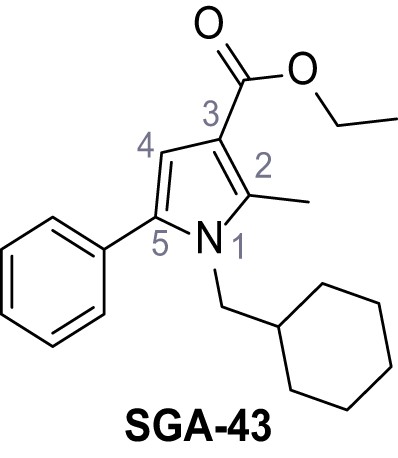
Ethyl 1-(cyclohexylmethyl)−2-methyl-5-phenyl-1H-pyrrole-3-carboxylate – SGA-43.
Following general procedure C, ethyl acetoacetate (42.0 µL, 3.30 mmol, 1.1 eq.) in dry THF (12 mL), NaH (180 mg, 4.50 mmol, 1.5 eq.) and a solution of 2-bromo-1-phenylethan-1-one (405 µL, 3.00 mmol, 1.0 eq.) and KI (996 mg, 6.00 mmol, 2.0 eq.) in dry THF (10 mL) were used. Then the residue was dissolved in acetic acid (10 mL) and cyclohexanemethanamine (781 µL, 6.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1) yielded SGA-43 as colorless solid (516 mg, 1.58 mmol, 53%). The compound is literature known, but no analytical data are available (Kang et al., 2010). Rf = 0.49 (9:1 hexanes/EtOAc). m.p.: 91°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.42–7.36 (m, 2H, Ph), 7.35–7.29 (m, 3H, Ph), 6.53 (s, 1H, 4 hr), 4.27 (q, J = 7.1 Hz, 2H, CH2CH3), 3.78 (d, J = 7.1 Hz, 2H, CH2-cy), 2.60 (s, 3H, CH3), 1.58–1.49 (m, 3H, cy), 1.41–1.31 (m, 6H, cy, CH2CH3), 1.06–0.95 (m, 3H, cy), 0.69–0.57 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 165.9 (COOEt), 137.0 (C-2), 134.1 (qPh), 133.7 (C-5), 129.6 (Ph), 128.5 (Ph), 127.4 (Ph), 112.0 (C-3), 110.0 (C-4), 59.4 (CH2CH3), 50.2 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=2975, 2926, 2850, 1738, 1698, 1420, 1242, 1224, 1191, 1062, 772, 702. HRMS (ESI): calcd. for C21H28NO2 (M+H)+ 326.21146; found 326.21121. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
According to general procedure D, LiOH (38.7 mg, 1.54 mmol, 10 eq.) and a solution of SGA-43 (50.0 mg, 0.154 mmol, 1.0 eq.) in dioxane/H2O (1.3 mL) were used. After 1 hr the reaction was completed and gave SGA-53 as a colorless solid (40.4 mg, 0.136 mmol, 88%). The compound is literature known, but no analytical data are available (Kang et al., 2010). Rf = 0.18 (6:1 hexanes/EtOAc). m.p.: 196°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 7.42–7.38 (m, 2H, Ph), 7.36–7.32 (m, 3H, Ph), 6.59 (s, 1H, 4 hr), 3.80 (d, J = 7.2 Hz, 2H, CH2), 2.62 (s, 3H, CH3), 1.59–1.52 (m, 3H, cy), 1.42–1.33 (m, 3H, cy), 1.06–0.97 (m, 3H, cy), 0.68–0.59 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 170.8 (COOH), 138.3 (C-2), 134.5 (C-5), 133.5 (qPh), 129.7 (Ph), 128.5 (Ph), 127.5 (Ph), 111.2 (C-3), 110.7 (C-4), 50.3 (CH2), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 12.3 (CH3). IR (ATR) max/cm−1=3030, 2971, 2921, 2848, 1738, 1660, 1533, 1435, 1364, 1267, 1227, 1205, 778, 768, 712, 703. HRMS (ESI): calcd. for C19H22NO2 (M-H)- 296.16572; found 296.16560. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
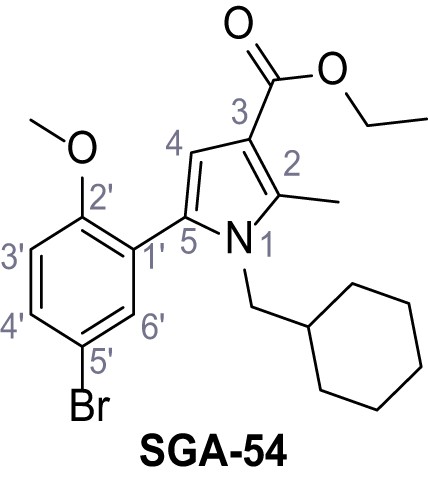
Ethyl 5-(5-bromo-2-methoxyphenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-54.
Following general procedure C, ethyl acetoacetate (143 µL, 1.10 mmol, 1.1 eq.) in dry THF (5.0 mL), NaH (60.0 mg, 1.50 mmol, 1.5 eq.) and a solution of 2-bromo-1-(5-bromo-2-methoxyphenyl)ethan-1-one (308 mg, 1.00 mmol, 1.0 eq.) in dry THF (1.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (0.260 mL, 2.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1) yielded SGA-54 as colorless solid (411 mg, 0.947 mmol, 95%). Rf = 0.37 (6:1 hexanes/EtOAc). m.p.: 83°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.45 (dd, J = 8.7, 2.5 Hz, 1H, 4’-H), 7.37 (d, J = 2.5 Hz, 1H, 6’-H), 6.81 (d, J = 8.7 Hz, 1H, 3’-H), 6.48 (s, 1H, 4 hr), 4.25 (q, J = 7.1 Hz, 2H, CH2CH3), 3.76 (s, 3H, OCH3), 3.56 (d, J = 7.2 Hz, 2H, CH2-cy), 2.58 (s, 3H, CH3), 1.60–1.54 (m, 3H, cy), 1.41–1.29 (m, 6H, cy, CH2CH3), 1.08–0.97 (m, 3H, cy), 0.68–0.57 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 165.7 (COOEt), 156.6 (C-2’), 136.9 (C-2), 135.2 (C-6’), 132.3 (C-4’), 128.9 (C-5), 124.8 (C-1’), 112.8 (C-5’), 112.6 (C-3’), 112.1 (C-3), 110.6 (C-4), 59.3 (CH2CH3), 55.9 (OCH3), 50.9 (CH2-cy), 39.0 (cy), 30.7 (cy), 26.3 (cy), 25.8 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=2979, 2928, 2849, 1695, 1676, 1473, 1461, 1434, 1253, 1234, 1187, 1176, 1060, 1048, 1027, 774, 619. HRMS (ESI): calculated for C22H2979BrNO3 (M+H)+ 434.13253; found 434.13229. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
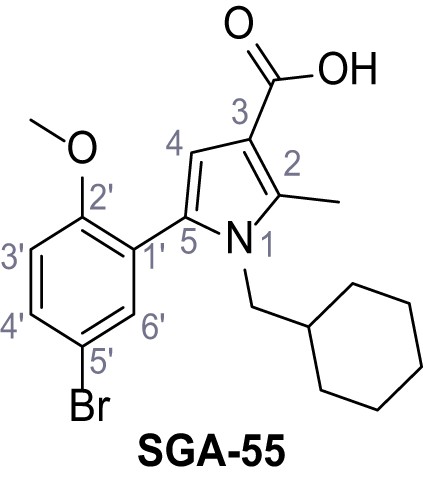
5-(5-Bromo-2-methoxyphenyl)−1-(cyclohexylmethyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-55.
According to general procedure D, LiOH (88.4 mg, 3.51 mmol, 10 eq.) and a solution of SGA-54 (152 mg, 0.351 mmol, 1.0 eq.) in dioxane/H2O (1.3 mL) were used. After 1 hr the reaction was completed and gave SGA-55 as a colorless solid (90.0 mg, 0.222 mmol, 63%). Rf = 0.72 (1:1 hexanes/EtOAc). m.p.: 224°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.46 (dd, J = 8.8, 2.5 Hz, 1H, 4’-H), 7.38 (d, J = 2.5 Hz, 1H, 6’-H), 6.82 (d, J = 8.8 Hz, 1H, 3’-H), 6.54 (s, 1H, 4 hr), 3.77 (s, 3H, OCH3), 3.57 (d, J = 7.2 Hz, 2H, CH2-cy), 2.59 (s, 3H, CH3), 1.62–1.52 (m, 3H, cy), 1.43–1.33 (m, 3H, cy), 1.10–0.98 (m, 3H, cy), 0.69–0.58 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 170.4 (COOH), 156.6 (C-2’), 138.2 (C-2), 135.2 (C-6’), 132.4 (C-4’), 129.3 (C-5), 124.6 (C-1’), 112.8 (C-5’), 112.6 (C-3’), 111.4 (C-4), 111.3 (C-3), 55.9 (OCH3), 51.0 (CH2-cy), 39.0 (cy), 30.7 (cy), 26.3 (cy), 25.8 (cy), 12.3 (CH3). IR (ATR) max/cm−1=3027, 2969, 2926, 2850, 1739, 1658, 1476, 1462, 1442, 1362, 1274, 1244, 1205, 1018, 808, 782, 619. HRMS (ESI): calculated for C20H2379BrNO3 (M-H)- 404.08668; found 404.08697. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
Ethyl 1-(cyclohexylmethyl)−5-(2,5-dichlorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-48.
Following general procedure C, ethyl acetoacetate (208 µL, 1.65 mmol, 1.1 eq.) in dry THF (5.0 mL), NaH (90.0 mg, 2.25 mmol, 1.5 eq.) and a solution of 2-bromo-1-(2,5-dichlorophenyl)ethan-1-one (402 mg, 1.50 mmol, 1.0 eq.) in dry THF (1.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (390 µL, 3.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1), followed by recrystallization from EtOH yielded SGA-48 as colorless solid (271 mg, 0.686 mmol, 46%). Rf = 0.48 (9:1 hexanes/EtOAc). m.p.: 98°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.48 (d, J = 2.0 Hz, 1H, 6’-H), 7.29 (dd, J = 8.2, 2.0 Hz, 1H, 4’-H), 7.25 (d, J = 9.1 Hz, 1H, 3’-H, collapses with chloroform), 6.51 (s, 1H, 4 hr), 4.27 (q, J = 7.0 Hz, 2H, CH2CH3), 3.55 (d, J = 7.0 Hz, 2H, CH2-cy), 2.59 (s, 3H, CH3), 1.61–1.56 (m, 3H, cy), 1.40–1.31 (m, 6H, cy, CH2CH3), 1.08–0.99 (m, 3H, cy), 0.68–0.58 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 165.6 (COOEt), 137.0 (C-2), 135.9 (C-1’ or C-5’), 134.8 (C-1’ or C-5’), 134.0 (C-3’), 131.2 (C-2’), 129.6 (C-6’), 129.2 (C-5), 127.2 (C-4’), 112.2 (C-3), 111.1 (C-4), 59.5 (CH2CH3), 50.8 (CH2-cy), 39.1 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=2981, 2923, 2845, 1739, 1723, 1695, 1565, 1454, 1262, 1238, 1201, 1159, 1076, 1066, 800, 771. HRMS (ESI): calculated for C21H2635Cl2NO2 (M+H)+ 394.13351; found 394.13343. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
1-(Cyclohexylmethyl)−5-(2,5-dichlorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-52.
According to general procedure D, LiOH (63.9 mg, 2.54 mmol, 10 eq.) and a solution of SGA-48 (100 mg, 0.254 mmol, 1.0 eq.) in dioxane/H2O (1.3 mL) were used. After 1 hr the reaction was completed and gave SGA-52 as a colorless solid (75.7 mg, 0.207 mmol, 81%). Rf = 0.18 (6:1 hexanes/EtOAc). m.p.: 180°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 7.48 (d, J = 2.0 Hz, 1H, 6’-H), 7.30 (dd, J = 8.2, 2.0 Hz, 1H, 4’-H), 7.26 (d, J = 8.2 Hz, 1H, 3’-H, collapses with chloroform), 6.57 (s, 1H, 4 hr), 3.57 (d, J = 6.4 Hz, 2H, CH2-cy), 2.60 (s, 3H, CH3), 1.63–1.57 (m, 3H, cy), 1.42–1.36 (m, 3H, cy), 1.08–1.00 (m, 3H, cy), 0.68–0.59 (m, 2H, cy). 13C NMR (126 MHz, CDCl3) δ/ppm = 170.7 (COOH), 138.3 (C-2), 136.0 (C-1’ or C-5’), 135.0 (C-1’ or C-5’), 134.0 (C-3’), 131.0 (C-2’), 129.7 (C-6’), 129.6 (C-5), 127.2 (C-4’), 111.8 (C-4), 111.4 (C-3), 50.9 (CH2-cy), 39.0 (cy), 30.7 (cy), 26.2 (cy), 25.8 (cy), 12.3 (CH3). IR (ATR) max/cm−1=3014, 2970, 2926, 2851, 1739, 1659, 1449, 1365, 1270, 1228, 1217, 1204, 814, 776. HRMS (ESI): calculated for C19H2035Cl2NO2 (M-H)- 364.08766; found 364.08783. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
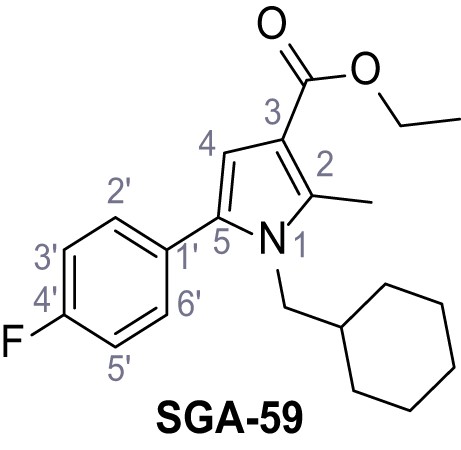
Ethyl 1-(cyclohexylmethyl)−5-(4-fluorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-59.
Following general procedure C, ethyl acetoacetate (208 µL, 1.65 mmol, 1.1 eq.) in dry THF (5.0 mL), NaH (90.0 mg, 2.25 mmol, 1.5 eq.) and a solution of 2-chloro-1-(4-fluorophenyl)ethan-1-one (259 mg, 1.50 mmol, 1.0 eq.) and KI (249 mg, 1.50 mmol, 1.0 eq.) in dry THF (3.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (0.390 mL, 3.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1) yielded SGA-59 as yellow oil (499 mg, 1.45 mmol, 97%). Rf = 0.43 (9:1 hexanes/EtOAc). 1H NMR (500 MHz, CDCl3) δ/ppm = 7.28–7.23 (m, 2H, 2’-H, 6’-H), 7.10–7.01 (m, 2H, 3’-H, 5’-H), 6.48 (s, 1H, 4 hr), 4.25 (q, J = 7.1 Hz, 2H, CH2CH3), 3.71 (d, J = 7.2 Hz, 2H, CH2-cy), 2.57 (s, 3H, CH3), 1.58–1.48 (m, 3H, cy), 1.38–1.28 (m, 6H, cy, CH2CH3), 1.06–0.89 (m, 3H, cy), 0.68–0.53 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 165.7 (COOH), 162.2 (d, JCF = 247.0 Hz, C-4’), 136.9 (C-2), 132.8 (C-5), 131.3 (d, JCF = 8.0 Hz, C-2’, C-6’), 129.7 (d, JCF = 3.3 Hz, C-1’), 115.4 (d, JCF = 21.3 Hz, C-3’, C-5’), 111.9 (C-3), 110.0 (C-4), 59.3 (CH2CH3), 50.1 (CH2-cy), 39.0 (cy), 30.5 (cy), 26.1 (cy), 25.7 (cy), 14.6 (CH2CH3), 12.0 (CH3). IR (ATR) max/cm−1=2977, 2925, 2853, 1693, 1242, 1227, 1218, 1195, 1152, 1062, 844, 811, 774. HRMS (ESI): calculated for C21H27FNO2 (M+H)+ 344.20203; found 344.20193. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
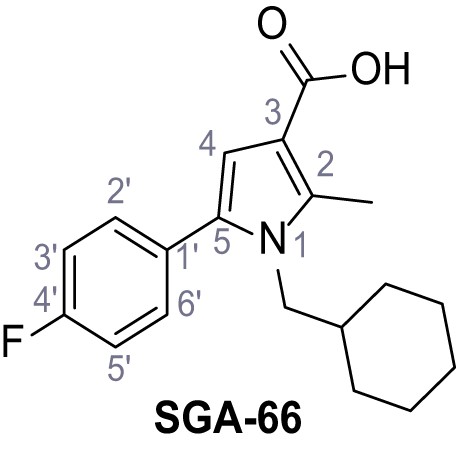
1-(Cyclohexylmethyl)−5-(4-fluorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-66.
According to general procedure D, LiOH (169 mg, 6.71 mmol, 10 eq.) and a solution of SGA-59 (230 mg, 0.671 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 2 hr the reaction was completed and gave SGA-66 as a colorless solid (186 mg, 0.590 mmol, 88%). Rf = 0.24 (6:1 hexanes/EtOAc). m.p.: 171°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.35–7.27 (m, 2H, 2’-H, 6’-H), 7.14–7.04 (m, 2H, 3’-H, 5’-H), 6.57 (s, 1H, 4 hr), 3.75 (d, J = 7.1 Hz, 2H, CH2-cy), 2.61 (s, 3H, CH3), 1.63–1.51 (m, 3H, cy), 1.41–1.32 (m, 3H, cy), 1.08–0.97 (m, 3H, cy), 0.70–0.57 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 171.2 (COOH), 162.37 (d, JCF = 247.1 Hz, C-4’), 138.3 (C-2), 133.3 (C-5), 131.42 (d, JCF = 8.1 Hz, C-2’, C-6’), 129.6 (d, JCF = 3.4 Hz, C-1’), 115.57 (d, JCF = 21.5 Hz, C-3’, C-5’), 111.2 (C-3), 110.8 (C-4), 50.3 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 12.3 (CH3). IR (ATR) max/cm−1=2927, 2854, 1739, 1652, 1568, 1494, 1449, 1265, 1223, 1203, 1158, 840, 776, 582. HRMS (ESI): calculated for C19H21FNO2 (M-H)- 314.15618; found 314.15635. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
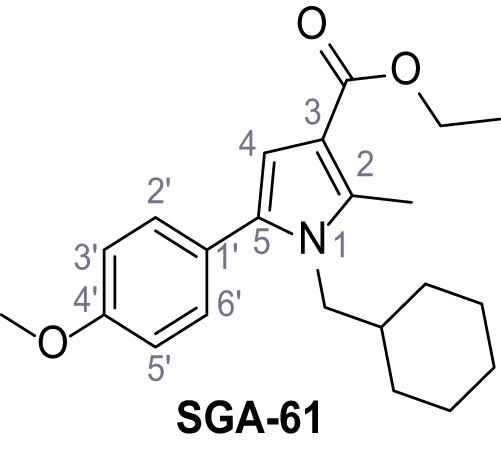
Ethyl 1-(cyclohexylmethyl)−5-(4-methoxyphenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-61.
Following general procedure C, ethyl acetoacetate (208 µL, 1.65 mmol, 1.1 eq.) in dry THF (5.0 mL), NaH (90.0 mg, 2.25 mmol, 1.5 eq.) and a solution of 2-bromo-1-(4-methoxyphenyl)ethan-1-one (344 mg, 1.50 mmol, 1.0 eq.) and KI (249 mg, 1.50 mmol, 1.0 eq.) in dry THF (3.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (390 µL, 3.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1), followed by recrystallization from EtOH yielded SGA-61 as colorless solid (274 mg, 0.770 mmol, 51%). Rf = 0.37 (9:1 hexanes/EtOAc). m.p.: 88°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 7.25–7.21 (m, 2H, 2’-H, 6’-H), 6.95–6.89 (m, 2H, 3’-H, 5’-H), 6.47 (s, 1H, 4 hr), 4.27 (q, J = 7.1 Hz, 2H, CH2CH3), 3.84 (s, 3H, OCH3), 3.73 (d, J = 7.3 Hz, 2H, CH2-cy), 2.58 (s, 3H, CH3), 1.60–1.51 (m, 3H, cy), 1.42–1.31 (m, 6H, cy, CH2CH3), 1.07–0.96 (m, 3H, cy), 0.70–0.59 (m, 2H, cy). 13C NMR (126 MHz, CDCl3) δ/ppm = 165.9 (COOH), 159.0 (C-4’), 136.6 (C-2), 133.8 (C-5), 130.9 (C-2’, C-6’), 126.1 (C-1’), 113.9 (C-3’, C-5’), 111.7 (C-3), 109.5 (C-4), 59.3 (CH2CH3), 55.4 (OCH3), 50.1 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=3016, 2970, 2928, 2847, 1739, 1693, 1568, 1532, 1496, 1443, 1424, 1373, 1243, 1226, 1195, 1175, 1064, 1031, 835, 817, 795, 774. HRMS (ESI): calculated for C22H30NO3 (M+H)+ 356.22202; found 356.22192. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
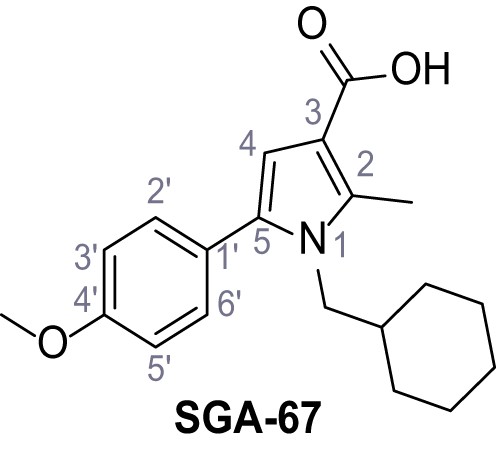
1-(Cyclohexylmethyl)−5-(4-methoxyphenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-67.
According to general procedure D, LiOH (78.7 mg, 3.12 mmol, 10 eq.) and a solution of SGA-61 (111 mg, 0.312 mmol, 1.0 eq.) in dioxane/H2O (1.3 mL) were used. After 2 hr the reaction was completed and gave SGA-67 as a colorless solid (95.3 mg, 0.291 mmol, 90%). Rf = 0.20 (6:1 hexanes/EtOAc). m.p.: 198°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.27–7.23 (m, 2H, 2’-H, 6’-H), 6.95–6.91 (m, 2H, 3’-H, 5’-H), 6.54 (s, 1H, 4 hr), 3.85 (s, 3H, OCH3), 3.75 (d, J = 7.2 Hz, 2H, CH2-cy), 2.60 (s, 3H, CH3), 1.62–1.53 (m, 3H, cy), 1.43–1.33 (m, 3H, cy), 1.09–0.98 (m, 3H, cy), 0.70–0.59 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 171.3 (COOH), 159.0 (C-4’), 137.8 (C-2), 134.1 (C-5), 131.0 (C-2’, C-6’), 125.9 (C-1’), 113.9 (C-3’, C-5’), 111.0 (C-3), 110.2 (C-4), 55.3 (OCH3), 50.2 (CH2-cy), 38.9 (cy), 30.5 (cy), 26.2 (cy), 25.8 (cy), 12.3 (CH3). IR (ATR) max/cm−1=3027, 3002, 2970, 2925, 2849, 1738, 1652, 1569, 1535, 1494, 1435, 1364, 1266, 1247, 1228, 1203, 840, 778. HRMS (ESI): calculated for C20H24NO3 (M-H)- 326.17617; found 326.17633. Purity (HPLC): 93% (λ = 210 nm),>96% (λ = 254 nm).
Ethyl 1-(cyclohexylmethyl)−5-(2,4-difluorophenyl)−2-methyl-1H-pyrrole-3-carboxylate – SGA-62.
Following general procedure C, ethyl acetoacetate (208 µL, 1.65 mmol, 1.1 eq.) in dry THF (5.0 mL), NaH (90.0 mg, 2.25 mmol, 1.5 eq.) and a solution of 2-chloro-1-(2,4-difluorophenyl)ethan-1-one (286 mg, 1.50 mmol, 1.0 eq.) and KI (249 mg, 1.50 mmol, 1.0 eq.) in dry THF (3.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (390 µL, 3.00 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1) yielded SGA-62 as yellow solid (424 mg, 1.17 mmol, 78%). Rf = 0.43 (9:1 hexanes/EtOAc). m.p.: 94°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 7.27 (td, J = 8.4, 6.5 Hz, 1H, 6’-H), 6.96–6.85 (m, 2H, 3’-H, 5’-H), 6.53 (s, 1H, 4 hr), 4.26 (q, J = 7.1 Hz, 2H, CH2CH3), 3.60 (d, J = 7.1 Hz, 2H, CH2-cy), 2.58 (s, 3H, CH3), 1.59–1.53 (m, 3H, cy), 1.38–1.30 (m, 6H, cy, CH2CH3), 1.06–0.98 (m, 3H, cy), 0.68–0.58 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 165.5 (COOEt), 162.9 (dd, JCF = 250.1, 11.6 Hz, C-4’), 160.2 (dd, JCF = 248.9, 12.0 Hz, C-2’), 137.2 (C-2), 133.5 (dd, JCF = 9.4, 4.1 Hz, C-6’), 126.2 (C-5), 117.6 (dd, JCF = 15.7, 3.9 Hz, C-1’), 112.3 (C-3), 111.6 (dd, JCF = 21.1, 3.7 Hz, C-5’), 111.3 (C-4), 104.20 (t, JCF = 25.8 Hz, C-3’), 59.4 (CH2CH3), 50.6 (d, JCF = 3.0 Hz, CH2-cy), 39.0 (cy), 30.5 (cy), 26.1 (cy), 25.7 (cy), 14.6 (CH2CH3), 12.0 (CH3). IR (ATR) max/cm−1=2971, 2929, 2848, 1739, 1698, 1571, 1426, 1371, 1235, 1199, 1067, 851, 834. HRMS (ESI): calculated for C21H26F2NO2 (M+H)+ 362.19261; found 362.19247. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
1-(Cyclohexylmethyl)−5-(2,4-difluorophenyl)−2-methyl-1H-pyrrole-3-carboxylic acid - SGA-68.
According to general procedure D, LiOH (167 mg, 6.61 mmol, 10 eq.) and a solution of SGA-62 (239 mg, 0.661 mmol, 1.0 eq.) in dioxane/H2O (1.3 mL) were used. After 1 hr the reaction was completed and gave SGA-68 as a colorless solid (170 mg, 0.511 mmol, 77%). Rf = 0.29 (6:1 hexanes/EtOAc). m.p.: 168°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.33–7.25 (m, 1H, 6’-H, collapses with chloroform), 6.98–6.86 (m, 2H, 3’-H, 5’-H), 6.60 (s, 1H, 4 hr), 3.62 (d, J = 7.1 Hz, 2H, CH2-cy), 2.61 (s, 3H, CH3), 1.65–1.51 (m, 3H, cy), 1.49–1.32 (m, 3H, cy), 1.14–0.94 (m, 3H, cy), 0.72–0.56 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 171.0 (COOH), 163.0 (dd, JCF = 250.3, 11.6 Hz, C-4’), 160.3 (dd, JCF = 249.0, 12.0 Hz, C-2’), 138.5 (C-2), 133.6 (dd, JCF = 9.5, 4.0 Hz, C-6’), 126.7 (C-5), 117.5 (dd, JCF = 15.8, 3.8 Hz, C-1’), 112.0 (C-3), 111.7 (dd, JCF = 21.3, 3.8 Hz, C-5’), 111.6 (C-4), 104.3 (t, JCF = 25.9 Hz, C-3’), 50.72 (d, JCF = 3.0 Hz, CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.7 (cy), 12.2 (CH3). IR (ATR) max/cm−1=2970, 2926, 2854, 1739, 1666, 1573, 1450, 1433, 1364, 1265, 1239, 1200, 1140, 778. HRMS (ESI): calculated for C19H20F2NO2 (M-H)- 332.14676; found 332.14697. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
Ethyl 1-(cyclohexylmethyl)−2-methyl-5-(2-(trifluoromethoxy)phenyl)−1H-pyrrole-3-carboxylate (25).
Following general procedure C, ethyl acetoacetate (123 µL, 0.972 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (53.0 mg, 1.32 mmol, 1.5 eq.) and a solution of ketone 24 (250 mg, 0.883 mmol, 1.0 eq.) and KI (147 mg, 0.883 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (230 µL, 1.77 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 9:1) yielded ester 25 as colorless solid (345 mg, 0.845 mmol, 95%). Rf = 0.46 (9:1 hexanes/EtOAc). m.p.: 66°C. 1H NMR (100 MHz, CDCl3) δ/ppm = 7.44–7.29 (m, 4H, 3’-H, 4’-H, 5’-H, 6’-H), 6.53 (s, 1H, 4 hr), 4.27 (q, J = 7.1 Hz, 2H, CH2CH3), 3.59 (d, J = 7.1 Hz, 2H, CH2-cy), 2.59 (s, 3H, CH3), 1.59–1.52 (m, 3H, cy), 1.40–1.32 (m, 6H, cy, CH2CH3), 1.06–0.94 (m, 3H, cy), 0.67–0.54 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 165.8 (COOEt), 147.4 (C-2’), 137.1 (C-2), 133.4 (C-4’, C-5’ or C-6’), 129.6 (C-4’, C-5’ or C-6’), 127.9 (C-5), 126.9 (C-1’), 126.7 (C-4’, C-5’ or C-6’), 120.5 (q, JCF = 257.4 Hz, OCF3), 120.4 (C-3’), 112.2 (C-3), 111.4 (C-4), 59.5 (CH2CH3), 50.7 (CH2-cy), 39.0 (cy), 30.6 (cy), 26.2 (cy), 25.8 (cy), 14.7 (CH2CH3), 12.1 (CH3). IR (ATR) max/cm−1=1934, 1692, 1447, 1422, 1242, 1192, 1155, 1059, 769. HRMS (ESI): calculated for C22H27F3NO3 (M+H)+ 410.19375; found 410.19336. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
1-(Cyclohexylmethyl)−2-methyl-5-(2-(trifluoromethoxy)phenyl)−1H-pyrrole-3-carboxylic acid – SGA-162.
According to general procedure D, LiOH (142 mg, 5.62 mmol, 10 eq.) and a solution of ester 25 (230 mg, 0.562 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 2 hr the reaction was completed and gave SGA-162 as a colorless solid (199 mg, 0.523 mmol, 93%). Rf = 0.11 (9:1 hexanes/EtOAc). m.p.: 170°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 11.13 (s, 1H, COOH), 7.46–7.30 (m, 4H, 3’-H, 4’-H, 5’-H, 6’-H), 6.59 (s, 1H, 4 hr), 3.60 (d, J = 7.1 Hz, 2H, CH2-cy), 2.61 (s, 3H, CH3), 1.61–1.52 (m, 3H, cy), 1.41–1.31 (m, 3H, cy), 1.09–0.96 (m, 3H, cy), 0.67–0.55 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 169.9 (COOH), 147.4 (C-2’), 138.3 (C-2), 133.4 (C-4’, C-5’ or C-6’), 129.8 (C-4’, C-5’ or C-6’), 128.3 (C-5), 126.8 (C-4’, C-5’ or C-6’), 126.7 (C-1’), 120.5 (q, JCF = 260.2 Hz, OCF3), 120.4 (C-4’), 112.1 (C-4), 111.2 (C-3), 50.8 (CH2-cy), 38.9 (cy), 30.6 (cy), 26.2 (cy), 25.7 (cy), 12.3 (CH3). IR (ATR) max/cm−1=2927, 1642, 1473, 1444, 1249, 1199, 1156, 776, 759. HRMS (ESI): calculated for C20H21F3NO3 (M-H)- 380.14790; found 380.14805. Purity (HPLC):>96% (λ = 210 nm).
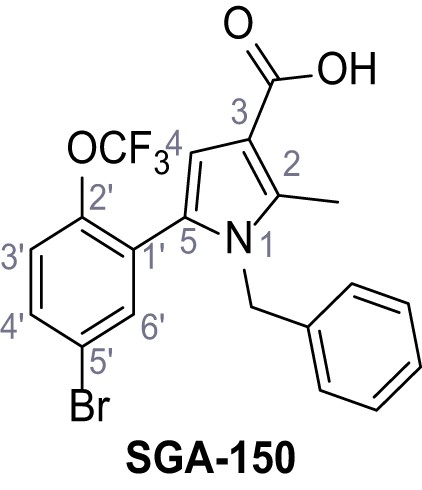
1-Benzyl-5-(5-bromo-2-(trifluoromethoxy)phenyl)−2-methyl-1H-pyrrole-3-carboxylic acid – SGA-150.
Following general procedure C, ethyl acetoacetate (104 µL, 0.821 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (44.8 mg, 1.12 mmol, 1.5 eq.) and a solution of ketone 23 (237 mg, 0.746 mmol, 1.0 eq.) and KI (124 mg, 0.746 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and benzylamine (204 µL, 1.87 mmol, 2.5 eq.) was added. FCC (hexanes/EtOAc 97:3) yielded ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-isopropyl-2-methyl-1H-pyrrole-3-carboxylate (26) as colorless oil (107 mg, 0.222 mmol). This product was used without further purification or characterization for the next step. Rf = 0.44 (9:1 hexanes/EtOAc). According to general procedure D, LiOH (55.9 mg, 2.22 mmol, 10 eq.) and a solution of ester 26 (107 mg, 0.222 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 16 hr the reaction was completed and gave SGA-150 as a colorless solid (34.4 mg, 0.0757 mmol, 10% over two steps). Rf = 0.27 (9:1 hexanes/EtOAc). m.p.: 185°C. 1H NMR (400 MHz, CD2Cl2) δ/ppm = 11.12 (s, 1H, COOH), 7.51 (dd, J = 8.8, 2.5 Hz, 1H, 4’-H), 7.39 (d, J = 2.5 Hz, 1H, 6’-H), 7.30–7.18 (m, 4H, 3’-H, Ph), 6.83–6.77 (m, 2H, Ph), 6.70 (s, 1H, 4 hr), 5.00 (s, 2H, CH2), 2.49 (s, 3H, CH3). 13C NMR (121 MHz, CD2Cl2) δ/ppm = 169.9 (COOH), 147.0 (C-2’), 139.6 (C-2), 137.4 (C-1’), 136.3 (C-6’), 133.4 (C-4’), 129.3 (Ph), 128.5 (qPh), 128.0 (Ph), 127.3 (C-5), 126.2 (Ph), 122.8 (C-3’), 120.8 (q, JCF = 258.7 Hz, OCF3), 120.3 (C-5’), 113.1 (C-4), 112.3 (C-3), 48.7 (CH2), 12.2 (CH3). IR (ATR) max/cm−1=2925, 2360, 1670, 1249, 1223, 1198, 1171, 733. HRMS (ESI): calculated for C20H1479BrF3NO3 (M-H)- 452.01146; found 452.01168. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
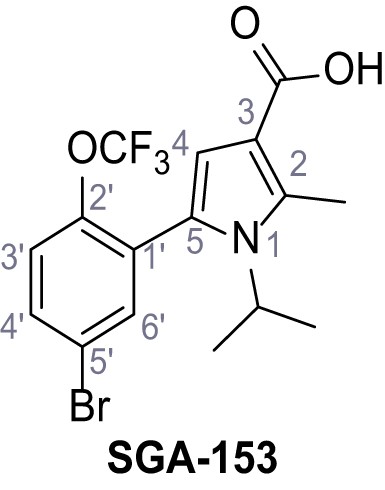
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-isopropyl-2-methyl-1H-pyrrole-3-carboxylic acid – SGA-153.
Following general procedure C, ethyl acetoacetate (87.6 µL, 0.693 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (37.8 mg, 0.945 mmol, 1.5 eq.) and a solution of ketone 23 (200 mg, 0.630 mmol, 1.0 eq.) and KI (105 mg, 0.630 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and isopropylamine (108 µL, 1.26 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 99:1) yielded ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-isopropyl-2-methyl-1H-pyrrole-3-carboxylate (27) as colorless oil (65.9 mg, 0.152 mmol). This product was used without further purification or characterization for the next step. Rf = 0.55 (9:1 hexanes/EtOAc). According to general procedure D, LiOH (38.3 mg, 1.52 mmol, 10 eq.) and a solution of ester 27 (65.9 mg, 0.152 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 18 hr the reaction was completed and gave SGA-153 as a colorless solid (32.4 mg, 0.0798 mmol, 12% over two steps). Rf = 0.11 (9:1 hexanes/EtOAc). m.p.: 192°C. 1H NMR (400 MHz, C3D6O) δ/ppm = 7.75 (dd, J = 8.8, 2.6 Hz, 1H, 4’-H), 7.65 (d, J = 2.6 Hz, 1H, 6’-H), 7.44 (dq, J = 8.8, 1.5 Hz, 1H, 3’-H), 6.47 (s, 1H, 4 hr), 4.27 (p, J = 7.0 Hz, 1H, CH), 2.72 (s, 3H, CH3), 1.45 (d, J = 7.0 Hz, 6H, CH(CH3)2). 13C NMR (101 MHz, C3D6O) δ/ppm = 166.4 (COOH), 147.7 (C-2’), 137.5 (C-2), 137.0 (C-6’), 134.0 (C-4’), 130.5 (C-1’), 126.4 (C-5), 123.7 (q, J = 257.6 Hz, OCF3), 123.5 (C-3’), 120.6 (C-5’), 113.9 (C-3), 113.0 (C-4), 50.2 (CH), 22.0 (CH(CH3)2), 13.0 (CH3). IR (ATR) max/cm−1=2936, 2358, 1672, 1248, 1214, 1200, 1166. HRMS (ESI): calculated for C16H1479BrF3NO3 (M-H)- 404.01146; found 404.01164. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
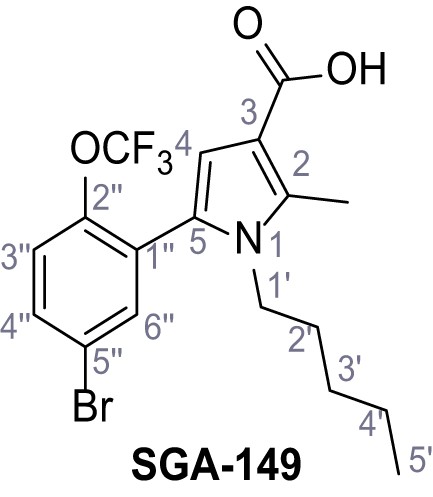
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−2-methyl-1-pentyl-1H-pyrrole-3-carboxylic acid – SGA-149.
Following general procedure C, ethyl acetoacetate (87.6 µL, 0.693 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (37.8 mg, 0.945 mmol, 1.5 eq.) and a solution of ketone 23 (200 mg, 0.630 mmol, 1.0 eq.) and KI (105 mg, 0.630 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and n-pentylamine (146 µL, 1.26 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 99:1) yielded ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−2-methyl-1-pentyl-1H-pyrrole-3-carboxylate (28) as colorless oil (81.5 mg, 0.176 mmol). The product was used without further purification or characterization for the next step. Rf = 0.51 (9:1 hexanes/EtOAc). According to general procedure D, LiOH (44.4 mg, 1.76 mmol, 10 eq.) and a solution of ester 28 (81.5 mg, 0.176 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 18 hr the reaction was completed and gave SGA-149 as a colorless solid (40.5 mg, 0.0933 mmol, 15% over two steps). Rf = 0.06 (9:1 hexanes/EtOAc). m.p.: 121°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.57–7.51 (m, 2H, 4’’-H, 6’’-H), 7.24–7.20 (m, 1H, 3’’-H), 6.61 (s, 1H, 4 hr), 3.74–3.68 (m, 2H, 1’-H), 2.61 (s, 3H, CH3), 1.49–1.43 (m, 2H, 2’-H), 1.19–1.06 (m, 4H, 3’-H, 4’-H), 0.80 (t, J = 7.2 Hz, 3H, 5’-H). 13C NMR (101 MHz, CDCl3) δ/ppm = 169.4 (COOH), 146.6 (C-2’’), 138.3 (C-2), 135.9 (C-6’’), 132.8 (C-4’’), 128.7 (C-5’’), 126.1 (C-5), 122.5 (q, J = 279.8 Hz, OCF3), 122.4 (C-3’’), 120.0 (C-1’’), 112.7 (C-4), 111.4 (C-3), 44.7 (C-1’), 30.2 (C-2’), 28.8 (C-3’), 22.1 (C-4’), 13.9 (C-5’), 11.9 (CH3). IR (ATR) max/cm−1=2929, 1663, 1471, 1436, 1247, 1212, 1194, 1160, 781. HRMS (ESI): calculated for C18H2079BrF3NO3 (M+H)+ 434.05732; found 434.05765. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
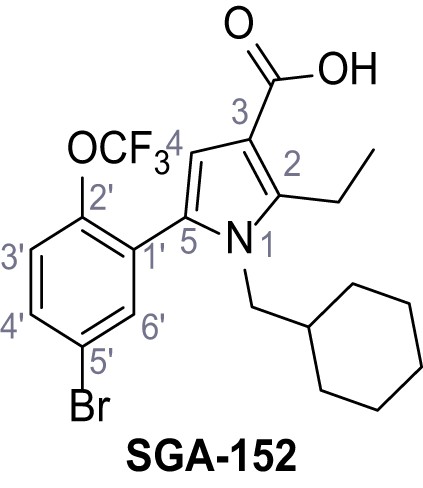
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-ethyl-1H-pyrrole-3-carboxylic acid – SGA-152.
Following general procedure C, ethyl propionylacetate (99.9 µL, 0.693 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (37.8 mg, 0.945 mmol, 1.5 eq.) and a solution of ketone 23 (200 mg, 0.630 mmol, 1.0 eq.) and KI (105 mg, 0.630 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (164 µL, 1.26 mmol, 2.0 eq.) was added. FCC (hexanes/EtOAc 99:1) yielded ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-ethyl-1H-pyrrole-3-carboxylate (29) as colorless oil (98.0 mg, 0.195 mmol). This product was used without further purification or characterization for the next step. Rf = 0.58 (9:1 hexanes/EtOAc). According to general procedure D, LiOH (49.2 mg, 1.95 mmol, 10 eq.) and a solution of ester 29 (98.0 mg, 0.195 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 18 hr the reaction was completed and gave SGA-152 as yellow solid (30.8 mg, 0.0649 mmol, 9% over two steps). Rf = 0.24 (9:1 hexanes/EtOAc). m.p.: 152°C. 1H NMR (500 MHz, CDCl3) δ/ppm = 11.35 (s, 1H, COOH), 7.57–7.50 (m, 2H, 4’-H, 6’-H), 7.20 (dd, J = 8.6, 1.4 Hz, 1H, 3’-H), 6.61 (s, 1H, 4 hr), 3.61 (d, J = 7.2 Hz, 2H, CH2-cy), 3.04 (q, J = 7.4 Hz, 2H, CH2CH3), 1.64–1.54 (m, 3H, cy), 1.39–1.31 (m, 3H, cy), 1.23 (t, J = 7.4 Hz, 3H, CH2CH3), 1.08–0.99 (m, 3H, cy), 0.70–0.60 (m, 2H, cy). 13C NMR (121 MHz, CDCl3) δ/ppm = 169.8 (COOH), 146.3 (C-2’), 145.1 (C-2), 135.5 (C-4’ or C-6’), 132.5 (C-4’ or C-6’), 129.1 (C-1’), 126.7 (C-5), 122.3 (C-3’), 120.3 (q, JCF = 259.2 Hz, OCF3), 120.0 (C-5’), 113.2 (C-4), 110.8 (C-3), 50.9 (CH2-cy), 39.6 (cy), 30.7 (cy), 26.2 (cy), 25.8 (cy), 19.2 (CH2CH3), 14.5 (CH2CH3). IR (ATR) max/cm−1=2927, 2359, 1659, 1469, 1441, 1250, 1212, 1192, 1172. HRMS (ESI): calculated for C21H2279BrF3NO3 (M-H)- 472.07406; found 472.07418. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
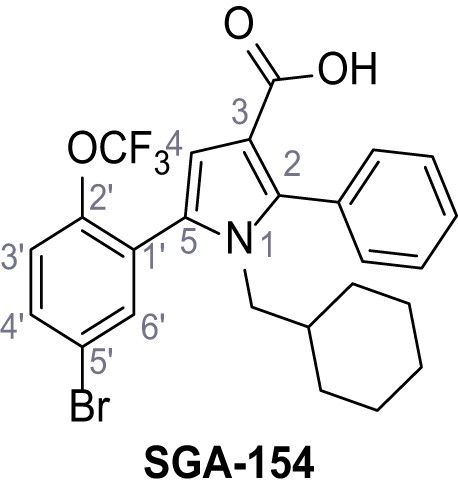
5-(5-Bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-phenyl-1H-pyrrole-3-carboxylic acid – SGA-154.
Following general procedure C, ethyl benzoylacetate (134 µL, 0.693 mmol, 1.1 eq.) in dry THF (4.0 mL), NaH (37.8 mg, 0.945 mmol, 1.5 eq.) and a solution of ketone 23 (200 mg, 0.630 mmol, 1.0 eq.) and KI (105 mg, 0.630 mmol, 1.0 eq.) in dry THF (2.0 mL) were used. Then the residue was dissolved in acetic acid (5.0 mL) and cyclohexanemethanamine (328 µL, 2.52 mmol, 4.0 eq.) was added. FCC (hexanes/EtOAc 99:1) yielded ethyl 5-(5-bromo-2-(trifluoromethoxy)phenyl)−1-(cyclohexylmethyl)−2-phenyl-1H-pyrrole-3-carboxylate (30) as yellow solid (151 mg, 0.274 mmol). This product was used without further purification or characterization for the next step. Rf = 0.50 (9:1 hexanes/EtOAc). According to general procedure D, LiOH (69.2 mg, 2.74 mmol, 10 eq.) and a solution of ester 30 (151 mg, 0.274 mmol, 1.0 eq.) in dioxane/H2O (3.0 mL) were used. After 18 hr the reaction was completed and gave SGA-154 as yellow solid (91.5 mg, 0.175 mmol, 28% over two steps). Rf = 0.12 (9:1 hexanes/EtOAc). m.p.: 170°C. 1H NMR (400 MHz, CDCl3) δ/ppm = 7.61 (d, J = 2.5 Hz, 1H, 6’-H), 7.55 (dd, J = 8.7, 2.5 Hz, 1H, 4’-H), 7.47–7.41 (m, 3H, Ph), 7.39–7.35 (m, 2H, Ph), 7.23 (dq, J = 8.7, 1.3 Hz, 1H, 3’-H), 6.74 (s, 1H, 4 hr), 3.56 (d, J = 7.4 Hz, 2H, CH2-cy), 1.48–1.42 (m, 3H, cy), 1.11–1.05 (m, 2H, cy), 1.03–0.95 (m, 1H, cy), 0.90–0.82 (m, 3H, cy), 0.43–0.32 (m, 2H, cy). 13C NMR (101 MHz, CDCl3) δ/ppm = 167.7 (COOH), 146.1 (C-2’), 141.7 (C-2), 135.2 (C-6’), 132.7 (C-4’), 131.7 (C-5, C-1’, C-5’ or qPh), 131.0 (Ph), 128.8 (C-5, C-1’, C-5’ or qPh), 128.7 (Ph), 128.3 (Ph), 122.4 (C-3’), 120.4 (q, JCF = 266.3 Hz, OCF3), 120.2 (C-5, C-1’, C-5’ or qPh), 113.8 (C-4), 112.7 (C-3), 52.0 (CH2-cy), 38.7 (cy), 30.3 (cy), 26.1 (cy), 25.6 (cy). One quaternary carbon is missing. IR (ATR) max/cm−1=2915, 2335, 1667, 1487, 1248, 1208, 1169, 1127, 796, 697. HRMS (ESI): calculated for C25H2279BrF3NO3 (M-H)- 520.07406; found 520.07418. Purity (HPLC):>96% (λ = 210 nm),>96% (λ = 254 nm).
Statistical analysis
Request a detailed protocolAll error bars are depicted as SEM. Statistical significance was determined via Student’s t-test, one-way ANOVA, or two-way ANOVA followed by either Tukey’s or Bonferroni’s post hoc test. Significance is denoted on figures with asterisks as outlined in the legends. All data presented are representative of three or more independent experiments.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
-
Essential requirement for two-pore channel 1 in NAADP-mediated calcium signalingThe Journal of Cell Biology 186:201–209.https://doi.org/10.1083/jcb.200904073
-
An NAADP-gated two-pore channel targeted to the plasma membrane uncouples triggering from amplifying Ca2+ signalsJournal of Biological Chemistry 285:38511–38516.https://doi.org/10.1074/jbc.M110.162073
-
The voltage-gated sodium channel TPC1 confers endolysosomal excitabilityNature Chemical Biology 10:463–469.https://doi.org/10.1038/nchembio.1522
-
Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenasesJournal of Medicinal Chemistry 52:2683–2693.https://doi.org/10.1021/jm800963t
-
2-Imino 2H-chromene and 2-(phenylimino) 2H-chromene 3-aryl carboxamide derivatives as novel cytotoxic agents: synthesis, biological assay, and molecular docking studyJournal of the Iranian Chemical Society 13:2163–2171.https://doi.org/10.1007/s13738-016-0934-7
-
The ionic requirements for the production of action potentials in crustacean muscle fibresThe Journal of Physiology 142:516–543.https://doi.org/10.1113/jphysiol.1958.sp006034
-
Small molecule activators of TRPML3Chemistry & Biology 17:135–148.https://doi.org/10.1016/j.chembiol.2009.12.016
-
Cell-permeable iminocoumarine-based fluorescent dyes for mitochondriaOrganic Letters 13:2884–2887.https://doi.org/10.1021/ol200908r
-
Arylpiperazine-containing pyrrole 3-carboxamide derivatives targeting serotonin 5-HT(2A), 5-HT(2C), and the serotonin transporter as a potential antidepressantBioorganic & Medicinal Chemistry Letters 20:1705–1711.https://doi.org/10.1016/j.bmcl.2010.01.093
-
Phosphatidylinositol-3,5-bisphosphate lipid-binding-induced activation of the human two-pore channel 2Cellular and Molecular Life Sciences 75:3803–3815.https://doi.org/10.1007/s00018-018-2829-5
-
Physical basis of apparent pore dilation of ATP-activated P2X receptor channelsNature Neuroscience 18:1577–1583.https://doi.org/10.1038/nn.4120
-
Lysosomal two-pore channel subtype 2 (TPC2) regulates skeletal muscle autophagic signalingJournal of Biological Chemistry 290:3377–3389.https://doi.org/10.1074/jbc.M114.608471
-
Photoaffinity labeling of nicotinic acid adenine dinucleotide phosphate (NAADP) targets in mammalian cellsJournal of Biological Chemistry 287:2296–2307.https://doi.org/10.1074/jbc.M111.305813
-
Understanding the mechanism of IL-1β secretionCytokine & Growth Factor Reviews 22:189–195.https://doi.org/10.1016/j.cytogfr.2011.10.001
-
Regulated lysosomal exocytosis mediates Cancer progressionScience Advances 1:e1500603.https://doi.org/10.1126/sciadv.1500603
-
Mechanosynthesis of amides in the total absence of organic solvent from reaction to product recoveryChemical Communications 48:11781–11783.https://doi.org/10.1039/c2cc36352f
-
3-(3,5-Dimethyl-1 H -Pyrazol-1-yl)-3-Oxopropanenitrile as Precursor for Some New Mono-Heterocyclic and Bis-Heterocyclic CompoundsJournal of Heterocyclic Chemistry 54:289–294.https://doi.org/10.1002/jhet.2578
-
NAADP induces pH changes in the lumen of acidic Ca2+ storesBiochemical Journal 402:301–310.https://doi.org/10.1042/BJ20060759
-
mTORC1 controls lysosomal Ca2+ release through the two-pore channel TPC2Science Signaling 11:eaao5775.https://doi.org/10.1126/scisignal.aao5775
-
Function and dysfunction of two-pore channelsScience Signaling 8:re7.https://doi.org/10.1126/scisignal.aab3314
-
Two-pore channels and diseaseBiochimica Et Biophysica Acta (BBA) - Molecular Cell Research 1865:1678–1686.https://doi.org/10.1016/j.bbamcr.2018.05.004
-
Mining of ebola virus entry inhibitors identifies approved drugs as two-pore channel pore blockersBiochimica Et Biophysica Acta (BBA) - Molecular Cell Research 1866:1151–1161.https://doi.org/10.1016/j.bbamcr.2018.10.022
-
TPC2 is a novel NAADP-sensitive Ca2+ release channel, operating as a dual sensor of luminal pH and Ca2+The Journal of Biological Chemistry 285:35039–35046.https://doi.org/10.1074/jbc.M110.156927
-
Characterization of two-pore channel 2 (TPCN2)-mediated Ca2+ currents in isolated lysosomesThe Journal of Biological Chemistry 285:21219–21222.https://doi.org/10.1074/jbc.C110.143123
-
Synthesis characterization and biological evaluation of some novel amino pyrazole derivativesAsian Journal of Chemistry 30:2647–2651.https://doi.org/10.14233/ajchem.2018.21499
-
Identification and validation of larixyl acetate as a potent TRPC6 inhibitorMolecular Pharmacology 89:197–213.https://doi.org/10.1124/mol.115.100792
-
Lysosomal physiologyAnnual Review of Physiology 77:57–80.https://doi.org/10.1146/annurev-physiol-021014-071649
-
Transient receptor potential mucolipin 1 (TRPML1) and two-pore channels are functionally independent organellar ion channelsJournal of Biological Chemistry 286:22934–22942.https://doi.org/10.1074/jbc.M110.210930
-
TRPM7 kinase activity regulates murine mast cell degranulationThe Journal of Physiology 594:2957–2970.https://doi.org/10.1113/JP271564
-
The two-pore channel TPCN2 mediates NAADP-dependent Ca2+-release from lysosomal storesPflügers Archiv - European Journal of Physiology 458:891–899.https://doi.org/10.1007/s00424-009-0690-y
Article and author information
Author details
Funding
Mucolipidosis IV Foundation (MDBR-17-120- ML4)
- Christian Grimm
Deutsche Forschungsgemeinschaft (SFB/TRR152 P04)
- Christian Grimm
Deutsche Forschungsgemeinschaft (SFB/TRR152 P06)
- Christian Wahl-Schott
Deutsche Forschungsgemeinschaft (SFB/TRR152 P12)
- Martin Biel
Deutsche Forschungsgemeinschaft (BR 1034/7-1)
- Franz Bracher
Biotechnology and Biological Sciences Research Council (BB/N01524X/1)
- Sandip Patel
Deutsche Forschungsgemeinschaft (239283807)
- Christian Grimm
Deutsche Forschungsgemeinschaft (SFB/TRR152 P14)
- Christian Grimm
Deutsche Forschungsgemeinschaft (GR 4315/2-1)
- Christian Grimm
University of Pennsylvania (Orphan Disease Center)
- Christian Grimm
Mucolipidosis IV Foundation (404.510.2577)
- Christian Grimm
NCL Foundation (2018-Grimm-Paquet)
- Christian Grimm
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Christopher Wolf and Elisabeth Butz for their assistance with pilot studies, Dr. Youxing Jiang (University of Texas Southwestern Medical Center) for plasmid gifts, and Dr. Herman van der Putten (NCL Foundation) for valuable discussions. This work was supported, in part, by funding from the German Research Foundation, DFG (project number 239283807, SFB/TRR152 projects P04 to CG, P06 to CW-S, P12 to MB, and P14 to SZ; BR 1034/7–1 to FB; GR 4315/2–1 to CG), the University of Pennsylvania Orphan Disease Center and the Mucolipidosis IV Foundation grant MDBR-17–120 ML4 to CG, the Mucolipidosis IV Foundation grant (project number 404.510.2577) to CG, the NCL Foundation grant (project 2018-Grimm-Paquet) to CG, and the BBSRC grant (BB/N01524X/1) to SP.
Ethics
Animal experimentation: This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Bavarian Government and the European Union. All of the animals were handled according to approved institutional animal care protocols of the University of Munich. The protocol was approved by the Bavarian Government (AZ55.2-1-54-2532-170-17).
Copyright
© 2020, Gerndt et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 6,119
- views
-
- 1,075
- downloads
-
- 147
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 147
- citations for umbrella DOI https://doi.org/10.7554/eLife.54712