A Mendelian randomization study of the role of lipoprotein subfractions in coronary artery disease
Figures
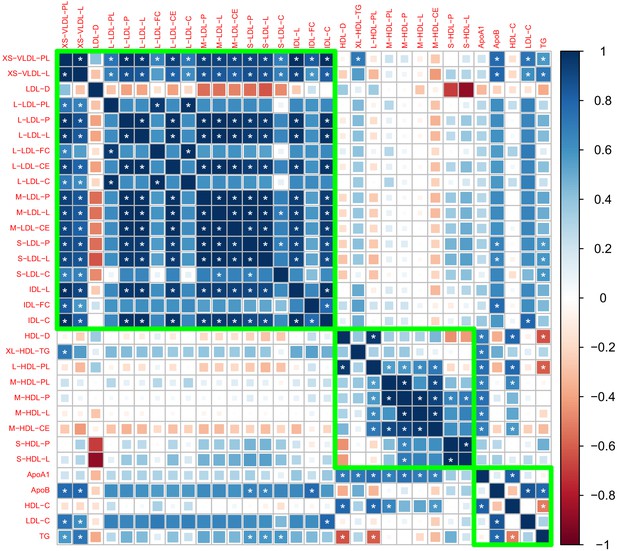
Genetic correlation matrix of the 27 lipoprotein subfraction traits selected in phenotypic screening and five traditional lipid traits.
White asterisk indicates the correlation is statistically significant after Bonferroni correction for multiple comparisons at level 0.05.
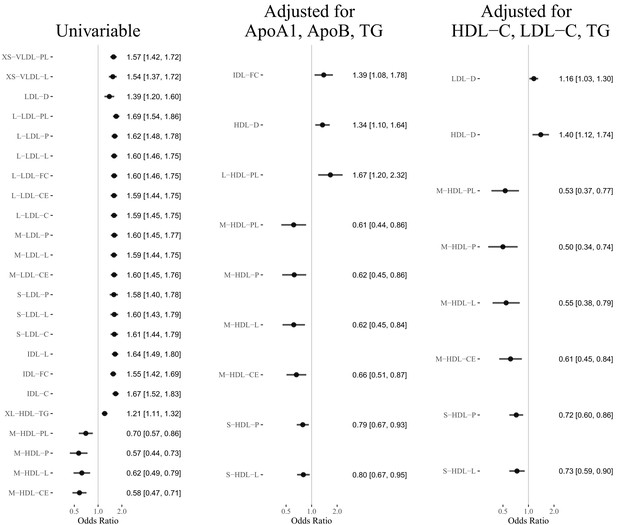
Results of the Mendelian randomization analyses (false discover rate = 0.05): Estimated odds ratio [95% confidence interval] per standard deviation increase of the selected lipoprotein measurements on MI or CAD.
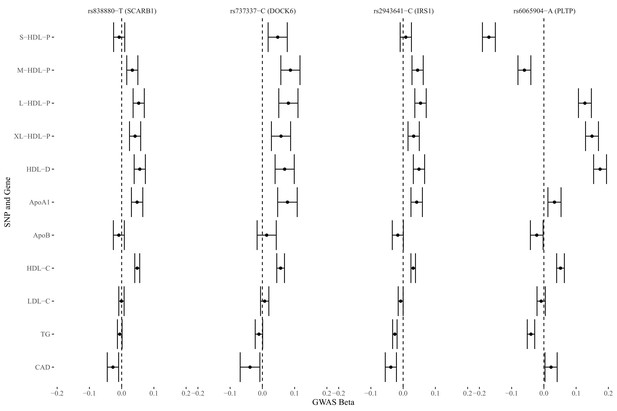
Genetic markers for HDL size (with risk alleles) and their associations with various lipid traits.
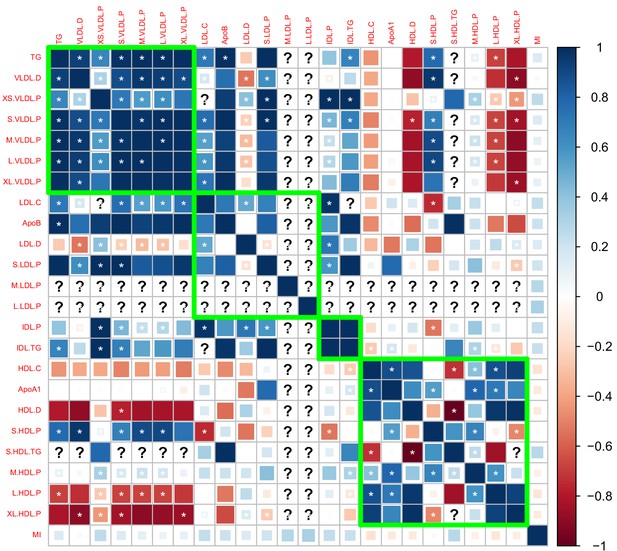
Genetic correlations computed using the Davis et al., 2017 GWAS summary dataset.
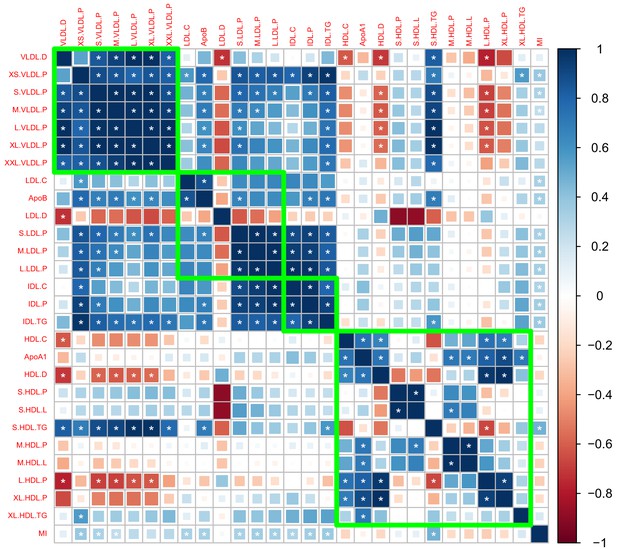
Genetic correlations computed using the Kettunen et al., 2016 GWAS summary dataset.
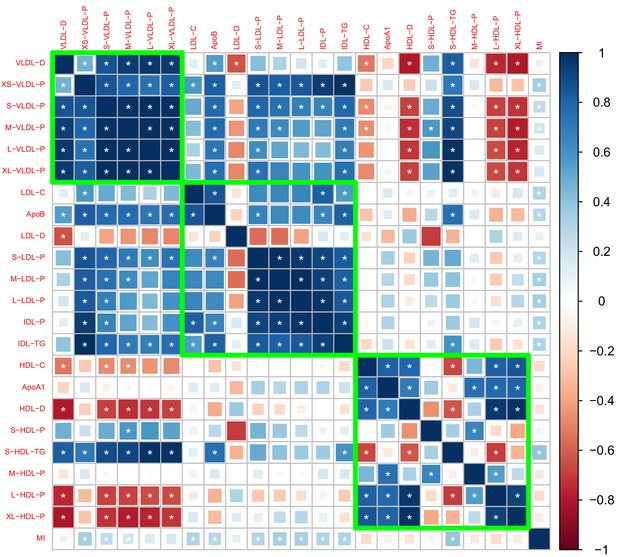
Genetic correlations computed by meta-analyzing the results in Appendix 2—figures 1 and 2.
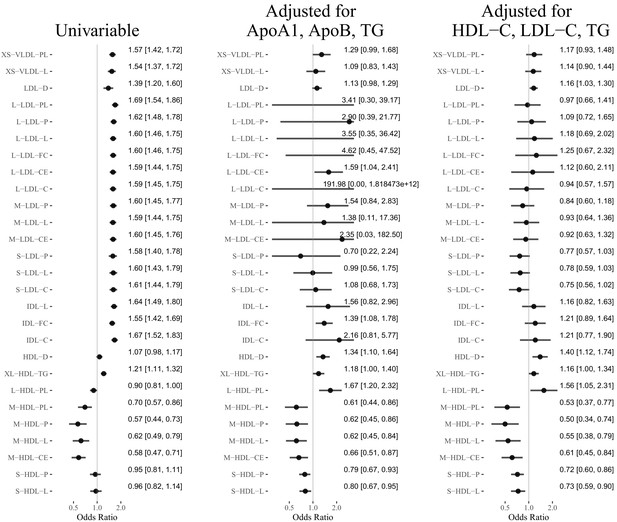
Mendelian randomization results for the 27 lipoprotein measurements selected in phenotypic screening.
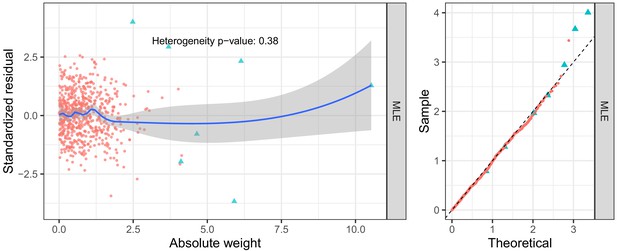
Diagnostic plots for S-HDL-P (selection: Davis; exposure: Kettunen; outcome: UK Biobank).
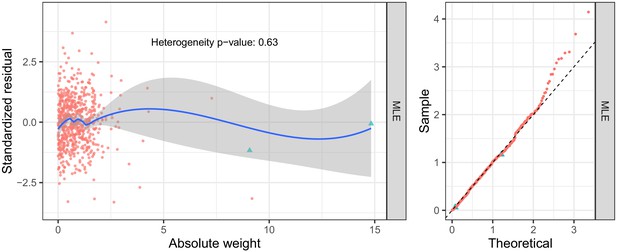
Diagnostic plots for M-HDL-P (selection: Davis; exposure: Kettunen; outcome: UK Biobank).
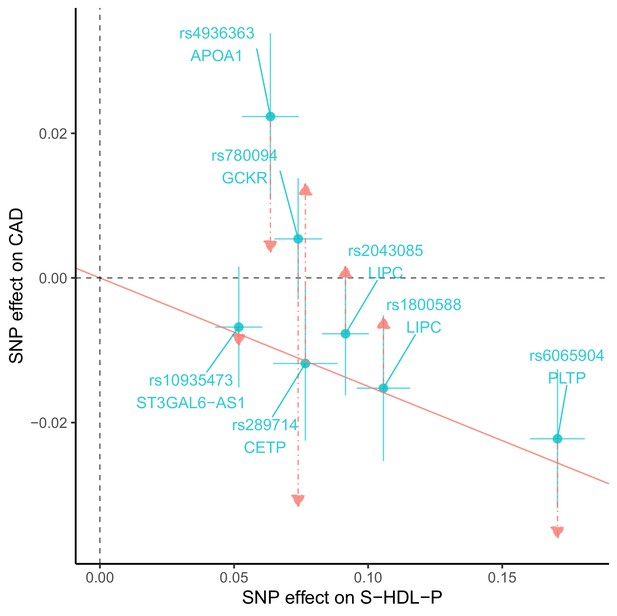
Scatter-plots for S-HDL-P with the effects on CAD adjusted for LDL-C and TG.
Red lines correspond the fitted effects of S-HDL-P in multivariable MR.
Tables
Information about the GWAS summary datasets used in this article.
The columns are the phenotypes reported by the GWAS studies, the consortium or name of the first author of the publication, PubMed ID, population, sample size, other GWAS datasets with other lapping sample, and URLs we used to download the datasets.
| Phenotype | Dataset name | PubMed ID | Population | Sample size | Sample overlap with other datasets | URL to summary dataset |
|---|---|---|---|---|---|---|
| Traditional lipid traits | GERA | 29507422 Hoffmann et al., 2018 | Multi-ethnic | 94,674 | ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/ | |
| GLGC | 24097068 Willer et al., 2013 | European | 188,578 | Kettunen, CARDIoGRAMplusC4D | http://csg.sph.umich.edu/abecasis/public/lipids2013/ | |
| Lipoprotein subfraction traits | Davis | 29084231 Davis et al., 2017 | Finnish | 8372 | http://csg.sph.umich.edu/boehnke/public/metsim-2017-lipoproteins/ | |
| Kettunen | 27005778 Kettunen et al., 2016 | European | 24,925 | GLGC, CARDIoGRAMplusC4D | http://www.computationalmedicine.fi/data#NMR_GWAS | |
| Heart disease traits | CARDIoGRAMplusC4D (CAD) | 26343387 Nikpay et al., 2015 | Mostly European | 185,000 | GLGC, Kettunen | http://www.cardiogramplusc4d.org/data-downloads/ |
| CARDIoGRAMplusC4D + UK Biobank (CAD) | 28714975 Nelson et al., 2017 | Mostly European | ||||
| UK Biobank (MI) | Interim round two release Abbott et al., 2018 | European | 360,420 | http://www.nealelab.is/uk-biobank/ |
Results of some multivariable Mendelian randomization analyses.
Each row in the table corresponds to a multivariable MR analysis with traditional lipid profile and the specified lipoprotein subfraction or particle size trait. Reported numbers are the point estimates and 95% confidence intervals of the exposure effect.
| Trait | Effect of TG | Effect of LDL-C | Effect of HDL-C | Effect of subfraction/particle size |
|---|---|---|---|---|
| None | 0.19 [0.09,0.29] | 0.38 [0.33,0.44] | −0.053 [-0.13,0.03] | |
| M-HDL-P | 0.37 [0.22,0.52] | 0.39 [0.32,0.45] | 0.30 [0.08,0.52] | −0.69 [-1.09,–0.3] |
| S-HDL-P | 0.23 [0.12,0.33] | 0.45 [0.38,0.52] | −0.11 [-0.2,–0.02] | −0.33 [-0.52,–0.15] |
| HDL-D | 0.11 [0.00,0.22] | 0.42 [0.36,0.49] | −0.44 [-0.69,–0.2] | 0.33 [0.11,0.56] |
| Effect of TG | Effect of ApoB | Effect of ApoA1 | Effect of Subfraction/Particle size | |
| None | 0.05 [-0.05,0.14] | 0.49 [0.38,0.60] | −0.095 [-0.21,0.02] | |
| M-HDL-P | −0.00 [-0.18,0.17] | 0.50 [0.31,0.69] | 0.13 [-0.06,0.32] | −0.47 [-0.80,–0.15] |
| S-HDL-P | 0.07 [-0.03,0.17] | 0.53 [0.41,0.65] | −0.13 [-0.25,–0.02] | −0.24 [-0.40,–0.08] |
| HDL-D | 0.06 [-0.04,0.15] | 0.61 [0.47,0.76] | −0.46 [-0.73,–0.19] | 0.30 [0.08,0.52] |
All 82 traits included in this study and whether they are measured in the Kettunen and Davis GWAS (NA means not available).
| Trait | Description | Kettunen | Davis |
|---|---|---|---|
| VLDL traits and total triglycerides | |||
| TG | Total triglycerides | ||
| VLDL-D | VLDL diameter | ||
| XS-VLDL-L | Total lipids in very small VLDL | NA | |
| XS-VLDL-P | Concentration of very small VLDL particles | ||
| XS-VLDL-PL | Phospholipids in very small VLDL | ||
| XS-VLDL-TG | Triglycerides in very small VLDL | ||
| S-VLDL-C | Total cholesterol in small VLDL | ||
| S-VLDL-FC | Free cholesterol in small VLDL | ||
| S-VLDL-L | Total lipids in small VLDL | NA | |
| S-VLDL-P | Concentration of small VLDL particles | ||
| S-VLDL-PL | Phospholipids in small VLDL | ||
| S-VLDL-TG | Triglycerides in small VLDL | ||
| M-VLDL-C | Total cholesterol in medium VLDL | ||
| M-VLDL-CE | Cholesterol esters in medium VLDL | ||
| M-VLDL-FC | Free cholesterol in medium VLDL | ||
| M-VLDL-L | Total lipids in medium VLDL | NA | |
| M-VLDL-P | Concentration of medium VLDL particles | ||
| M-VLDL-PL | Phospholipids in medium VLDL | ||
| M-VLDL-TG | Triglycerides in medium VLDL | ||
| L-VLDL-C | Total cholesterol in large VLDL | ||
| L-VLDL-CE | Cholesterol esters in large VLDL | ||
| L-VLDL-FC | Free cholesterol in large VLDL | ||
| L-VLDL-L | Total lipids in large VLDL | NA | |
| L-VLDL-P | Concentration of large VLDL particles | ||
| L-VLDL-PL | Phospholipids in large VLDL | ||
| L-VLDL-TG | Triglycerides in large VLDL | ||
| XL-VLDL-L | Total lipids in very large VLDL | NA | |
| XL-VLDL-P | Concentration of very large VLDL particles | ||
| XL-VLDL-PL | Phospholipids in very large VLDL | ||
| XL-VLDL-TG | Triglycerides in very large VLDL | ||
| XXL-VLDL-L | Total lipids in chylomicrons and extremely very large VLDL | NA | |
| XXL-VLDL-P | Concentration of chylomicrons and extremely very large VLDL particles | ||
| XXL-VLDL-PL | Phospholipids in chylomicrons and extremely very large | ||
| XXL-VLDL-TG | Triglycerides in chylomicrons and extremely very large | ||
| LDL and IDL traits | |||
| LDL-C | Total cholesterol in LDL | ||
| ApoB | Apolipoprotein B | ||
| LDL-D | LDL diameter | ||
| S-LDL-C | Total cholesterol in small LDL | ||
| S-LDL-L | Total lipids in small LDL | NA | |
| S-LDL-P | Phospholipids in small LDL | ||
| M-LDL-C | Total cholesterol in medium LDL | ||
| M-LDL-CE | Cholesterol esters in medium LDL | ||
| M-LDL-L | Total lipids in medium LDL | NA | |
| M-LDL-P | Concentration of medium LDL particles | ||
| M-LDL-PL | Phospholipids in medium LDL | ||
| L-LDL-C | Total cholesterol in large LDL | ||
| L-LDL-CE | Cholesterol esters in large LDL | ||
| L-LDL-FC | Free cholesterol in large LDL | ||
| L-LDL-L | Total lipids in large LDL | NA | |
| L-LDL-P | Concentration of large LDL particles | ||
| L-LDL-PL | Phospholipids in large LDL | ||
| IDL-C | Total cholesterol in IDL | ||
| IDL-FC | Free cholesterol in IDL | ||
| IDL-L | Total lipids in IDL | NA | |
| IDL-P | Concentration of IDL particles | ||
| IDL-PL | Phospholipids in IDL | ||
| IDL-TG | Triglycerides in IDL | ||
| HDL traits | |||
| HDL-C | Total cholesterol in HDL | ||
| ApoA1 | Apolipoprotein A1 | ||
| HDL-D | HDL diameter | ||
| S-HDL-L | Total lipids in small HDL | NA | |
| S-HDL-P | Concentration of small HDL particles | ||
| S-HDL-TG | Triglycerides in small HDL | ||
| M-HDL-C | Total cholesterol in medium HDL | ||
| M-HDL-CE | Cholesterol esters in medium HDL | ||
| M-HDL-FC | Free cholesterol in medium HDL | ||
| M-HDL-L | Total lipids in medium HDL | NA | |
| M-HDL-P | Concentration of medium HDL particles | ||
| M-HDL-PL | Phospholipids in medium HDL | ||
| L-HDL-C | Total cholesterol in large HDL | ||
| L-HDL-CE | Cholesterol esters in large HDL | ||
| L-HDL-FC | Free cholesterol in large HDL | ||
| L-HDL-L | Total lipids in large HDL | NA | |
| L-HDL-P | Concentration of large HDL particles | ||
| L-HDL-PL | Phospholipids in large HDL | ||
| XL-HDL-C | Total cholesterol in very large HDL | ||
| XL-HDL-CE | Cholesterol esters in very large HDL | ||
| XL-HDL-FC | Free cholesterol in very large HDL | ||
| XL-HDL-L | Total lipids in very large HDL | NA | |
| XL-HDL-P | Concentration of very large HDL particles | ||
| XL-HDL-PL | Phospholipids in very large HDL | ||
| XL-HDL-TG | Triglycerides in very large HDL | ||
Estimated genetic correlation (standard error) of the lipoprotein subfractions with the traditional lipid risk factors using the Davis GWAS.
Bolded estimates are above 0.8 and the corresponding traits were removed in phenotypic screening.
| Trait | ApoA1 | ApoB | HDL-C | LDL-C | TG |
|---|---|---|---|---|---|
| S-HDL-L | 0.31 (0.28) | 0.34 (0.25) | 0.13 (0.26) | 0.27 (0.3) | 0.2 (0.22) |
| S-HDL-P | 0.36 (0.24) | 0.27 (0.22) | −0.01 (0.22) | 0.1 (0.31) | 0.48 (0.17) |
| S-HDL-TG | −0.13 (0.25) | 0.77 (0.13) | −0.66 (0.15) | 0.13 (0.28) | 1.03 (0.07) |
| M-HDL-C | 0.65 (0.14) | −0.18 (0.2) | 0.81 (0.09) | −0.09 (0.25) | −0.34 (0.17) |
| M-HDL-CE | 0.68 (0.14) | −0.23 (0.21) | 0.57 (0.12) | −0.24 (0.24) | −0.32 (0.18) |
| M-HDL-FC | 0.67 (0.12) | −0.08 (0.21) | 0.83 (0.08) | 0.04 (0.24) | −0.28 (0.18) |
| M-HDL-L | 0.71 (0.15) | 0.02 (0.27) | 0.52 (0.17) | −0.03 (0.29) | −0.19 (0.25) |
| M-HDL-P | 0.75 (0.12) | 0.15 (0.23) | 0.46 (0.14) | 0.08 (0.26) | 0 (0.19) |
| M-HDL-PL | 0.69 (0.13) | 0.04 (0.22) | 0.65 (0.11) | 0.02 (0.25) | −0.04 (0.19) |
| L-HDL-C | 0.76 (0.11) | −0.42 (0.13) | 0.95 (0.02) | −0.1 (0.18) | −0.62 (0.09) |
| L-HDL-CE | 0.82 (0.1) | −0.4 (0.12) | 0.93 (0.04) | −0.16 (0.17) | −0.62 (0.09) |
| L-HDL-FC | 0.66 (0.12) | −0.46 (0.13) | 0.92 (0.03) | −0.13 (0.18) | −0.7 (0.08) |
| L-HDL-L | 0.81 (0.11) | −0.29 (0.15) | 0.74 (0.07) | −0.15 (0.18) | −0.56 (0.12) |
| L-HDL-P | 0.79 (0.09) | −0.35 (0.13) | 0.82 (0.05) | −0.12 (0.16) | −0.61 (0.09) |
| L-HDL-PL | 0.77 (0.09) | −0.34 (0.13) | 0.79 (0.05) | −0.12 (0.17) | −0.61 (0.09) |
| XL-HDL-C | 0.75 (0.16) | −0.25 (0.19) | 0.9 (0.1) | 0.4 (0.27) | −0.63 (0.13) |
| XL-HDL-CE | 0.82 (0.16) | −0.17 (0.19) | 0.82 (0.09) | 0.41 (0.27) | −0.54 (0.12) |
| XL-HDL-FC | 0.72 (0.14) | −0.37 (0.18) | 0.94 (0.08) | 0.17 (0.23) | −0.71 (0.11) |
| XL-HDL-L | 0.93 (0.16) | −0.08 (0.25) | 0.68 (0.14) | 0.1 (0.27) | −0.35 (0.2) |
| XL-HDL-P | 0.81 (0.13) | −0.32 (0.16) | 0.86 (0.08) | 0.17 (0.21) | −0.69 (0.11) |
| XL-HDL-PL | 0.76 (0.12) | −0.41 (0.15) | 0.83 (0.07) | −0.09 (0.18) | −0.7 (0.09) |
| XL-HDL-TG | 0.72 (0.13) | 0.49 (0.17) | 0.33 (0.13) | 0.13 (0.26) | 0.3 (0.15) |
| HDL-D | 0.7 (0.11) | −0.36 (0.13) | 0.8 (0.06) | −0.08 (0.17) | −0.64 (0.09) |
| IDL-C | 0.38 (0.21) | 0.58 (0.19) | 0.07 (0.19) | 0.8 (0.14) | 0.39 (0.17) |
| IDL-FC | 0.23 (0.2) | 0.78 (0.12) | −0.05 (0.17) | 0.61 (0.19) | 0.44 (0.15) |
| IDL-L | 0.38 (0.23) | 0.65 (0.18) | 0.05 (0.2) | 0.64 (0.2) | 0.47 (0.17) |
| IDL-P | 0.31 (0.2) | 0.66 (0.14) | −0.04 (0.17) | 0.82 (0.13) | 0.49 (0.14) |
| IDL-PL | 0.25 (0.23) | 0.83 (0.1) | −0.12 (0.19) | 0.7 (0.19) | 0.64 (0.15) |
| IDL-TG | 0.22 (0.18) | 0.82 (0.08) | −0.2 (0.13) | 0.56 (0.15) | 0.67 (0.08) |
| S-LDL-C | 0.11 (0.28) | 0.66 (0.18) | −0.16 (0.22) | 0.44 (0.34) | 0.58 (0.14) |
| S-LDL-L | 0.26 (0.23) | 0.66 (0.17) | −0.06 (0.21) | 0.62 (0.21) | 0.58 (0.13) |
| S-LDL-P | 0.34 (0.2) | 0.68 (0.15) | −0.02 (0.19) | 0.63 (0.18) | 0.58 (0.13) |
| M-LDL-C | 0.15 (0.26) | 0.63 (0.18) | 0.22 (0.22) | 0.87 (0.08) | 0.13 (0.23) |
| M-LDL-CE | 0.3 (0.23) | 0.61 (0.2) | 0.05 (0.21) | 0.65 (0.2) | 0.45 (0.16) |
| M-LDL-L | 0.29 (0.22) | 0.63 (0.18) | 0.01 (0.21) | 0.66 (0.19) | 0.5 (0.15) |
| M-LDL-P | 0.29 (0.23) | 0.63 (0.18) | −0.01 (0.21) | 0.65 (0.21) | 0.51 (0.15) |
| M-LDL-PL | 0.2 (0.24) | 0.69 (0.16) | 0.11 (0.2) | 0.89 (0.06) | 0.18 (0.22) |
| L-LDL-C | 0.25 (0.24) | 0.58 (0.21) | 0.25 (0.22) | 0.68 (0.19) | 0.23 (0.21) |
| L-LDL-CE | 0.3 (0.23) | 0.58 (0.22) | 0.05 (0.21) | 0.65 (0.21) | 0.41 (0.17) |
| L-LDL-FC | 0.31 (0.24) | 0.57 (0.22) | 0.33 (0.23) | 0.7 (0.18) | 0.13 (0.23) |
| L-LDL-L | 0.31 (0.23) | 0.61 (0.2) | 0.04 (0.21) | 0.65 (0.21) | 0.44 (0.17) |
| L-LDL-P | 0.31 (0.23) | 0.63 (0.19) | 0.02 (0.21) | 0.65 (0.21) | 0.47 (0.16) |
| L-LDL-PL | 0.27 (0.25) | 0.61 (0.2) | 0.24 (0.22) | 0.67 (0.2) | 0.27 (0.2) |
| LDL-D | −0.33 (0.25) | −0.22 (0.23) | −0.15 (0.21) | −0.15 (0.29) | −0.37 (0.16) |
| XS-VLDL-L | 0.25 (0.23) | 0.8 (0.08) | −0.2 (0.17) | 0.61 (0.14) | 0.73 (0.09) |
| XS-VLDL-P | 0.17 (0.18) | 0.83 (0.07) | −0.26 (0.13) | 0.57 (0.13) | 0.71 (0.07) |
| XS-VLDL-PL | 0.21 (0.19) | 0.78 (0.09) | −0.15 (0.15) | 0.74 (0.14) | 0.57 (0.11) |
| XS-VLDL-TG | 0.06 (0.18) | 0.83 (0.08) | −0.37 (0.11) | 0.56 (0.13) | 0.85 (0.04) |
| S-VLDL-FC | −0.08 (0.2) | 0.94 (0.05) | −0.49 (0.12) | 0.59 (0.12) | 0.92 (0.03) |
| S-VLDL-L | −0.12 (0.24) | 0.7 (0.08) | −0.46 (0.15) | 0.5 (0.14) | 0.8 (0.05) |
| S-VLDL-P | −0.09 (0.19) | 0.78 (0.07) | −0.48 (0.11) | 0.5 (0.14) | 0.95 (0.02) |
| S-VLDL-PL | −0.03 (0.2) | 0.82 (0.08) | −0.43 (0.12) | 0.44 (0.17) | 0.92 (0.03) |
| S-VLDL-TG | −0.1 (0.2) | 0.9 (0.08) | −0.49 (0.11) | 0.49 (0.15) | 0.98 (0.01) |
| S-VLDL-C | 0.01 (0.2) | 0.9 (0.06) | −0.39 (0.13) | 0.61 (0.15) | 0.89 (0.05) |
| M-VLDL-C | −0.01 (0.2) | 0.8 (0.09) | −0.47 (0.12) | 0.41 (0.18) | 0.95 (0.02) |
| M-VLDL-CE | 0.01 (0.19) | 0.78 (0.08) | −0.43 (0.12) | 0.5 (0.15) | 0.9 (0.03) |
| M-VLDL-FC | 0 (0.21) | 0.83 (0.09) | −0.48 (0.12) | 0.4 (0.18) | 0.97 (0.01) |
| M-VLDL-L | −0.1 (0.24) | 0.66 (0.11) | −0.48 (0.15) | 0.4 (0.18) | 0.8 (0.05) |
| M-VLDL-P | −0.06 (0.19) | 0.78 (0.1) | −0.46 (0.12) | 0.43 (0.16) | 0.98 (0.02) |
| M-VLDL-PL | 0.03 (0.21) | 0.85 (0.09) | −0.48 (0.12) | 0.4 (0.18) | 0.98 (0.01) |
| M-VLDL-TG | −0.02 (0.21) | 0.82 (0.11) | −0.5 (0.13) | 0.33 (0.19) | 0.98 (0.02) |
| L-VLDL-C | −0.05 (0.2) | 0.83 (0.12) | −0.55 (0.12) | 0.36 (0.19) | 1 (0.02) |
| L-VLDL-CE | 0 (0.19) | 0.78 (0.12) | −0.44 (0.12) | 0.43 (0.19) | 0.93 (0.03) |
| L-VLDL-FC | −0.03 (0.2) | 0.84 (0.12) | −0.53 (0.13) | 0.36 (0.19) | 1 (0.02) |
| L-VLDL-L | −0.06 (0.24) | 0.66 (0.14) | −0.47 (0.16) | 0.36 (0.2) | 0.86 (0.05) |
| L-VLDL-P | −0.02 (0.21) | 0.72 (0.12) | −0.44 (0.13) | 0.33 (0.18) | 0.98 (0.02) |
| L-VLDL-PL | 0.01 (0.21) | 0.86 (0.12) | −0.53 (0.13) | 0.3 (0.2) | 1.04 (0.03) |
| L-VLDL-TG | −0.06 (0.21) | 0.78 (0.12) | −0.54 (0.13) | 0.26 (0.19) | 1 (0.02) |
| XL-VLDL-L | −0.08 (0.24) | 0.7 (0.15) | −0.52 (0.16) | 0.43 (0.2) | 0.85 (0.05) |
| XL-VLDL-P | −0.06 (0.2) | 0.76 (0.12) | −0.48 (0.13) | 0.44 (0.18) | 0.95 (0.03) |
| XL-VLDL-PL | −0.09 (0.23) | 0.82 (0.13) | −0.62 (0.15) | 0.32 (0.21) | 1.06 (0.04) |
| XL-VLDL-TG | −0.14 (0.21) | 0.86 (0.13) | −0.65 (0.13) | 0.34 (0.19) | 1.03 (0.04) |
| XXL-VLDL-L | −0.07 (0.25) | 0.65 (0.16) | −0.5 (0.17) | 0.38 (0.22) | 0.83 (0.06) |
| XXL-VLDL-P | 0.17 (0.2) | 0.72 (0.15) | −0.3 (0.15) | 0.39 (0.21) | 0.86 (0.07) |
| XXL-VLDL-PL | −0.3 (0.24) | 0.66 (0.17) | −0.8 (0.16) | 0.22 (0.21) | 1.06 (0.06) |
| XXL-VLDL-TG | −0.21 (0.25) | 0.64 (0.16) | −0.7 (0.15) | 0.22 (0.22) | 1.08 (0.05) |
| VLDL-D | −0.22 (0.2) | 0.55 (0.14) | −0.53 (0.12) | 0.12 (0.19) | 0.86 (0.04) |
Three-sample Mendelian randomization designs.
| MR design | Selection | Exposure | Outcome | Reported in |
|---|---|---|---|---|
| Univariable(traditional selection) | GERA | Davis | CARDIoGRAMplusC4D | Appendix 3—table 2–4 |
| GERA | Davis | UK Biobank | Appendix 3—table 2–4 | |
| GERA | Kettunen | UK Biobank | Appendix 3—table 2–4 | |
| GLGC | Davis | UK Biobank | Appendix 3—table 2–4 | |
| Univariable(subfraction selection) | Davis | Kettunen | UK Biobank | Figure 2; Appendix 3—figure 1 and Appendix 3—table 2–4 |
| Kettunen | Davis | UK Biobank | Appendix 3—figure 1 and Appendix 3—table 2–4 | |
| Multivariable | Davis, GERA | Kettunen, GLGC | CARDIoGRAMplusC4D+ UK Biobank | Figure 2, Table 2; Appendix 3—figure 1 and Appendix 3—table 2–4 |
Mendelian randomization results using all selected SNPs (univariable MR using RAPS and multivariable MR using GRAPPLE).
| Method: RAPS/GRAPPLE + All SNPs | ||||||||
|---|---|---|---|---|---|---|---|---|
| Screening | GERA | GERA | GERA | GLGC | Davis | Kettunen | GERA + Davis | GERA + Davis |
| Exposure | Davis | Davis | Kettunen | Davis | Kettunen | Davis | GLGC + Kettunen | GLGC + Kettunen |
| Outcome | CAD | UKB | UKB | UKB | UKB | UKB | CAD + UKB | CAD + UKB |
| Adjusted | HDL-C + LDL-C + TG | ApoA1 + ApoB + TG | ||||||
| VLDL traits | ||||||||
| TG | NA | NA | NA | NA | ||||
| VLDL-D | 0.028 (0.074) | 0.072 (0.073) | 0.116 (0.065) | |||||
| XS-VLDL-L | NA | NA | NA | NA | 0.132 (0.119) | 0.084 (0.141) | ||
| XS-VLDL-P | 0.118 (0.125) | 0.061 (0.158) | ||||||
| XS-VLDL-PL | 0.159 (0.12) | 0.253 (0.135) | ||||||
| XS-VLDL-TG | -0.157 (0.187) | -0.248 (0.15) | ||||||
| S-VLDL-C | NA | NA | ||||||
| S-VLDL-FC | ||||||||
| S-VLDL-L | NA | NA | NA | NA | ||||
| S-VLDL-P | ||||||||
| S-VLDL-PL | ||||||||
| S-VLDL-TG | NaN | |||||||
| M-VLDL-C | 0.304 (0.078) | 0.312 (0.079) | ||||||
| M-VLDL-CE | ||||||||
| M-VLDL-FC | ||||||||
| M-VLDL-L | NA | NA | 0.311 (0.079) | NA | NA | |||
| M-VLDL-P | ||||||||
| M-VLDL-PL | ||||||||
| M-VLDL-TG | -0.806 (0.455) | |||||||
| L-VLDL-C | -0.609 (0.337) | |||||||
| L-VLDL-CE | 0.282 (0.072) | |||||||
| L-VLDL-FC | ||||||||
| L-VLDL-L | NA | NA | NA | NA | -0.532 (0.278) | |||
| L-VLDL-P | -1.357 (0.344) | |||||||
| L-VLDL-PL | 0.346 (0.089) | 0.397 (0.101) | ||||||
| L-VLDL-TG | ||||||||
| XL-VLDL-L | NA | NA | NA | NA | ||||
| XL-VLDL-P | ||||||||
| XL-VLDL-PL | ||||||||
| XL-VLDL-TG | ||||||||
| XXL-VLDL-L | NA | NA | NA | NA | ||||
| XXL-VLDL-P | ||||||||
| XXL-VLDL-PL | ||||||||
| XXL-VLDL-TG | ||||||||
| IDL/LDL traits | ||||||||
| LDL-C | NA | 0.319 (0.182) | ||||||
| ApoB | NA | |||||||
| LDL-D | 0.271 (0.215) | 0.452 (0.299) | 0.831 (0.684) | 0.119 (0.071) | ||||
| S-LDL-C | -0.282 (0.152) | 0.08 (0.238) | ||||||
| S-LDL-L | NA | NA | NA | NA | -0.251 (0.145) | -0.005 (0.29) | ||
| S-LDL-P | -0.266 (0.151) | -0.362 (0.596) | ||||||
| M-LDL-C | -0.271 (0.162) | -0.169 (0.909) | ||||||
| M-LDL-CE | -0.088 (0.188) | NaN | ||||||
| M-LDL-L | NA | NA | NA | NA | -0.069 (0.191) | NaN | ||
| M-LDL-P | -0.179 (0.174) | 0.432 (0.31) | ||||||
| M-LDL-PL | -0.566 (0.839) | |||||||
| L-LDL-C | -0.059 (0.261) | NaN | ||||||
| L-LDL-CE | 0.116 (0.321) | |||||||
| L-LDL-FC | 0.223 (0.315) | NaN | ||||||
| L-LDL-L | NA | NA | NA | NA | 0.167 (0.273) | NaN | ||
| L-LDL-P | 0.084 (0.213) | NaN | ||||||
| L-LDL-PL | -0.036 (0.195) | NaN | ||||||
| IDL-C | 0.192 (0.229) | 0.769 (0.501) | ||||||
| IDL-FC | 0.19 (0.156) | |||||||
| IDL-L | NA | NA | NA | NA | 0.148 (0.175) | 0.444 (0.328) | ||
| IDL-P | 0.153 (0.148) | 0.292 (0.173) | ||||||
| IDL-PL | 0.153 (0.18) | |||||||
| IDL-TG | 0.11 (0.103) | 0.047 (0.135) | ||||||
| HDL traits | ||||||||
| HDL-C | -0.045 (0.059) | NA | NaN | |||||
| ApoA1 | 0.023 (0.058) | 0.075 (0.064) | -0.13 (0.068) | -0.481 (0.271) | NA | |||
| HDL-D | -0.008 (0.027) | 0.004 (0.046) | -0.092 (0.048) | 0.067 (0.045) | 0.007 (0.041) | |||
| S-HDL-L | NA | NA | -0.098 (0.095) | NA | -0.037 (0.085) | NA | ||
| S-HDL-P | -0.13 (0.092) | -0.053 (0.081) | -0.08 (0.094) | |||||
| S-HDL-TG | 0.253 (0.637) | -0.044 (0.466) | ||||||
| M-HDL-C | ||||||||
| M-HDL-CE | ||||||||
| M-HDL-FC | ||||||||
| M-HDL-L | NA | NA | NA | NA | ||||
| M-HDL-P | -0.307 (0.079) | |||||||
| M-HDL-PL | ||||||||
| L-HDL-C | -0.049 (0.045) | |||||||
| L-HDL-CE | ||||||||
| L-HDL-FC | -0.03 (0.047) | |||||||
| L-HDL-L | NA | NA | NA | NA | ||||
| L-HDL-P | -0.083 (0.05) | |||||||
| L-HDL-PL | ||||||||
| XL-HDL-C | 0.055 (0.046) | -0.013 (0.068) | 0.11 (0.066) | 0.064 (0.073) | 0.048 (0.069) | 0.112 (0.068) | ||
| XL-HDL-CE | 0.064 (0.044) | 0.006 (0.066) | 0.08 (0.07) | 0.057 (0.068) | 0.046 (0.075) | |||
| XL-HDL-FC | 0.009 (0.039) | -0.05 (0.059) | 0.066 (0.058) | -0.026 (0.067) | 0.102 (0.06) | 0.049 (0.066) | ||
| XL-HDL-L | NA | NA | 0.073 (0.055) | NA | 0.038 (0.058) | NA | ||
| XL-HDL-P | 0.038 (0.033) | -0.022 (0.049) | 0.112 (0.057) | 0.017 (0.056) | 0.083 (0.055) | 0.023 (0.057) | ||
| XL-HDL-PL | 0.029 (0.031) | -0.031 (0.046) | 0.037 (0.05) | 0.005 (0.055) | 0.038 (0.052) | 0.013 (0.046) | ||
| XL-HDL-TG | 0.165 (0.086) | |||||||
Mendelian randomization results using genome-wide significant SNPs and inverse variance weighted (IVW) estimator.
| Method: IVW + Significant SNPs | ||||||
|---|---|---|---|---|---|---|
| Selection | GERA | GERA | GERA | GLGC | Davis | Kettunen |
| Exposure | Davis | Davis | Kettunen | Davis | Kettunen | Davis |
| Outcome | CAD | UKB | UKB | UKB | UKB | UKB |
| VLDL traits | ||||||
| TG | NA | NA | ||||
| VLDL-D | 0.044 (0.06) | 0.052 (0.09) | 0.038 (0.102) | 0.118 (0.091) | -0.083 (0.16) | -0.083 (0.138) |
| XS-VLDL-L | NA | NA | NA | NA | ||
| XS-VLDL-P | ||||||
| XS-VLDL-PL | ||||||
| XS-VLDL-TG | ||||||
| S-VLDL-C | NA | NA | ||||
| S-VLDL-FC | ||||||
| S-VLDL-L | NA | NA | NA | NA | ||
| S-VLDL-P | ||||||
| S-VLDL-PL | ||||||
| S-VLDL-TG | ||||||
| M-VLDL-C | ||||||
| M-VLDL-CE | ||||||
| M-VLDL-FC | ||||||
| M-VLDL-L | NA | NA | NA | NA | ||
| M-VLDL-P | ||||||
| M-VLDL-PL | ||||||
| M-VLDL-TG | ||||||
| L-VLDL-C | 0.108 (0.223) | |||||
| L-VLDL-CE | 0.182 (0.187) | |||||
| L-VLDL-FC | ||||||
| L-VLDL-L | NA | NA | NA | 0.125 (0.232) | NA | |
| L-VLDL-P | ||||||
| L-VLDL-PL | ||||||
| L-VLDL-TG | ||||||
| XL-VLDL-L | NA | NA | NA | 0.365 (0.286) | NA | |
| XL-VLDL-P | 0.346 (0.28) | |||||
| XL-VLDL-PL | 0.333 (0.265) | |||||
| XL-VLDL-TG | 0.323 (0.272) | |||||
| XXL-VLDL-L | NA | NA | NA | -0.165 (0.425) | NA | |
| XXL-VLDL-P | -0.12 (0.389) | 0.006 (0.153) | ||||
| XXL-VLDL-PL | -0.145 (0.395) | 0.071 (0.191) | ||||
| XXL-VLDL-TG | 0.09 (0.36) | 0.349 (0.303) | ||||
| IDL/LDL traits | ||||||
| LDL-C | ||||||
| ApoB | ||||||
| LDL-D | 0.217 (0.151) | 0.271 (0.143) | ||||
| S-LDL-C | ||||||
| S-LDL-L | NA | NA | NA | NA | ||
| S-LDL-P | ||||||
| M-LDL-C | ||||||
| M-LDL-CE | ||||||
| M-LDL-L | NA | NA | NA | NA | ||
| M-LDL-P | ||||||
| M-LDL-PL | ||||||
| L-LDL-C | ||||||
| L-LDL-CE | ||||||
| L-LDL-FC | ||||||
| L-LDL-L | NA | NA | NA | NA | ||
| L-LDL-P | ||||||
| L-LDL-PL | ||||||
| IDL-C | ||||||
| IDL-FC | ||||||
| IDL-L | NA | NA | NA | NA | ||
| IDL-P | ||||||
| IDL-PL | ||||||
| IDL-TG | ||||||
| HDL traits | ||||||
| HDL-C | -0.085 (0.044) | -0.146 (0.085) | -0.082 (0.159) | -0.015 (0.109) | ||
| ApoA1 | -0.072 (0.054) | -0.036 (0.09) | 0.001 (0.192) | 0.066 (0.158) | ||
| HDL-D | -0.027 (0.042) | -0.071 (0.058) | -0.052 (0.073) | -0.092 (0.063) | 0.073 (0.098) | 0.074 (0.074) |
| S-HDL-L | NA | NA | -0.064 (0.148) | NA | -0.033 (0.092) | NA |
| S-HDL-P | -0.117 (0.087) | -0.172 (0.116) | -0.13 (0.146) | -0.033 (0.09) | -0.115 (0.174) | |
| S-HDL-TG | 0.286 (0.17) | |||||
| M-HDL-C | ||||||
| M-HDL-CE | -0.341 (0.238) | |||||
| M-HDL-FC | -0.288 (0.218) | -0.278 (0.144) | ||||
| M-HDL-L | NA | NA | NA | NA | ||
| M-HDL-P | -0.386 (0.307) | -0.18 (0.118) | ||||
| M-HDL-PL | -0.419 (0.301) | |||||
| L-HDL-C | -0.047 (0.044) | -0.097 (0.059) | -0.124 (0.08) | 0.022 (0.106) | 0.021 (0.105) | |
| L-HDL-CE | -0.049 (0.044) | -0.098 (0.059) | -0.12 (0.079) | 0.023 (0.112) | 0.004 (0.106) | |
| L-HDL-FC | -0.044 (0.046) | -0.094 (0.062) | -0.106 (0.082) | -0.127 (0.067) | 0.038 (0.103) | 0.017 (0.109) |
| L-HDL-L | NA | NA | -0.106 (0.077) | NA | 0.034 (0.102) | NA |
| L-HDL-P | -0.045 (0.043) | -0.097 (0.058) | -0.102 (0.077) | 0.009 (0.111) | 0.025 (0.11) | |
| L-HDL-PL | -0.054 (0.044) | -0.11 (0.06) | -0.115 (0.079) | 0.006 (0.115) | 0.016 (0.115) | |
| XL-HDL-C | 0.03 (0.06) | -0.012 (0.084) | 0.014 (0.099) | -0.05 (0.088) | -0.015 (0.165) | 0.161 (0.101) |
| XL-HDL-CE | 0.03 (0.059) | -0.009 (0.081) | 0.025 (0.098) | -0.042 (0.086) | -0.001 (0.166) | |
| XL-HDL-FC | -0.003 (0.056) | -0.05 (0.076) | -0.001 (0.089) | -0.077 (0.081) | 0.072 (0.11) | 0.057 (0.092) |
| XL-HDL-L | NA | NA | 0.001 (0.085) | NA | -0.009 (0.138) | NA |
| XL-HDL-P | 0.015 (0.049) | -0.021 (0.067) | 0.013 (0.088) | -0.042 (0.071) | 0.103 (0.1) | 0.135 (0.093) |
| XL-HDL-PL | 0 (0.047) | -0.037 (0.065) | -0.026 (0.079) | -0.055 (0.069) | 0.081 (0.088) | 0.071 (0.069) |
| XL-HDL-TG | 0.103 (0.059) | 0.14 (0.075) | ||||
Mendelian randomization results using genome-wide significant SNPs and the weighted median estimator.
| Method: Weighted median + Significant SNPs | ||||||
|---|---|---|---|---|---|---|
| Selection | GERA | GERA | GERA | GLGC | Davis | Kettunen |
| Exposure | Davis | Davis | Kettunen | Davis | Kettunen | Davis |
| Outcome | CAD | UKB | UKB | UKB | UKB | UKB |
| VLDL traits | ||||||
| TG | 0.042 (0.055) | NA | NA | |||
| VLDL-D | -0.098 (0.052) | 0.039 (0.095) | 0.057 (0.11) | 0.058 (0.093) | -0.107 (0.099) | -0.052 (0.115) |
| XS-VLDL-L | NA | NA | NA | NA | ||
| XS-VLDL-P | ||||||
| XS-VLDL-PL | ||||||
| XS-VLDL-TG | ||||||
| S-VLDL-C | NA | NA | ||||
| S-VLDL-FC | ||||||
| S-VLDL-L | NA | NA | NA | NA | ||
| S-VLDL-P | ||||||
| S-VLDL-PL | ||||||
| S-VLDL-TG | ||||||
| M-VLDL-C | ||||||
| M-VLDL-CE | ||||||
| M-VLDL-FC | 0.081 (0.058) | |||||
| M-VLDL-L | NA | NA | NA | NA | ||
| M-VLDL-P | 0.047 (0.06) | |||||
| M-VLDL-PL | 0.103 (0.056) | |||||
| M-VLDL-TG | -0.005 (0.06) | |||||
| L-VLDL-C | 0.109 (0.068) | |||||
| L-VLDL-CE | ||||||
| L-VLDL-FC | 0.045 (0.065) | |||||
| L-VLDL-L | NA | NA | NA | NA | ||
| L-VLDL-P | 0.041 (0.064) | |||||
| L-VLDL-PL | 0.08 (0.063) | |||||
| L-VLDL-TG | -0.008 (0.061) | |||||
| XL-VLDL-L | NA | NA | NA | NA | NA | |
| XL-VLDL-P | -0.026 (0.063) | NA | ||||
| XL-VLDL-PL | -0.006 (0.067) | NA | ||||
| XL-VLDL-TG | -0.026 (0.064) | NA | ||||
| XXL-VLDL-L | NA | NA | NA | -0.156 (0.22) | NA | |
| XXL-VLDL-P | 0.091 (0.071) | -0.104 (0.173) | 0.185 (0.098) | |||
| XXL-VLDL-PL | 0.153 (0.082) | -0.139 (0.178) | 0.126 (0.124) | |||
| XXL-VLDL-TG | 0.126 (0.078) | 0.227 (0.171) | 0.23 (0.123) | |||
| IDL/LDL traits | ||||||
| LDL-C | ||||||
| ApoB | ||||||
| LDL-D | 0.271 (0.142) | |||||
| S-LDL-C | ||||||
| S-LDL-L | NA | NA | NA | NA | ||
| S-LDL-P | ||||||
| M-LDL-C | ||||||
| M-LDL-CE | ||||||
| M-LDL-L | NA | NA | NA | NA | ||
| M-LDL-P | ||||||
| M-LDL-PL | ||||||
| L-LDL-C | ||||||
| L-LDL-CE | ||||||
| L-LDL-FC | ||||||
| L-LDL-L | NA | NA | NA | NA | ||
| L-LDL-P | ||||||
| L-LDL-PL | ||||||
| IDL-C | ||||||
| IDL-FC | ||||||
| IDL-L | NA | NA | NA | NA | ||
| IDL-P | ||||||
| IDL-PL | ||||||
| IDL-TG | ||||||
| HDL traits | ||||||
| HDL-C | -0.017 (0.04) | -0.096 (0.077) | -0.085 (0.07) | |||
| ApoA1 | 0.094 (0.049) | -0.06 (0.076) | -0.069 (0.087) | 0.005 (0.083) | -0.051 (0.121) | |
| HDL-D | 0.062 (0.061) | 0.102 (0.064) | 0.088 (0.061) | 0.099 (0.061) | 0.096 (0.058) | |
| S-HDL-L | NA | NA | -0.174 (0.113) | NA | NA | NA |
| S-HDL-P | 0.018 (0.106) | -0.171 (0.109) | NA | -0.049 (0.108) | ||
| S-HDL-TG | ||||||
| M-HDL-C | ||||||
| M-HDL-CE | ||||||
| M-HDL-FC | ||||||
| M-HDL-L | NA | NA | NA | NA | ||
| M-HDL-P | -0.165 (0.131) | |||||
| M-HDL-PL | -0.203 (0.12) | |||||
| L-HDL-C | -0.009 (0.066) | 0.031 (0.083) | -0.032 (0.08) | 0.003 (0.09) | 0.006 (0.068) | |
| L-HDL-CE | -0.011 (0.067) | 0.075 (0.077) | -0.037 (0.076) | 0.015 (0.091) | -0.006 (0.068) | |
| L-HDL-FC | -0.005 (0.067) | 0.079 (0.081) | -0.019 (0.076) | 0.041 (0.078) | 0.027 (0.074) | |
| L-HDL-L | NA | NA | 0.074 (0.077) | NA | 0.068 (0.084) | NA |
| L-HDL-P | 0.046 (0.062) | 0.075 (0.074) | -0.01 (0.066) | 0.066 (0.07) | 0.078 (0.064) | |
| L-HDL-PL | 0 (0.067) | 0.051 (0.082) | -0.021 (0.071) | 0.054 (0.075) | 0.074 (0.071) | |
| XL-HDL-C | 0.122 (0.091) | 0.136 (0.087) | 0.132 (0.09) | 0.02 (0.098) | 0.161 (0.096) | |
| XL-HDL-CE | 0.106 (0.088) | 0.122 (0.09) | 0.148 (0.085) | 0.038 (0.091) | ||
| XL-HDL-FC | 0.065 (0.079) | 0.133 (0.081) | 0.027 (0.077) | 0.052 (0.086) | ||
| XL-HDL-L | NA | NA | 0.119 (0.075) | NA | 0.023 (0.078) | NA |
| XL-HDL-P | 0.087 (0.07) | 0.12 (0.073) | 0.129 (0.067) | |||
| XL-HDL-PL | 0.064 (0.07) | 0.11 (0.072) | 0.121 (0.069) | 0.088 (0.065) | ||
| XL-HDL-TG | ||||||
Multivariable Mendelian randomization results (adjusted for HDL-C, LDL-C, and TG).
| Trait | HDL-C | LDL-C | TG | Subfraction |
|---|---|---|---|---|
| VLDL traits | ||||
| VLDL-D | -0.251 (0.052) | 0.29 (0.037) | 0.6 (0.087) | -0.588 (0.094) |
| XS-VLDL-L | -0.086 (0.046) | 0.286 (0.077) | 0.089 (0.099) | 0.132 (0.119) |
| XS-VLDL-P | -0.083 (0.045) | 0.299 (0.078) | 0.093 (0.106) | 0.118 (0.125) |
| XS-VLDL-PL | -0.083 (0.046) | 0.249 (0.098) | 0.112 (0.076) | 0.159 (0.12) |
| XS-VLDL-TG | -0.114 (0.046) | 0.463 (0.079) | 0.286 (0.173) | -0.157 (0.187) |
| S-VLDL-C | -0.267 (0.084) | 0.754 (0.112) | 1.033 (0.28) | -1.035 (0.323) |
| S-VLDL-FC | -0.195 (0.068) | 0.898 (0.163) | 0.935 (0.26) | -1.027 (0.337) |
| S-VLDL-L | -0.25 (0.072) | 0.755 (0.112) | 0.876 (0.233) | -0.898 (0.28) |
| S-VLDL-P | -0.31 (0.101) | 0.819 (0.157) | 1.209 (0.4) | -1.245 (0.463) |
| S-VLDL-PL | -0.168 (0.051) | 0.673 (0.074) | 0.626 (0.159) | -0.613 (0.182) |
| S-VLDL-TG | -0.499 (0.305) | 0.906 (0.34) | 2.532 (1.57) | -2.628 (1.741) |
| M-VLDL-C | -0.201 (0.068) | 0.808 (0.127) | 1.472 (0.424) | -1.433 (0.451) |
| M-VLDL-CE | -0.168 (0.061) | 0.799 (0.111) | 0.996 (0.249) | -1.035 (0.293) |
| M-VLDL-FC | -0.2 (0.072) | 0.658 (0.089) | 1.469 (0.417) | -1.412 (0.444) |
| M-VLDL-L | -0.355 (0.139) | 0.602 (0.096) | 1.787 (0.654) | -1.878 (0.75) |
| M-VLDL-P | -0.362 (0.124) | 0.569 (0.08) | 1.889 (0.676) | -1.974 (0.745) |
| M-VLDL-PL | -0.332 (0.141) | 0.722 (0.159) | 1.996 (0.869) | -2.012 (0.943) |
| M-VLDL-TG | -0.408 (0.153) | 0.432 (0.061) | 1.974 (0.772) | -2.133 (0.879) |
| L-VLDL-C | -0.216 (0.063) | 0.509 (0.046) | 1.163 (0.254) | -1.254 (0.297) |
| L-VLDL-CE | -0.272 (0.072) | 0.465 (0.04) | 1.038 (0.242) | -1.081 (0.282) |
| L-VLDL-FC | -0.144 (0.059) | 0.493 (0.044) | 1.233 (0.27) | -1.274 (0.308) |
| L-VLDL-L | -0.228 (0.066) | 0.414 (0.045) | 1.17 (0.263) | -1.277 (0.313) |
| L-VLDL-P | -0.115 (0.056) | 0.442 (0.046) | 1.351 (0.317) | -1.357 (0.344) |
| L-VLDL-PL | -0.221 (0.111) | 0.473 (0.07) | 2.135 (0.948) | -2.316 (1.112) |
| L-VLDL-TG | -0.196 (0.066) | 0.355 (0.05) | 1.357 (0.322) | -1.428 (0.372) |
| XL-VLDL-L | -0.126 (0.049) | 0.451 (0.04) | 0.896 (0.159) | -1.069 (0.203) |
| XL-VLDL-P | -0.127 (0.053) | 0.474 (0.043) | 1.038 (0.183) | -1.209 (0.238) |
| XL-VLDL-PL | -0.138 (0.055) | 0.5 (0.044) | 1.052 (0.204) | -1.214 (0.257) |
| XL-VLDL-TG | -0.129 (0.049) | 0.424 (0.04) | 0.944 (0.167) | -1.071 (0.205) |
| XXL-VLDL-L | -0.228 (0.067) | 0.444 (0.043) | 0.978 (0.207) | -1.355 (0.318) |
| XXL-VLDL-P | 0.063 (0.076) | 0.452 (0.05) | 1.371 (0.384) | -1.639 (0.502) |
| XXL-VLDL-PL | -0.185 (0.056) | 0.371 (0.042) | 0.997 (0.185) | -1.259 (0.262) |
| XXL-VLDL-TG | -0.152 (0.059) | 0.41 (0.04) | 0.966 (0.19) | -1.202 (0.262) |
| LDL/IDL traits | ||||
| ApoB | -0.084 (0.046) | 0.8 (0.146) | 0.427 (0.101) | -0.532 (0.191) |
| LDL-D | -0.057 (0.042) | 0.367 (0.03) | 0.21 (0.053) | 0.145 (0.061) |
| S-LDL-C | -0.062 (0.043) | 0.614 (0.126) | 0.261 (0.062) | -0.282 (0.152) |
| S-LDL-L | -0.06 (0.044) | 0.584 (0.118) | 0.266 (0.068) | -0.251 (0.145) |
| S-LDL-P | -0.033 (0.047) | 0.589 (0.119) | 0.29 (0.078) | -0.266 (0.151) |
| M-LDL-C | -0.082 (0.044) | 0.623 (0.146) | 0.203 (0.054) | -0.271 (0.162) |
| M-LDL-CE | -0.074 (0.043) | 0.485 (0.167) | 0.169 (0.059) | -0.088 (0.188) |
| M-LDL-L | -0.071 (0.044) | 0.444 (0.171) | 0.19 (0.063) | -0.069 (0.191) |
| M-LDL-P | -0.054 (0.044) | 0.539 (0.153) | 0.213 (0.063) | -0.179 (0.174) |
| M-LDL-PL | -0.081 (0.045) | 0.747 (0.134) | 0.232 (0.062) | -0.407 (0.162) |
| L-LDL-C | -0.071 (0.049) | 0.437 (0.242) | 0.167 (0.054) | -0.059 (0.261) |
| L-LDL-CE | -0.07 (0.048) | 0.277 (0.301) | 0.149 (0.065) | 0.116 (0.321) |
| L-LDL-FC | -0.112 (0.057) | 0.184 (0.304) | 0.163 (0.053) | 0.223 (0.315) |
| L-LDL-L | -0.075 (0.049) | 0.229 (0.26) | 0.146 (0.068) | 0.167 (0.273) |
| L-LDL-P | -0.083 (0.046) | 0.33 (0.2) | 0.128 (0.064) | 0.084 (0.213) |
| L-LDL-PL | -0.101 (0.046) | 0.446 (0.177) | 0.155 (0.057) | -0.036 (0.195) |
| IDL-C | -0.108 (0.057) | 0.231 (0.215) | 0.128 (0.064) | 0.192 (0.229) |
| IDL-FC | -0.107 (0.05) | 0.23 (0.147) | 0.123 (0.056) | 0.19 (0.156) |
| IDL-L | -0.1 (0.05) | 0.274 (0.161) | 0.123 (0.069) | 0.148 (0.175) |
| IDL-P | -0.101 (0.047) | 0.269 (0.134) | 0.109 (0.071) | 0.153 (0.148) |
| IDL-PL | -0.076 (0.048) | 0.25 (0.162) | 0.134 (0.071) | 0.153 (0.18) |
| IDL-TG | -0.083 (0.046) | 0.314 (0.069) | 0.103 (0.089) | 0.11 (0.103) |
| HDL traits | ||||
| ApoA1 | 0.345 (0.25) | 0.544 (0.081) | 0.334 (0.109) | -0.481 (0.271) |
| HDL-D | -0.442 (0.124) | 0.421 (0.033) | 0.111 (0.055) | 0.333 (0.114) |
| S-HDL-L | -0.117 (0.046) | 0.488 (0.044) | 0.189 (0.054) | -0.312 (0.106) |
| S-HDL-P | -0.112 (0.046) | 0.453 (0.035) | 0.225 (0.056) | -0.331 (0.095) |
| S-HDL-TG | 0.002 (0.145) | 0.314 (0.156) | -0.007 (0.469) | 0.253 (0.637) |
| M-HDL-C | 0.179 (0.097) | 0.36 (0.038) | 0.147 (0.054) | -0.508 (0.165) |
| M-HDL-CE | 0.167 (0.087) | 0.319 (0.036) | 0.166 (0.055) | -0.487 (0.157) |
| M-HDL-FC | 0.339 (0.141) | 0.436 (0.04) | 0.247 (0.059) | -0.649 (0.225) |
| M-HDL-L | 0.27 (0.108) | 0.362 (0.032) | 0.299 (0.063) | -0.606 (0.188) |
| M-HDL-P | 0.302 (0.112) | 0.386 (0.033) | 0.371 (0.075) | -0.694 (0.204) |
| M-HDL-PL | 0.311 (0.117) | 0.402 (0.033) | 0.333 (0.07) | -0.632 (0.191) |
| L-HDL-C | -0.589 (0.211) | 0.469 (0.039) | 0.146 (0.055) | 0.516 (0.213) |
| L-HDL-CE | -0.602 (0.239) | 0.477 (0.042) | 0.137 (0.056) | 0.519 (0.23) |
| L-HDL-FC | -0.573 (0.177) | 0.437 (0.034) | 0.171 (0.054) | 0.518 (0.181) |
| L-HDL-L | -0.556 (0.193) | 0.437 (0.034) | 0.142 (0.055) | 0.457 (0.189) |
| L-HDL-P | -0.515 (0.198) | 0.417 (0.03) | 0.133 (0.056) | 0.422 (0.191) |
| L-HDL-PL | -0.53 (0.201) | 0.415 (0.034) | 0.152 (0.055) | 0.443 (0.202) |
| XL-HDL-C | -0.447 (0.182) | 0.342 (0.036) | 0.071 (0.079) | 0.474 (0.223) |
| XL-HDL-CE | -0.425 (0.146) | 0.366 (0.038) | 0.051 (0.069) | 0.426 (0.177) |
| XL-HDL-FC | -0.459 (0.147) | 0.377 (0.031) | 0.097 (0.062) | 0.433 (0.16) |
| XL-HDL-L | -0.405 (0.146) | 0.364 (0.031) | 0.077 (0.068) | 0.358 (0.154) |
| XL-HDL-P | -0.451 (0.134) | 0.374 (0.03) | 0.078 (0.064) | 0.41 (0.139) |
| XL-HDL-PL | -0.422 (0.119) | 0.412 (0.033) | 0.115 (0.055) | 0.343 (0.118) |
| XL-HDL-TG | -0.186 (0.073) | 0.336 (0.035) | 0.045 (0.086) | 0.147 (0.074) |
Multivariable Mendelian randomization results (adjusted for ApoA1, ApoB, and TG).
| Trait | ApoA1 | ApoB | TG | Subfraction |
|---|---|---|---|---|
| VLDL traits | ||||
| VLDL-D | -0.227 (0.067) | 0.545 (0.092) | 0.208 (0.139) | -0.32 (0.112) |
| XS-VLDL-L | -0.123 (0.063) | 0.53 (0.163) | -0.121 (0.085) | 0.084 (0.141) |
| XS-VLDL-P | -0.121 (0.064) | 0.553 (0.17) | -0.123 (0.088) | 0.061 (0.158) |
| XS-VLDL-PL | -0.147 (0.066) | 0.273 (0.138) | 0.028 (0.05) | 0.253 (0.135) |
| XS-VLDL-TG | -0.102 (0.06) | 0.762 (0.168) | 0.069 (0.055) | -0.248 (0.15) |
| S-VLDL-C | -0.384 (0.141) | 1.426 (0.354) | 0.606 (0.351) | -1.265 (0.568) |
| S-VLDL-FC | -0.188 (0.077) | 1.001 (0.235) | 0.081 (0.053) | -0.489 (0.213) |
| S-VLDL-L | -0.46 (0.146) | 1.776 (0.417) | 0.7 (0.316) | -1.629 (0.586) |
| S-VLDL-P | -0.494 (0.159) | 1.677 (0.386) | 0.825 (0.372) | -1.644 (0.606) |
| S-VLDL-PL | -0.262 (0.097) | 1.41 (0.343) | 0.532 (0.261) | -1.213 (0.478) |
| S-VLDL-TG | -0.18 (0.069) | 0.792 (0.121) | 0.078 (0.051) | -0.301 (0.108) |
| M-VLDL-C | -0.157 (0.062) | 0.867 (0.132) | 0.085 (0.051) | -0.373 (0.118) |
| M-VLDL-CE | -0.221 (0.069) | 1.224 (0.223) | 0.47 (0.21) | -0.995 (0.338) |
| M-VLDL-FC | -0.222 (0.074) | 0.902 (0.133) | 0.482 (0.251) | -0.799 (0.311) |
| M-VLDL-L | -0.174 (0.065) | 0.76 (0.104) | 0.073 (0.05) | -0.298 (0.098) |
| M-VLDL-P | -0.181 (0.065) | 0.764 (0.1) | 0.077 (0.051) | -0.312 (0.096) |
| M-VLDL-PL | -0.159 (0.065) | 0.776 (0.116) | 0.08 (0.051) | -0.297 (0.106) |
| M-VLDL-TG | -0.263 (0.106) | 0.724 (0.094) | 0.547 (0.406) | -0.806 (0.455) |
| L-VLDL-C | -0.218 (0.084) | 0.732 (0.101) | 0.352 (0.278) | -0.609 (0.337) |
| L-VLDL-CE | -0.293 (0.079) | 0.781 (0.096) | 0.405 (0.189) | -0.673 (0.217) |
| L-VLDL-FC | -0.197 (0.069) | 0.737 (0.094) | 0.365 (0.25) | -0.619 (0.291) |
| L-VLDL-L | -0.194 (0.071) | 0.666 (0.087) | 0.289 (0.234) | -0.532 (0.278) |
| L-VLDL-P | -0.184 (0.061) | 0.677 (0.086) | 0.415 (0.217) | -0.617 (0.229) |
| L-VLDL-PL | -0.155 (0.063) | 0.715 (0.095) | 0.075 (0.051) | -0.287 (0.104) |
| L-VLDL-TG | -0.154 (0.062) | 0.67 (0.083) | 0.073 (0.05) | -0.252 (0.091) |
| XL-VLDL-L | -0.186 (0.066) | 0.694 (0.088) | 0.263 (0.19) | -0.577 (0.249) |
| XL-VLDL-P | -0.167 (0.061) | 0.742 (0.088) | 0.075 (0.05) | -0.373 (0.109) |
| XL-VLDL-PL | -0.191 (0.068) | 0.712 (0.092) | 0.271 (0.197) | -0.583 (0.268) |
| XL-VLDL-TG | -0.195 (0.068) | 0.666 (0.087) | 0.334 (0.21) | -0.603 (0.248) |
| XXL-VLDL-L | -0.173 (0.066) | 0.732 (0.098) | 0.088 (0.052) | -0.402 (0.144) |
| XXL-VLDL-P | -0.071 (0.065) | 0.705 (0.097) | 0.607 (0.321) | -1.089 (0.449) |
| XXL-VLDL-PL | -0.244 (0.082) | 0.666 (0.091) | 0.414 (0.257) | -0.814 (0.344) |
| XXL-VLDL-TG | -0.3 (0.091) | 0.694 (0.095) | 0.627 (0.306) | -1.075 (0.402) |
| IDL/LDL traits | ||||
| LDL-C | -0.119 (0.062) | 0.247 (0.167) | 0.066 (0.054) | 0.319 (0.182) |
| LDL-D | -0.123 (0.06) | 0.544 (0.091) | -0.036 (0.087) | 0.119 (0.071) |
| S-LDL-C | -0.097 (0.06) | 0.438 (0.216) | 0.044 (0.051) | 0.08 (0.238) |
| S-LDL-L | -0.097 (0.063) | 0.503 (0.268) | 0.043 (0.051) | -0.005 (0.29) |
| S-LDL-P | -0.059 (0.103) | 0.932 (0.597) | -0.122 (0.112) | -0.362 (0.596) |
| M-LDL-C | -0.099 (0.065) | 0.78 (1.034) | -0.172 (0.425) | -0.169 (0.909) |
| M-LDL-CE | -0.157 (0.128) | -0.346 (2.587) | 0.195 (0.855) | 0.854 (2.221) |
| M-LDL-L | -0.123 (0.095) | 0.247 (1.479) | -0.001 (0.445) | 0.32 (1.293) |
| M-LDL-P | -0.134 (0.07) | 0.13 (0.286) | 0.053 (0.052) | 0.432 (0.31) |
| M-LDL-PL | -0.075 (0.077) | 1.165 (0.868) | -0.248 (0.253) | -0.566 (0.839) |
| L-LDL-C | -0.855 (1.68) | -5.337 (13.402) | 2.405 (5.735) | 5.257 (11.72) |
| L-LDL-CE | -0.151 (0.065) | 0.129 (0.193) | 0.061 (0.052) | 0.461 (0.213) |
| L-LDL-FC | -0.397 (0.219) | -1.139 (1.395) | 0.786 (0.711) | 1.531 (1.189) |
| L-LDL-L | -0.265 (0.148) | -0.854 (1.42) | 0.41 (0.51) | 1.266 (1.188) |
| L-LDL-P | -0.258 (0.153) | -0.607 (1.225) | 0.276 (0.402) | 1.064 (1.029) |
| L-LDL-PL | -0.312 (0.187) | -0.741 (1.411) | 0.39 (0.518) | 1.227 (1.245) |
| IDL-C | -0.3 (0.123) | -0.334 (0.616) | 0.276 (0.254) | 0.769 (0.501) |
| IDL-FC | -0.199 (0.069) | 0.247 (0.118) | 0.044 (0.049) | 0.33 (0.127) |
| IDL-L | -0.215 (0.089) | 0.021 (0.409) | 0.101 (0.15) | 0.444 (0.328) |
| IDL-P | -0.175 (0.075) | 0.214 (0.172) | 0.04 (0.051) | 0.292 (0.173) |
| IDL-PL | -0.183 (0.07) | 0.159 (0.172) | 0.031 (0.049) | 0.406 (0.184) |
| IDL-TG | -0.143 (0.075) | 0.565 (0.146) | -0.119 (0.087) | 0.047 (0.135) |
| HDL traits | ||||
| HDL-C | -1.513 (1.109) | 0.982 (0.314) | 0.27 (0.291) | 1.446 (1.112) |
| HDL-D | -0.457 (0.138) | 0.613 (0.073) | 0.056 (0.049) | 0.296 (0.1) |
| S-HDL-L | -0.128 (0.059) | 0.524 (0.062) | 0.067 (0.05) | -0.224 (0.087) |
| S-HDL-P | -0.132 (0.059) | 0.531 (0.059) | 0.071 (0.05) | -0.24 (0.083) |
| S-HDL-TG | -0.11 (0.113) | 0.595 (0.221) | -0.057 (0.297) | -0.044 (0.466) |
| M-HDL-C | 0.091 (0.084) | 0.459 (0.101) | -0.1 (0.083) | -0.442 (0.143) |
| M-HDL-CE | 0.09 (0.078) | 0.291 (0.083) | 0.082 (0.05) | -0.413 (0.137) |
| M-HDL-FC | 0.148 (0.11) | 0.378 (0.063) | 0.066 (0.049) | -0.408 (0.166) |
| M-HDL-L | 0.133 (0.091) | 0.491 (0.097) | -0.029 (0.086) | -0.485 (0.155) |
| M-HDL-P | 0.129 (0.097) | 0.501 (0.097) | -0.004 (0.09) | -0.472 (0.166) |
| M-HDL-PL | 0.162 (0.107) | 0.519 (0.096) | -0.037 (0.087) | -0.486 (0.171) |
| L-HDL-C | -0.724 (0.232) | 0.856 (0.132) | 0.032 (0.093) | 0.575 (0.204) |
| L-HDL-CE | -0.761 (0.236) | 0.899 (0.145) | 0.004 (0.084) | 0.61 (0.206) |
| L-HDL-FC | -0.749 (0.174) | 0.842 (0.102) | 0.094 (0.05) | 0.59 (0.148) |
| L-HDL-L | -0.717 (0.217) | 0.815 (0.12) | 0.023 (0.089) | 0.541 (0.184) |
| L-HDL-P | -0.653 (0.191) | 0.749 (0.104) | 0.057 (0.049) | 0.476 (0.155) |
| L-HDL-PL | -0.679 (0.201) | 0.774 (0.109) | 0.05 (0.049) | 0.51 (0.169) |
| XL-HDL-C | -0.639 (0.194) | 0.692 (0.095) | -0.058 (0.086) | 0.565 (0.196) |
| XL-HDL-CE | -0.576 (0.2) | 0.667 (0.096) | -0.077 (0.086) | 0.511 (0.206) |
| XL-HDL-FC | -0.734 (0.174) | 0.674 (0.073) | 0.094 (0.052) | 0.609 (0.159) |
| XL-HDL-L | -0.652 (0.168) | 0.733 (0.097) | -0.06 (0.084) | 0.481 (0.141) |
| XL-HDL-P | -0.52 (0.147) | 0.691 (0.094) | -0.075 (0.084) | 0.39 (0.135) |
| XL-HDL-PL | -0.652 (0.151) | 0.687 (0.076) | 0.079 (0.051) | 0.466 (0.12) |
| XL-HDL-TG | -0.281 (0.111) | 0.539 (0.09) | -0.152 (0.092) | 0.165 (0.086) |
Modified Cochran’s Q-statistics (p-values) for the multivariable Mendelian randomization analyses (adjusted for HDL-C, LDL-C, and TG).
DF is short for degrees of freedom.
| Trait | DF | HDL-C | LDL-C | TG | Subfraction |
|---|---|---|---|---|---|
| VLDL traits | |||||
| VLDL-D | 432 | 7640.8 (0) | 1918.9 (7.9e-186) | 877.6 (1.4e-32) | 840.2 (1.6e-28) |
| XS-VLDL-L | 436 | 7983.9 (0) | 1104.9 (1.1e-59) | 1935.8 (2.2e-187) | 926 (1.9e-37) |
| XS-VLDL-P | 436 | 7927.8 (0) | 1066.6 (1.1e-54) | 1814 (4.8e-167) | 893.6 (9.6e-34) |
| XS-VLDL-PL | 435 | 8291.5 (0) | 968.1 (1.4e-42) | 2771.5 (0) | 849.8 (4.3e-29) |
| XS-VLDL-TG | 431 | 7549.8 (0) | 894.4 (1.3e-34) | 739.5 (1.3e-18) | 682.5 (1.2e-13) |
| S-VLDL-C | 429 | 8598.1 (0) | 652.6 (1.7e-11) | 1220.7 (4.6e-77) | 541.3 (0.00018) |
| S-VLDL-FC | 434 | 7861.2 (0) | 576 (5.4e-06) | 519.4 (0.003) | 507.9 (0.0082) |
| S-VLDL-L | 438 | 7105.3 (0) | 626 (8.5e-09) | 525.2 (0.0026) | 514.3 (0.0069) |
| S-VLDL-P | 438 | 6686.5 (0) | 616.5 (3.6e-08) | 515.6 (0.0061) | 507.3 (0.012) |
| S-VLDL-PL | 437 | 7589.1 (0) | 702.8 (1e-14) | 591.5 (1.1e-06) | 555.1 (0.00011) |
| S-VLDL-TG | 437 | 7658.7 (0) | 612.7 (5.3e-08) | 498.9 (0.021) | 494.5 (0.03) |
| M-VLDL-C | 432 | 9167.8 (0) | 740.8 (1.3e-18) | 558.9 (3.5e-05) | 551.5 (8.3e-05) |
| M-VLDL-CE | 432 | 8055.2 (0) | 705.9 (1.6e-15) | 556.6 (4.6e-05) | 539.7 (0.00031) |
| M-VLDL-FC | 436 | 8272.8 (0) | 814.8 (2.7e-25) | 528.3 (0.0016) | 519.1 (0.0037) |
| M-VLDL-L | 429 | 7109.2 (0) | 1269.2 (5.5e-84) | 532.6 (0.00047) | 515.9 (0.0025) |
| M-VLDL-P | 436 | 8260.7 (0) | 2059.5 (2.1e-208) | 527.5 (0.0017) | 516.8 (0.0046) |
| M-VLDL-PL | 435 | 6849.2 (0) | 599.6 (2.6e-07) | 496.8 (0.021) | 493.5 (0.027) |
| M-VLDL-TG | 436 | 6123.7 (0) | 9854.8 (0) | 532.3 (0.0011) | 521 (0.0031) |
| L-VLDL-C | 435 | 8617.2 (0) | 8966 (0) | 654.7 (4.3e-11) | 561.5 (3.9e-05) |
| L-VLDL-CE | 434 | 6636.6 (0) | 11134 (0) | 581.6 (2.6e-06) | 539.5 (0.00041) |
| L-VLDL-FC | 431 | 7779.6 (0) | 6691 (0) | 595.1 (2.5e-07) | 562.7 (1.9e-05) |
| L-VLDL-L | 434 | 8104.9 (0) | 5191.4 (0) | 560.3 (3.9e-05) | 548.6 (0.00015) |
| L-VLDL-P | 435 | 2308 (5.1e-252) | 10360.3 (0) | 545.4 (0.00024) | 537.9 (0.00054) |
| L-VLDL-PL | 430 | 8155.4 (0) | 1310.8 (8.6e-90) | 491.8 (0.021) | 489.7 (0.024) |
| L-VLDL-TG | 438 | 8581.8 (0) | 4800.1 (0) | 569.1 (2.3e-05) | 559.2 (7.5e-05) |
| XL-VLDL-L | 437 | 8686.8 (0) | 8322.2 (0) | 674.7 (1.9e-12) | 620.2 (1.7e-08) |
| XL-VLDL-P | 431 | 8550.2 (0) | 2459.4 (2e-280) | 608.3 (3.6e-08) | 588.6 (6.3e-07) |
| XL-VLDL-PL | 431 | 7478.2 (0) | 5042.5 (0) | 613.3 (1.7e-08) | 591.6 (4.1e-07) |
| XL-VLDL-TG | 433 | 8237.3 (0) | 9628.9 (0) | 651.8 (4.6e-11) | 618.3 (1.1e-08) |
| XXL-VLDL-L | 439 | 8476.2 (0) | 10436.4 (0) | 652.9 (1.3e-10) | 570.7 (2.2e-05) |
| XXL-VLDL-P | 437 | 1291.3 (2.8e-85) | 9987.4 (0) | 540.3 (0.00053) | 529.5 (0.0016) |
| XXL-VLDL-PL | 436 | 9631.8 (0) | 11287.1 (0) | 641.6 (4.8e-10) | 595.5 (5.3e-07) |
| XXL-VLDL-TG | 429 | 7809.4 (0) | 9476.4 (0) | 595.6 (1.7e-07) | 564 (1.2e-05) |
| LDL/IDL traits | |||||
| ApoB | 435 | 9220.8 (0) | 550.1 (0.00014) | 1809.7 (1.2e-166) | 535.1 (0.00072) |
| LDL-D | 429 | 2909.2 (0) | 3918.8 (0) | 2706 (0) | 1426.1 (2.9e-107) |
| S-LDL-C | 431 | 8189.7 (0) | 569.8 (7.8e-06) | 4880.9 (0) | 564.1 (1.6e-05) |
| S-LDL-L | 435 | 8403.8 (0) | 574.4 (7.8e-06) | 3931.2 (0) | 564.3 (2.7e-05) |
| S-LDL-P | 431 | 7371.4 (0) | 547.1 (0.00012) | 3144.7 (0) | 537.9 (0.00034) |
| M-LDL-C | 430 | 9723.7 (0) | 570.9 (5.8e-06) | 6568.6 (0) | 562.9 (1.6e-05) |
| M-LDL-CE | 432 | 8442.1 (0) | 558.3 (3.8e-05) | 5773.6 (0) | 549.1 (0.00011) |
| M-LDL-L | 430 | 8801.7 (0) | 555.4 (4e-05) | 5176.1 (0) | 548.2 (9.5e-05) |
| M-LDL-P | 429 | 8798.9 (0) | 541.6 (0.00018) | 5049.7 (0) | 535.2 (0.00035) |
| M-LDL-PL | 436 | 7981.7 (0) | 573.9 (9.6e-06) | 4304.8 (0) | 558.9 (6e-05) |
| L-LDL-C | 432 | 8865.2 (0) | 567.7 (1.2e-05) | 6179.8 (0) | 567 (1.3e-05) |
| L-LDL-CE | 433 | 8464.3 (0) | 558.7 (4.1e-05) | 5731.3 (0) | 555.6 (5.9e-05) |
| L-LDL-FC | 431 | 7481.1 (0) | 580.6 (1.9e-06) | 6760.8 (0) | 580.2 (2e-06) |
| L-LDL-L | 433 | 8486.8 (0) | 604.5 (8.9e-08) | 5755.8 (0) | 601.8 (1.3e-07) |
| L-LDL-P | 434 | 8310.7 (0) | 592.1 (6.3e-07) | 5553.3 (0) | 584.9 (1.7e-06) |
| L-LDL-PL | 435 | 8341.4 (0) | 588.5 (1.2e-06) | 5327.8 (0) | 577.4 (5.3e-06) |
| IDL-C | 434 | 7873.9 (0) | 645.5 (1.7e-10) | 6336 (0) | 642.1 (2.9e-10) |
| IDL-FC | 432 | 8036 (0) | 729.5 (1.4e-17) | 6630.5 (0) | 725.6 (3e-17) |
| IDL-L | 434 | 7869.8 (0) | 694.5 (2.4e-14) | 5198.3 (0) | 689 (7e-14) |
| IDL-P | 436 | 9660.5 (0) | 736.7 (9e-18) | 5002 (0) | 726.6 (7.1e-17) |
| IDL-PL | 431 | 8432.6 (0) | 680.6 (1.7e-13) | 5023 (0) | 677.4 (3e-13) |
| IDL-TG | 436 | 7741.2 (0) | 1077.5 (4.2e-56) | 1992.9 (4.9e-197) | 931.6 (4.4e-38) |
| HDL traits | |||||
| ApoA1 | 434 | 494.1 (0.024) | 511.5 (0.006) | 932.1 (1.8e-38) | 492 (0.028) |
| HDL-D | 438 | 783.5 (6.6e-22) | 8500 (0) | 5713.2 (0) | 860.1 (9.4e-30) |
| S-HDL-L | 438 | 3067.3 (0) | 4414.6 (0) | 3763.2 (0) | 882.2 (3.7e-32) |
| S-HDL-P | 438 | 2592.4 (1.1e-301) | 7652.1 (0) | 3097.3 (0) | 951.1 (4.9e-40) |
| S-HDL-TG | 425 | 896.9 (6.9e-36) | 641.3 (5.2e-11) | 540.1 (0.00013) | 523 (8e-04) |
| M-HDL-C | 437 | 957.6 (5.5e-41) | 10172.4 (0) | 4875.5 (0) | 628.3 (4.9e-09) |
| M-HDL-CE | 434 | 955.3 (3.2e-41) | 1383.1 (1.7e-99) | 4355.4 (0) | 648.3 (1e-10) |
| M-HDL-FC | 432 | 759.4 (2.4e-20) | 2989.1 (0) | 3512.2 (0) | 538.2 (0.00037) |
| M-HDL-L | 435 | 914.2 (3e-36) | 11535.3 (0) | 2327.7 (1.7e-255) | 570.3 (1.3e-05) |
| M-HDL-P | 434 | 997.6 (2.3e-46) | 10709.6 (0) | 1942.9 (3.2e-189) | 561.3 (3.4e-05) |
| M-HDL-PL | 434 | 977.8 (6.3e-44) | 9439.9 (0) | 2566 (1.8e-298) | 581.3 (2.7e-06) |
| L-HDL-C | 434 | 580 (3.2e-06) | 1257.1 (4.4e-81) | 4502.7 (0) | 604.3 (1.1e-07) |
| L-HDL-CE | 434 | 549 (0.00014) | 930.2 (3e-38) | 5517.2 (0) | 557.2 (5.6e-05) |
| L-HDL-FC | 441 | 627.6 (1.2e-08) | 8415.3 (0) | 3594 (0) | 658.4 (7.9e-11) |
| L-HDL-L | 434 | 603.6 (1.2e-07) | 6743.8 (0) | 5314.7 (0) | 623.7 (5.7e-09) |
| L-HDL-P | 432 | 601.1 (1.2e-07) | 7769.3 (0) | 6024.6 (0) | 633.2 (8.6e-10) |
| L-HDL-PL | 434 | 584.5 (1.8e-06) | 9935.5 (0) | 3544.3 (0) | 611.3 (3.8e-08) |
| XL-HDL-C | 430 | 732.9 (3.9e-18) | 10426.6 (0) | 2077.7 (1.4e-213) | 686.9 (4e-14) |
| XL-HDL-CE | 430 | 771.4 (9.3e-22) | 8564.4 (0) | 2457 (2.2e-280) | 711.4 (3.3e-16) |
| XL-HDL-FC | 432 | 761.8 (1.4e-20) | 11265.2 (0) | 2549.4 (3.1e-296) | 770.9 (1.9e-21) |
| XL-HDL-L | 429 | 767.6 (1.6e-21) | 11490.7 (0) | 2355.7 (1.2e-262) | 784.6 (3.4e-23) |
| XL-HDL-P | 433 | 724.9 (4.6e-17) | 11372.5 (0) | 2539.9 (3.9e-294) | 798.5 (4.8e-24) |
| XL-HDL-PL | 443 | 809.7 (7.8e-24) | 10093.1 (0) | 5762 (0) | 895.4 (7.5e-33) |
| XL-HDL-TG | 432 | 1849.1 (3.9e-174) | 2635.9 (6.5e-312) | 2240.8 (2.9e-241) | 1267.8 (4.4e-83) |
Modified Cochran’s Q-statistics (p-values) for the multivariable Mendelian randomization analyses (adjusted for ApoA1, ApoB, and TG).
DF is short for degrees of freedom.
| Trait | DF | ApoA1 | ApoB | TG | Subfraction |
|---|---|---|---|---|---|
| VLDL traits | |||||
| VLDL-D | 297 | 1194.1 (9.1e-108) | 550 (2.4e-17) | 573.7 (8.2e-20) | 606.7 (2.1e-23) |
| XS-VLDL-L | 295 | 1185.1 (6.7e-107) | 927 (2e-66) | 1151.3 (2.2e-101) | 887.9 (1.1e-60) |
| XS-VLDL-P | 295 | 1194.9 (1.7e-108) | 900 (1.9e-62) | 895.5 (8.7e-62) | 826.7 (6.4e-52) |
| XS-VLDL-PL | 296 | 1148.5 (1.2e-100) | 973.9 (3.2e-73) | 2104.2 (1.4e-269) | 961.4 (2.5e-71) |
| XS-VLDL-TG | 302 | 1263.7 (1.1e-117) | 757.9 (4.7e-41) | 1308.1 (4.4e-125) | 976.5 (4.6e-72) |
| S-VLDL-C | 290 | 988.8 (4.4e-77) | 394 (4.5e-05) | 459.8 (7.8e-10) | 402.6 (1.3e-05) |
| S-VLDL-FC | 296 | 1092 (1.4e-91) | 904 (8.6e-63) | 1238.7 (2.1e-115) | 1010.4 (8.1e-79) |
| S-VLDL-L | 301 | 1107.9 (1.1e-92) | 412.3 (2.1e-05) | 420.8 (5.9e-06) | 384.7 (0.00078) |
| S-VLDL-P | 301 | 1116.6 (4.6e-94) | 424.8 (3.3e-06) | 401.3 (9.4e-05) | 380.6 (0.0013) |
| S-VLDL-PL | 299 | 1096 (2.3e-91) | 428.9 (1.2e-06) | 446 (7.1e-08) | 432.1 (7.1e-07) |
| S-VLDL-TG | 300 | 1152.4 (4.3e-100) | 908.5 (1.8e-62) | 1453.4 (1.8e-150) | 1303.1 (7.1e-125) |
| M-VLDL-C | 298 | 1171.2 (1e-103) | 824 (7.3e-51) | 1480 (8.9e-156) | 1212.5 (1.8e-110) |
| M-VLDL-CE | 298 | 1185.4 (4.9e-106) | 564.4 (1.1e-18) | 468.9 (9.2e-10) | 431.6 (6.3e-07) |
| M-VLDL-FC | 298 | 1190.4 (7.4e-107) | 899.8 (1.1e-61) | 415.2 (8.1e-06) | 398.8 (8.4e-05) |
| M-VLDL-L | 298 | 1144.1 (2.4e-99) | 869.8 (2.4e-57) | 1381 (1e-138) | 1237.4 (1.4e-114) |
| M-VLDL-P | 297 | 1121.3 (5.7e-96) | 821.1 (1.1e-50) | 1250.5 (4.6e-117) | 1206.7 (8.1e-110) |
| M-VLDL-PL | 298 | 1149.9 (2.8e-100) | 843.2 (1.5e-53) | 1391.8 (1.5e-140) | 1226.3 (9.8e-113) |
| M-VLDL-TG | 296 | 1187.4 (5.8e-107) | 717.3 (5.8e-37) | 366.3 (0.0033) | 360.6 (0.006) |
| L-VLDL-C | 295 | 1196.5 (9.1e-109) | 820 (5.6e-51) | 462.5 (1.5e-09) | 376.9 (0.00088) |
| L-VLDL-CE | 302 | 1183.1 (1.8e-104) | 844.6 (7.4e-53) | 541.8 (7.2e-16) | 441.7 (2.6e-07) |
| L-VLDL-FC | 295 | 1172.3 (8.2e-105) | 851.6 (1.9e-55) | 460.8 (2.1e-09) | 406.2 (1.8e-05) |
| L-VLDL-L | 295 | 1163.6 (2.2e-103) | 797 (8.8e-48) | 406.5 (1.7e-05) | 391.5 (0.00014) |
| L-VLDL-P | 293 | 1160.2 (2e-103) | 809.5 (5.9e-50) | 420.2 (1.5e-06) | 407.9 (1e-05) |
| L-VLDL-PL | 296 | 1292 (2.6e-124) | 833.4 (1.3e-52) | 1216.5 (9.7e-112) | 1098.9 (1.1e-92) |
| L-VLDL-TG | 294 | 1150.8 (1.3e-101) | 1213.6 (7e-112) | 1262.6 (5.2e-120) | 1162.8 (1.5e-103) |
| XL-VLDL-L | 294 | 1196 (5.4e-109) | 829.4 (1.6e-52) | 442 (4.9e-08) | 423.6 (1.1e-06) |
| XL-VLDL-P | 294 | 1265.9 (1.4e-120) | 1180.9 (1.6e-106) | 1202.2 (5.2e-110) | 982.1 (5.4e-75) |
| XL-VLDL-PL | 296 | 1199.1 (6.9e-109) | 874.2 (1.9e-58) | 421.2 (2.3e-06) | 405.6 (2.3e-05) |
| XL-VLDL-TG | 296 | 1184.3 (1.8e-106) | 828.6 (5.9e-52) | 430.8 (4.9e-07) | 430.1 (5.5e-07) |
| XXL-VLDL-L | 304 | 1119.2 (1.2e-93) | 1041.9 (1.6e-81) | 900.9 (2e-60) | 699.6 (3.2e-33) |
| XXL-VLDL-P | 303 | 1148 (1.7e-98) | 876.4 (4e-57) | 382.2 (0.0013) | 366 (0.0076) |
| XXL-VLDL-PL | 303 | 1203 (2.1e-107) | 775.1 (4e-43) | 438.1 (5.8e-07) | 376.5 (0.0025) |
| XXL-VLDL-TG | 303 | 1183 (3.7e-104) | 881.8 (6.6e-58) | 393.7 (0.00034) | 372.7 (0.0039) |
| LDL/IDL traits | |||||
| LDL-C | 293 | 1198.7 (9.6e-110) | 938.8 (1.1e-68) | 1060.2 (2.1e-87) | 917.6 (1.5e-65) |
| LDL-D | 296 | 1325.2 (6.7e-130) | 747.9 (5.9e-41) | 879.1 (3.7e-59) | 1163.5 (4.6e-103) |
| S-LDL-C | 296 | 1195.3 (2.9e-108) | 706 (1.6e-35) | 1426 (4.1e-147) | 686.4 (4.8e-33) |
| S-LDL-L | 296 | 1054.7 (1.1e-85) | 608 (1e-23) | 1519.6 (2.2e-163) | 586.4 (2.5e-21) |
| S-LDL-P | 297 | 852.9 (3.6e-55) | 438.7 (1.6e-07) | 954.7 (4.5e-70) | 440.1 (1.3e-07) |
| M-LDL-C | 296 | 1210.9 (8e-111) | 396.2 (8.6e-05) | 409 (1.4e-05) | 398.9 (6e-05) |
| M-LDL-CE | 295 | 1204.3 (4.8e-110) | 350.8 (0.014) | 361.7 (0.0048) | 351.3 (0.013) |
| M-LDL-L | 296 | 1212 (5.3e-111) | 370 (0.0022) | 392.3 (0.00015) | 371.6 (0.0019) |
| M-LDL-P | 297 | 1125.4 (1.2e-96) | 623.9 (2.3e-25) | 911.4 (1.3e-63) | 582.4 (9.6e-21) |
| M-LDL-PL | 299 | 1172.5 (1.2e-103) | 399.3 (9.1e-05) | 434.9 (4.5e-07) | 396.2 (0.00014) |
| L-LDL-C | 300 | 1174.6 (1.1e-103) | 325.5 (0.15) | 325.5 (0.15) | 325.5 (0.15) |
| L-LDL-CE | 299 | 1179.5 (9e-105) | 769.8 (3e-43) | 902.5 (7.7e-62) | 743.8 (8.4e-40) |
| L-LDL-FC | 295 | 1161 (5.8e-103) | 322.4 (0.13) | 323.2 (0.12) | 322.3 (0.13) |
| L-LDL-L | 300 | 1172.3 (2.6e-103) | 336.9 (0.07) | 349.6 (0.026) | 340.3 (0.055) |
| L-LDL-P | 300 | 1185.4 (2e-105) | 352.1 (0.021) | 378.4 (0.0014) | 355.4 (0.015) |
| L-LDL-PL | 296 | 1155.2 (9.8e-102) | 343.2 (0.031) | 360.1 (0.0063) | 344.5 (0.027) |
| IDL-C | 296 | 1181.7 (4.9e-106) | 426.5 (9.8e-07) | 427.6 (8.3e-07) | 427.7 (8.1e-07) |
| IDL-FC | 298 | 1096.5 (9.9e-92) | 986.9 (1.1e-74) | 1075.8 (1.9e-88) | 975.4 (6.1e-73) |
| IDL-L | 296 | 1176.1 (4e-105) | 516.7 (3.3e-14) | 531 (1.4e-15) | 521.4 (1.2e-14) |
| IDL-P | 297 | 1094.8 (9.5e-92) | 910.9 (1.5e-63) | 1103.9 (3.5e-93) | 890.2 (1.6e-60) |
| IDL-PL | 297 | 1107.8 (8.3e-94) | 798.9 (1.3e-47) | 931.6 (1.3e-66) | 785.6 (8.6e-46) |
| IDL-TG | 302 | 1060.8 (5.4e-85) | 1052.1 (1.2e-83) | 1092.6 (5.6e-90) | 1118.3 (4.7e-94) |
| HDL traits | |||||
| HDL-C | 298 | 318.7 (0.2) | 336.3 (0.063) | 329.1 (0.1) | 318.6 (0.2) |
| HDL-D | 300 | 637.4 (1.9e-26) | 1156.6 (9.1e-101) | 2305.2 (1.3e-305) | 1183.8 (3.5e-105) |
| S-HDL-L | 299 | 1597.7 (4.8e-176) | 1222.5 (8.2e-112) | 1916.4 (1.5e-233) | 1057 (3.1e-85) |
| S-HDL-P | 299 | 1666.8 (2.5e-188) | 1249.4 (2.9e-116) | 2146.5 (3.4e-276) | 1103.3 (1.6e-92) |
| S-HDL-TG | 299 | 899 (2.5e-61) | 464.9 (2.4e-09) | 464.5 (2.6e-09) | 457.6 (9.2e-09) |
| M-HDL-C | 299 | 1145.2 (3.2e-99) | 768.2 (4.9e-43) | 951.8 (4e-69) | 786.8 (1.5e-45) |
| M-HDL-CE | 299 | 1201.9 (2e-108) | 1183.9 (1.7e-105) | 2139.7 (6.4e-275) | 843.9 (1.9e-53) |
| M-HDL-FC | 298 | 881.1 (5.6e-59) | 1252 (5.5e-117) | 1989.1 (2.4e-247) | 660.1 (1.8e-29) |
| M-HDL-L | 299 | 1059 (1.5e-85) | 766.4 (8.7e-43) | 920.6 (1.7e-64) | 672.5 (8.6e-31) |
| M-HDL-P | 298 | 990.2 (3.5e-75) | 760.4 (3.4e-42) | 1027.6 (6.2e-81) | 613.7 (4.7e-24) |
| M-HDL-PL | 295 | 929.5 (8.3e-67) | 763.9 (2.7e-43) | 1057.2 (2.3e-86) | 588.3 (1.1e-21) |
| L-HDL-C | 299 | 579.3 (4.1e-20) | 623.2 (5.7e-25) | 639.6 (7.3e-27) | 617.8 (2.3e-24) |
| L-HDL-CE | 299 | 612.2 (1e-23) | 650.7 (3.6e-28) | 690.4 (5.5e-33) | 644 (2.2e-27) |
| L-HDL-FC | 308 | 581.7 (4.4e-19) | 857.5 (2.6e-53) | 1213.3 (1.4e-107) | 915.8 (1.3e-61) |
| L-HDL-L | 299 | 655.9 (8.7e-29) | 747.7 (2.6e-40) | 670.7 (1.4e-30) | 713.2 (7.5e-36) |
| L-HDL-P | 298 | 591.3 (1.5e-21) | 934 (9.9e-67) | 1269.7 (6.2e-120) | 956.8 (3.9e-70) |
| L-HDL-PL | 299 | 580 (3.4e-20) | 863.5 (3.3e-56) | 1262.4 (2.1e-118) | 891.8 (2.8e-60) |
| XL-HDL-C | 298 | 475.3 (2.7e-10) | 734 (1e-38) | 976.1 (4.9e-73) | 554 (1.3e-17) |
| XL-HDL-CE | 299 | 472.9 (5.4e-10) | 736.9 (6.7e-39) | 1117.4 (9e-95) | 517.5 (6.5e-14) |
| XL-HDL-FC | 295 | 527.8 (2.1e-15) | 1182.8 (1.6e-106) | 2169.4 (3.1e-282) | 677.3 (4.3e-32) |
| XL-HDL-L | 298 | 555.2 (9.6e-18) | 701.2 (1.6e-34) | 1014 (7.9e-79) | 775.3 (3.4e-44) |
| XL-HDL-P | 300 | 578.9 (6.3e-20) | 744.5 (1.1e-39) | 1015.5 (1.6e-78) | 751.3 (1.4e-40) |
| XL-HDL-PL | 306 | 604.9 (7.8e-22) | 1153.9 (1.4e-98) | 1899 (1.5e-227) | 909.3 (3.7e-61) |
| XL-HDL-TG | 300 | 702.2 (2.8e-34) | 779.8 (2.2e-44) | 1140.8 (3.2e-98) | 1399.2 (3.7e-141) |
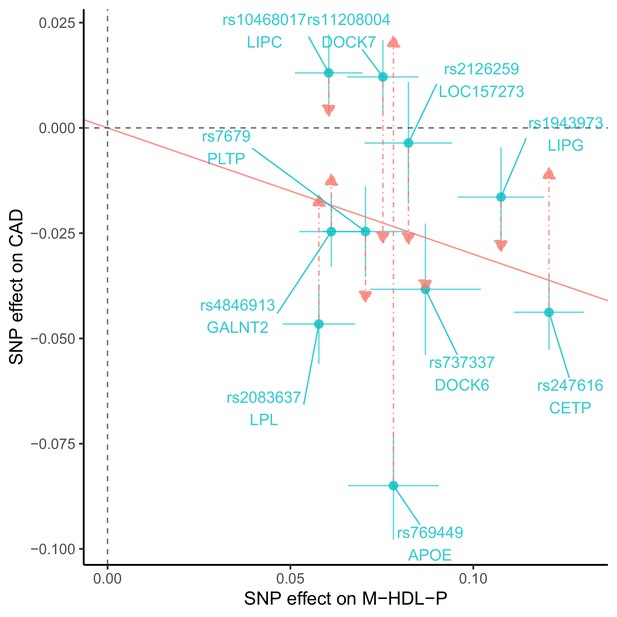
List of SNPs associated with M-HDL-P.
| SNP | Chr | Gene | S-HDL-P | M-HDL-P | L-HDL-P | XL-HDL-P | HDL-C | LDL-C | TG | CAD |
|---|---|---|---|---|---|---|---|---|---|---|
| rs11208004 | 1 | DOCK7 | -0.039 ** | -0.075 *** | -0.015 | -0.002 | -0.015 ** | -0.050 *** | -0.069 *** | -0.012 |
| rs4846913 | 1 | GALNT2 | -0.000 | -0.061 *** | -0.062 *** | -0.023 . | -0.055 *** | -0.006 | -0.044 *** | -0.025 . |
| rs2126259 | 8 | LOC157273 | -0.066 *** | -0.082 *** | -0.063 ** | -0.025 . | -0.075 *** | -0.063 *** | -0.016 . | -0.004 |
| rs2083637 | 8 | LPL | -0.001 | -0.058 *** | -0.092 *** | -0.053 ** | -0.105 *** | -0.008 | -0.108 *** | -0.047 ** |
| rs10468017 | 15 | ALDH1A2/LIPC | -0.096 *** | -0.060 *** | -0.209 *** | -0.202 *** | -0.118 *** | -0.002 | -0.038 *** | -0.013 |
| rs247616 | 16 | CETP | -0.058 *** | -0.121 *** | -0.198 *** | -0.129 *** | -0.243 *** | -0.055 *** | -0.039 *** | -0.044 ** |
| rs1943973 | 18 | LIPG | -0.022 | -0.108 *** | -0.104 *** | -0.078 *** | -0.077 *** | -0.024 ** | -0.009 | -0.016 |
| rs737337 | 19 | DOCK6 | -0.047 . | -0.087 *** | -0.081 ** | -0.058 * | -0.056 *** | -0.007 | -0.011 | -0.038 . |
| rs769449 | 19 | APOE | -0.016 | -0.078 *** | -0.071 *** | -0.015 | -0.064 *** | -0.214 *** | -0.042 *** | -0.085 *** |
| rs7679 | 20 | PCIF1/PLTP | -0.188 *** | -0.071 *** | -0.129 *** | -0.152 *** | -0.059 *** | -0.009 | -0.051 *** | -0.025 . |
List of SNPs associated with S-HDL-P.
| SNP | Chr | Gene | S-HDL-P | M-HDL-P | L-HDL-P | XL-HDL-P | HDL-C | LDL-C | TG | CAD |
|---|---|---|---|---|---|---|---|---|---|---|
| rs780094 | 2 | GCKR | -0.074 *** | -0.034 * | -0.04 ** | -0.034 * | -0.011 . | -0.021 ** | -0.110 *** | -0.005 |
| rs10935473 | 3 | ST3GAL6-AS1 | -0.052 *** | -0.014 | -0.029 . | -0.031 * | -0.009 . | -0.003 | -0.005 | -0.007 |
| rs4936363 | 11 | SIK3 | -0.064 *** | -0.046 ** | -0.019 | -0.006 | -0.034 ** | -0.018 . | -0.043 *** | -0.022 |
| rs2043085 | 15 | ALDH1A2/LIPC | -0.092 *** | -0.056 *** | -0.202 *** | -0.197 *** | -0.106 *** | -0.003 | -0.033 *** | -0.008 |
| rs1800588 | 15 | ALDH1A2/LIPC | -0.106 *** | -0.050 ** | -0.215 *** | -0.212 *** | -0.114 *** | -0.002 | -0.044 *** | -0.015 |
| rs289714 | 16 | CETP | -0.077 *** | -0.122 *** | -0.162 *** | -0.102 *** | -0.214 *** | -0.036 *** | -0.035 *** | -0.012 |
| rs6065904 | 20 | PLTP | -0.171 *** | -0.060 *** | -0.127 *** | -0.149 *** | -0.052 *** | -0.008 | -0.040 *** | -0.022 . |
Tissue-specific gene expressions associated with the 4 discovered genetic markers in the GTEx project.
| SNP.Id | Type | Gene.Symbol | Variant.Id | p value | Effect | Tissue |
|---|---|---|---|---|---|---|
| rs838880 | eQTL | SCARB1 | chr12_124777047_C_T_b38 | 1.5E-08 | -0.20 | Cells - Cultured fibroblasts |
| rs838880 | sQTL | SCARB1 | chr12_124777047_C_T_b38 | 4.1E-06 | -0.34 | Testis |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 3.8E-43 | 0.99 | Artery - Tibial |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 6.4E-35 | 0.93 | Adipose - Subcutaneous |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 6.4E-35 | 0.93 | Adipose - Subcutaneous |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 1.6E-27 | 0.95 | Esophagus - Muscularis |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 3.2E-20 | 1.10 | Colon - Sigmoid |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 1.1E-17 | 0.93 | Esophagus - Gastroesophageal Junction |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 1.8E-09 | 0.81 | Artery - Coronary |
| rs737337 | sQTL | DOCK6 | chr19_11236817_T_C_b38 | 1.2E-07 | -0.49 | Thyroid |
| rs737337 | sQTL | KANK2 | chr19_11236817_T_C_b38 | 4.4E-07 | 0.43 | Artery - Tibial |
| rs737337 | sQTL | KANK2 | chr19_11236817_T_C_b38 | 3.5E-06 | 0.55 | Heart - Left Ventricle |
| rs2943641 | eQTL | IRS1 | chr2_226229029_T_C_b38 | 1.4E-16 | -0.30 | Adipose - Subcutaneous |
| rs2943641 | eQTL | IRS1 | chr2_226229029_T_C_b38 | 6.1E-12 | -0.23 | Adipose - Visceral (Omentum) |
| rs2943641 | eQTL | RP11-395N3.2 | chr2_226229029_T_C_b38 | 3.5E-09 | -0.23 | Adipose - Subcutaneous |
| rs2943641 | eQTL | RP11-395N3.1 | chr2_226229029_T_C_b38 | 2.1E-07 | -0.23 | Adipose - Subcutaneous |
| rs2943641 | eQTL | RP11-395N3.2 | chr2_226229029_T_C_b38 | 2.3E-06 | -0.19 | Adipose - Visceral (Omentum) |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 4.4E-22 | -0.27 | Muscle - Skeletal |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.6E-16 | -0.27 | Adipose - Subcutaneous |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.2E-15 | -0.28 | Adipose - Visceral (Omentum) |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 3.2E-15 | -0.42 | Heart - Atrial Appendage |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 7.2E-14 | -0.25 | Artery - Tibial |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.8E-12 | -0.27 | Nerve - Tibial |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 7.3E-12 | -0.26 | Esophagus - Muscularis |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 2.0E-11 | -0.29 | Colon - Transverse |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 4.1E-11 | -0.32 | Colon - Sigmoid |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.2E-09 | -0.26 | Artery - Aorta |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 4.2E-09 | -0.29 | Heart - Left Ventricle |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 5.0E-09 | -0.22 | Thyroid |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.7E-08 | -0.29 | Stomach |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 4.3E-08 | -0.24 | Lung |
| rs6065904 | eQTL | NEURL2 | chr20_45906012_G_A_b38 | 6.6E-08 | -0.26 | Adipose - Subcutaneous |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 6.8E-08 | -0.33 | Liver |
| rs6065904 | eQTL | CTSA | chr20_45906012_G_A_b38 | 4.0E-07 | -0.14 | Nerve - Tibial |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 5.3E-07 | -0.37 | Spleen |
| rs6065904 | eQTL | NEURL2 | chr20_45906012_G_A_b38 | 5.6E-07 | -0.26 | Adipose - Visceral (Omentum) |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 8.9E-07 | -0.46 | Small Intestine - Terminal Ileum |
| rs6065904 | eQTL | RP3-337O18.9 | chr20_45906012_G_A_b38 | 1.8E-06 | -0.22 | Adipose - Subcutaneous |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 2.9E-06 | -0.31 | Nerve - Tibial |
| rs6065904 | eQTL | DNTTIP1 | chr20_45906012_G_A_b38 | 3.1E-06 | -0.17 | Artery - Tibial |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 4.5E-06 | -0.27 | Skin - Sun Exposed (Lower leg) |
| rs6065904 | eQTL | SNX21 | chr20_45906012_G_A_b38 | 4.8E-06 | -0.15 | Esophagus - Muscularis |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 8.9E-06 | -0.27 | Skin - Not Sun Exposed (Suprapubic) |
| rs6065904 | eQTL | DNTTIP1 | chr20_45906012_G_A_b38 | 1.0E-05 | -0.14 | Nerve - Tibial |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.1E-05 | -0.27 | Prostate |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.3E-05 | -0.26 | Pituitary |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.4E-05 | -0.21 | Esophagus - Gastroesophageal Junction |
| rs6065904 | eQTL | SNX21 | chr20_45906012_G_A_b38 | 1.5E-05 | -0.16 | Esophagus - Mucosa |
| rs6065904 | eQTL | SNX21 | chr20_45906012_G_A_b38 | 1.7E-05 | -0.23 | Colon - Sigmoid |
| rs6065904 | eQTL | SNX21 | chr20_45906012_G_A_b38 | 1.7E-05 | -0.17 | Thyroid |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 2.6E-05 | -0.21 | Breast - Mammary Tissue |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 2.9E-05 | -0.23 | Artery - Tibial |
| rs6065904 | eQTL | NEURL2 | chr20_45906012_G_A_b38 | 3.2E-05 | -0.21 | Thyroid |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 3.7E-05 | -0.17 | Testis |
| rs6065904 | eQTL | CTSA | chr20_45906012_G_A_b38 | 4.4E-05 | -0.11 | Skin - Not Sun Exposed (Suprapubic) |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 5.8E-05 | -0.23 | Muscle - Skeletal |
| rs6065904 | eQTL | NEURL2 | chr20_45906012_G_A_b38 | 8.2E-05 | -0.27 | Heart - Atrial Appendage |
| rs6065904 | eQTL | SNX21 | chr20_45906012_G_A_b38 | 8.4E-05 | -0.17 | Artery - Aorta |
| rs6065904 | eQTL | NEURL2 | chr20_45906012_G_A_b38 | 9.5E-05 | -0.24 | Artery - Aorta |
| rs6065904 | eQTL | WFDC3 | chr20_45906012_G_A_b38 | 9.5E-05 | -0.31 | Artery - Aorta |
| rs6065904 | eQTL | RP3-337O18.9 | chr20_45906012_G_A_b38 | 9.5E-05 | -0.29 | Heart - Atrial Appendage |
| rs6065904 | eQTL | PLTP | chr20_45906012_G_A_b38 | 1.2E-04 | -0.15 | Skin - Sun Exposed (Lower leg) |
| rs6065904 | eQTL | WFDC13 | chr20_45906012_G_A_b38 | 1.5E-04 | 0.28 | Esophagus - Muscularis |
| rs6065904 | eQTL | DNTTIP1 | chr20_45906012_G_A_b38 | 2.1E-04 | -0.12 | Cells - Cultured fibroblasts |
| rs6065904 | sQTL | ZNF335 | chr20_45906012_G_A_b38 | 3.3E-11 | -0.65 | Testis |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 1.3E-09 | 0.58 | Heart - Left Ventricle |
| rs6065904 | sQTL | PLTP | chr20_45906012_G_A_b38 | 4.5E-08 | -0.32 | Whole Blood |
| rs6065904 | sQTL | PLTP | chr20_45906012_G_A_b38 | 4.8E-08 | 0.53 | Spleen |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 1.3E-07 | 0.42 | Esophagus - Mucosa |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 2.6E-07 | 0.49 | Heart - Atrial Appendage |
| rs6065904 | sQTL | CTSA | chr20_45906012_G_A_b38 | 1.0E-06 | -0.41 | Artery - Aorta |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 1.2E-06 | 0.33 | Nerve - Tibial |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 1.2E-06 | 0.67 | Brain - Spinal cord (cervical c-1) |
| rs6065904 | sQTL | TNNC2 | chr20_45906012_G_A_b38 | 2.1E-06 | 0.54 | Brain - Cerebellum |
| rs6065904 | sQTL | ACOT8 | chr20_45906012_G_A_b38 | 2.1E-06 | 0.54 | Brain - Cerebellum |
| rs6065904 | sQTL | WFDC3 | chr20_45906012_G_A_b38 | 5.5E-06 | 0.23 | Skin - Sun Exposed (Lower leg) |
| rs6065904 | sQTL | WFDC3 | chr20_45906012_G_A_b38 | 9.4E-06 | -0.28 | Skin - Not Sun Exposed (Suprapubic) |